PD is caused by proteostasis dysfunction, specifically the misfolding and self-association of aberrant α-synuclein. ALS may have several causes, including protein mislocalization and faulty RNA quality control.
Abstract
Parkinson’s disease (PD) is a movement disorder that afflicts over one million in the U.S.; amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease) is less prevalent but also has a high incidence. The two disorders sometimes present together, making a comparative study of interest. Both ALS and PD are neurodegenerative diseases, and are characterized by the presence of intraneuronal inclusions; however, different classes of neurons are affected and the primary protein in the inclusions differs between the diseases, and in some cases is different in distinct forms of the same disease. These observations might suggest that the more general approach of proteostasis pathway alteration would be a powerful one in treating these disorders. Examining results from human genetics and studies in model organisms, as well as from biochemical and biophysical characterization of the proteins involved in both diseases, we find that most instances of PD can be considered as arising from the misfolding, and self-association to a toxic species, of the small neuronal protein α-synuclein, and that proteostasis strategies are likely to be of value for this disorder. For ALS, the situation is much more complex and less clear-cut; the available data are most consistent with a view that ALS may actually be a family of disorders, presenting similarly but arising from distinct and nonoverlapping causes, including mislocalization of some properly folded proteins and derangement of RNA quality control pathways. Applying proteostasis approaches to this disease may require rethinking or broadening the concept of what proteostasis means.
INTRODUCTION: NEUROLOGIC DISORDERS OF MOVEMENT
More than two million Americans are believed to suffer from some form of neurodegenerative movement disorder, the total cost of which is estimated to exceed $10 billion annually. Because no society on earth is spared the effects of these crippling diseases, the figures for other countries are similar, adjusted for population differences. The neurodegenerative diseases Parkinson’s disease (PD) and amyotrophic lateral sclerosis (ALS or Lou Gehrig’s disease) are the most important of the movement disorders from a proteostasis perspective. ALS is often classified separately, as a motor neuron disease, but there are a number of reasons for considering them together, of which perhaps the most important is that the two sometimes present together, as in the Parkinsonism-dementia complex of Guam (Lytico-bodig). This movement disorder occurs among the Chamorro populations of Guam and the Mariana Islands and is frequently accompanied by a motor neuron disease resembling ALS. The course of the disease is rapid, with death typically occurring within 5 years (Steele 2005).
PD is the second most common neurodegenerative disease after Alzheimer's disease (AD); estimates of prevalence range to over one million in the U.S. Although ALS is considered a so-called “orphan disease” because of its relatively low prevalence (around 30,000 cases in the United States), the annual incidence of ALS is actually closer to that of Parkinson’s: ∼6000 for ALS versus ∼60,000 for PD in the United States; the much lower prevalence of ALS is because of its rapid progression to mortality (typically 3–4 years postdiagnosis versus 20 years for PD). The major symptoms of both diseases arise from the degeneration of particular classes of neurons: the dopaminergic neurons of the substantia nigra pars compacta in the case of PD and the spinal motor neurons in the case of ALS. However, it is now clear that other neurons are also affected in both diseases, and there are even suggestions that PD may start in the brain stem or elsewhere in the CNS and spread to the nigral neurons by, perhaps, noncell autonomous processes (Ilieva et al. 2009).
An interesting commonality between the two disorders is that about 10% of all cases of PD and ALS are genetic, and run in families with strict Mendelian inheritance (the remaining 90% are termed sporadic and idiopathic, although some genetic and environmental risk factors have been identified (Shulman et al. 2011; Bosco and Landers 2010). The genetics of these rarer familial forms, some of which present with significantly earlier age of onset than the sporadic disease, have provided a rich source of information on the pathways and processes that may be involved. Some of these data suggest that dysregulation of proteostasis of particular proteins may be responsible for the initiation and/or progression of these diseases.
The other reason both ALS and PD are of interest to researchers in proteostasis is that the histopathological hallmarks of both diseases are usually (but not always) dense intraneuronal deposits of apparently misfolded protein, called Lewy bodies in PD and Bunina bodies in ALS. Although there is some evidence that the aggregates may have a defined supramolecular structure, and certainly contain more than one type of protein, in these movement disorders it is generally thought that the primary component of the aggregate is a single protein that has aberrantly adopted an amyloid-like fold (Lashuel and Lansbury 2006). Lewy bodies appear to comprise amyloid fibrils of a small protein called α-synuclein (Uversky and Eliezer 2009). Bunina bodies are eosinphilic inclusions that contain Cystatin C. Other types of aggregates detected in ALS include cytosolic inclusions containing TDP-43 (Neumann et al. 2006), which are prominent in most sporadic ALS cases, and aggregates containing the enzyme superoxide dismutase 1 (SOD1) in both familial ALS cases harboring SOD1 mutations (Brujin et al. 1998) and some cases of sporadic ALS (Forsberg et al. 2010).
It is not clear in any neurodegenerative disease whether macroscopic protein aggregates are the actual causes of cell death or an attempt by the cell to sequester damaged or useless macromolecules (Caughey and Lansbury 2003). If the former, then prevention of the formation of such aggregates or breaking them up once they have formed are possible therapeutic strategies, and both could be accomplished in theory by altered regulation of proteostasis pathways (chaperones, proteasomes, autophagy, etc.). However, if the aggregates are harmless or protective, then attempts to alter them might actually result in an increase in the true pathological species, and the current dogma is that the macroscopic aggregates are more likely protective than toxic. This would suggest that proteostasis strategies aimed at stabilizing the nonpathologic, physiological form(s) of the misfolded protein (pharmacological chaperones, inhibition of covalent modifications that destabilize the protein, etc.) need to be considered and perhaps emphasized.
In the following sections, we will review the evidence that links dysfunction in the proteostasis of certain proteins to the pathology of ALS and PD. Recent data support the notion that at least most forms of Parkinson’s disease may be amenable to therapeutic strategies aimed at proteostasis of α-synuclein or proteins called parkin or LRRK2, while the situation with ALS is still far from clear.
PARKINSON’S DISEASE
As mentioned above, 10% of PD cases are genetic. Depending on the genes involved, they may be inherited in either an autosomal dominant or recessive manner. In only some instances has postmortem examination been possible; so for many of the PD-associated genes, we do not yet know whether Lewy bodies containing aggregated α-synuclein are present. As we shall see, this point is of considerable significance.
α-Synuclein
Autosomal dominant PD of the PARK1 form is caused by mutations in the SNCA gene encoding α-synuclein (Polymeropoulos et al. 1997), the principal protein component of Lewy bodies. In neurons, α-synuclein is abundant in the presynaptic nerve terminal, and can be isolated both as a soluble and vesicle-associated molecule (Surguchov 2008). It is normally amino-terminally acetylated, and in vivo, there is often phosphorylation on Ser 87 and 129 (Oueslati et al. 2010). Its biochemical and cellular functions are still uncertain, although recent evidence suggests that it may play a role in SNARE-mediated vesicle fusion (Burré et al. 2010).
Most literature references describe α-synuclein as a monomeric protein of 140 amino acids that is natively unfolded, readily forms amyloid-like aggregates, and is toxic to cell membranes (e.g., Uversky and Eliezer 2009). Careful assessment of the evidence for these statements confirms that the protein indeed comprises 140 amino acids. The idea that it is natively unfolded is based primarily on experiments in which the protein has been treated harshly during purification; examination of the sequence by bioinformatics tools predicts that much of the protein should be structured (J Sussman, pers. comm.). α-Synuclein makes up about 1% of the total protein in a neuron, which also calls into question the notion that it has no persistent secondary or tertiary structure in vivo. In this discussion, we shall assume that there is a native, partially folded, nonpathological form of α-synuclein in the cell, and that mutations and posttranslational modifications shift a dynamic, context-dependent equilibrium between that state and an aggregation-prone form in favor of the latter. This is an important assumption, because it opens up a range of possible therapeutic approaches based on proteostasis considerations that would be unavailable if α-synuclein really were unstructured in its native state.
α-Synuclein Variants
Three PD-associated point mutations in α-synuclein have been identified thus far: A30P, A53T, and E46K. All present with autosomal dominant inheritance. The first two replace the strongest helix-forming amino acid, alanine, with a helix-breaking or β-sheet-forming residue, suggesting that they may simply increase the intrinsic β-sheet propensity of the amino-terminal portion of the α-synuclein sequence. The structural or functional consequences of the E46K mutation are harder to predict; one would not necessarily assume such a substitution to be of catastrophic consequence in a natively unfolded protein, so its disease association is one more suggestion that α-synuclein may be a structured protein in the cell.
The fact that α-synuclein is the major protein in Lewy bodies and that mutations in its gene give rise to inherited PD immediately suggested that α-synuclein aggregation might be the molecular cause of most forms of Parkinson’s disease. Consistent with this notion, mutant forms of α-synuclein are toxic to neurons when overexpressed (Kahle et al. 2000), although frequently it is necessary to supply additional stress in the form of, for example, chemical inhibitors of mitochondrial complex I for full toxicity to develop (e.g., Schneider et al. 2007). Although overexpression of mutant or wild-type α-synuclein produces striking toxic phenotypes in simple model organisms (e.g., Outeiro and Lindquist 2003), finding evidence that the protein is involved in neurodegeneration in higher animals has been more difficult. The loss of nigral dopaminergic neurons is rarely observed in synuclein transgenic mice (Schneider et al. 2007). A rat model for PD that recapitulates some of the histopathology of the disease, including Lewy body-like inclusions, requires considerable overexpression using lentivirus or adeno-associated virus vectors (Schneider et al. 2008). The α-synuclein misfolding/aggregation hypothesis for the etiology of PD has received perhaps its strongest support from human genetics, namely the discovery of families with inherited, early-onset Parkinson’s disease caused by duplications and triplications of the wild-type α-synuclein locus (Ross et al. 2008).
α-Synuclein Misfolding and Aggregation
α-Synuclein knockout mice lack a major disease-resembling phenotype, consistent with the notion of a toxic gain of function from misfolded α-synuclein rather than a loss of normal function as the major factor in neuronal cell death (Abeliovich et al. 2000). Overexpression of mutant α-synuclein does produce neurologic defects in mice and rats, but none of these models recapitulate the full panoply of PD phenotypes. In particular, it has proven remarkably difficult to observe either Lewy body-like inclusions or nigral degeneration in many models, and in most cases massive overdriving of synuclein expression or additional stresses are required to see any effects (Dawson et al. 2010). Cell culture models and studies of α-synuclein in vitro have met with more success, and much of what is currently assumed about the disease process in people is extrapolated from these. It is thought that neurodegeneration arises from the formation of toxic misfolded oligomers of α-synuclein that may exert their toxic effects by disrupting cell membranes such as those of the mitochondria. α-Synuclein proteostasis depends on both the ubiquitin-proteasome and lysosomal pathways of protein turnover, so disruption of either of these pathways by, e.g., proteasome inhibitors and lysosomal storage diseases, might be expected to lead to increased accumulation of α-synuclein and eventually to the formation of α-synuclein aggregates (Lopez and Sidransky 2010; Vernon et al. 2011). The role of the large Lewy body aggregates is controversial. Though their amyloid nature is not in doubt, whether they are protective or harmful to cells is not established; nor is the question of whether the toxic oligomeric species (assuming they exist) are on the pathway to Lewy body formation answered. Then there is the issue of reversibility. If the fibrils can be disaggregated, what species are formed? Are the aggregates in equilibrium with misfolded monomers or toxic β-sheet-containing oligomers, or can they give rise to properly folded, functional α-synuclein? Clearly, approaches to PD treatment aimed at α-synuclein proteostasis would greatly benefit from clarity here.
Posttranslational Modifications of α-Synuclein
In vitro, α-synuclein forms amyloid fibrils rather slowly, so efforts have been made to identify factors that might accelerate this process. Oxidation, metal ion binding, and partial proteolytic cleavage have all been found to promote aggregation of the wild-type protein (Fink 2006). Forms of α-synuclein that aggregate more readily have the remarkable property of being able to accelerate the conversion of unmodified, wild-type α-synuclein into an amyloid-like state (Murray et al. 2003; Li et al. 2005; Dufty et al. 2007).
Encouraging for models of PD etiology based on α-synuclein misfolding is the fact that most of these in vitro perturbations can be linked to known causes of, or risk factors for, the disease in people. α-Synuclein mutants are less stable than wild-type protein to β-sheet formation and aggregation; the same mutations cause familial PD (Q Hoang, pers. comm.). Overexpression of wild-type α-synuclein is toxic to yeast, and gene duplication and triplication of the wild-type SNCA locus leads to inherited Parkinson’s (Outeiro and Lindquist 2003). Oxidation of α-synuclein promotes its misfolding and aggregation; oxidative stress, especially by mitochondrial toxins, is a known risk factor for PD and can induce PD-like motor neuron defects in mice and primates, sometimes in the absence of α-synuclein mutations or overexpression (Dawson et al. 2010). Finally, carboxy-terminally truncated fragments of α-synuclein are found in Lewy bodies (Murray et al. 2003), and the same truncated protein aggregates rapidly in vitro, and can catalyze conversion of full-length α-synuclein to amyloid. The protease(s) responsible for these truncations has/have yet to be identified, but if α-synuclein fragmentation is, for example, an initiating or accelerating event in PD, these enzymes could be extremely attractive drug targets.
The role of other posttranslational modifications of α-synuclein in PD has not been established. In mammalian cells and when ectopically expressed in yeast, α-synuclein is amino-terminally acetylated; bacterially expressed recombinant α-synuclein, which is what has been used for most in vitro experiments, is not. Given the sensitivity of α-synuclein to even small perturbations in sequence and structure, it is important to determine whether this difference is significant for PD etiology. Another difference between recombinant and natural α-synuclein is phosphorylation. Lashuel and coworkers have shown that serine 87 and serine 129 are phosphorylated to a considerable extent in α-synuclein isolated from brain (Paleologou et al. 2010). The levels of S87-P are increased in brains of transgenic models of synucleinopathies and human brains from Alzheimer disease, Lewy body disease, and multiple system atrophy patients—all diseases in which synuclein aggregation has been observed—compared with normal controls. In brain homogenates from diseased human brains and transgenic animals, the majority of S87-P was detected in the membrane fractions. A battery of biophysical methods was used to dissect the effect of S87 phosphorylation on the structure, aggregation, and membrane-binding properties of monomeric α-synuclein; these showed that phosphorylation at S87 increases the protein’s conformational flexibility and blocks its fibrillization in vitro. Furthermore, phosphorylation at S87, but not S129, resulted in significant reduction of α-synuclein binding to membranes (Paleologou et al. 2010). How these effects correlate with α-synuclein homeostasis and with disease pathology in the various synucleinopathies remains to be determined, but therapeutic strategies aimed at manipulating the posttranslational modifications of α-synuclein would appear to be an underconsidered approach.
Parkin
Autosomal recessive mutations within the PARK2 locus are the most common cause of autosomal recessive Parkinson’s disease, and are perhaps second only to mutations in LRRK2 (see below) as contributors to the familial forms of PD (Kitada et al. 1998; Hatano et al. 2009; Dachsel and Farrer 2010). The PARK2 gene encodes a 465 amino acid ubiquitin E3 ligase named parkin (Shimura et al. 2000). Parkin belongs to the RING (really interesting new gene) family of E3 ligases and possesses a RING1, in-between-RING, and RING2 architecture within the carboxy-terminal half of the protein. A novel feature of parkin is the existence of a ubiquitin-like domain within its amino terminus, which is not commonly found within E3 ligases (thus, the importance or role of this ubiquitin-like domain has been the subject of much speculation). Because parkin clearly functions in the ubiquitin-mediated pathway of protein degradation, it provides a direct link between this proteostasis pathway and at least one genetic form of PD. A second property of parkin that may be particularly relevant to proteostasis is the unusually high number of cysteine residues within the protein. Parkin contains 35 cysteines, which constitute almost 8% of the protein. Cysteines are subject to a diverse array of posttranslational modifications and this particular aspect of parkin biochemistry has received a great deal of attention from the research community, as will be addressed in detail below.
The Impact of Disease-Linked Truncations and Mutations on Parkin Stability
The clinical mutations in Parkin associated with disease span from genomic deletions to premature truncations and subtle missense mutations in the coding sequence. For this reason, it has been widely accepted that the pathogenicity of these PARK2 mutations derives from a critical loss-of-function. Despite the predicted loss of E3 ligase activity of these parkin mutants, biochemical evidence of parkin E3 ligase activity and the impact of parkin deficiency has been notoriously difficult to establish. Although numerous putative substrates have been reported, many have failed to withstand independent confirmation. The lack of well-founded substrates has also made it difficult to examine the degree to which each mutation alters activity, precluding an analysis of the correlation between the biochemical impact of a given parkin mutation and the severity of the resultant disease in patients. Nonetheless, there have been many valuable observations made regarding the novel PD-associated parkin variants.
Many of the PD-linked mutations in parkin show reduced E3 ligase activity against putative substrates, but at the same time some of the hypomorphic PD-associated variants may show either normal levels of auto-ubiquitination or even enhanced self-ubiquitination (Sriram et al. 2005; Hampe et al. 2006; Matsuda et al. 2006). Differential effects on the association of the E3 ligase with substrate also have been observed, consistent with data on other E3 ligases in which catalytically inactive E3 ligase mutants bind substrate more avidly than the wild-type protein (Qiu and Goldberg 2002). These effects are likely attributed to a reduced catalysis or off-rate from the defective enzyme.
Pathogenic parkin mutations can be found throughout the coding sequence of the protein, not segregated to a single domain. Therefore, there must be multiple mechanisms through which sequence variation can inactivate parkin. Work from several groups has now established the model whereby several of the disease-linked mutations abrogate parkin function through a destabilization of the protein within the cellular environment. It was first shown by the Winklhofer group that the pathogenic W453X carboxy-terminal truncation of parkin was associated with reduced solubility in a variety of cell types (Winklhofer et al. 2003). Further work examined many other PD-linked variants, and reported complementary findings but also extended these observations to include aberrant localization of disease-linked parkin (Sriram et al. 2005) or cytoplasmic aggregation (Henn et al. 2005). Other findings included the enhanced proteasomal degradation of mutant parkin as a mechanism for inactivating some parkin species (Henn et al. 2005; Schlehe et al. 2008) as well.
Stress-Induced Parkin Misfolding and Aggregation
Thus, the subtle pathogenic mutations in parkin can promote the misfolding and aggregation of the protein. However, this is not only true of mutant parkin but is also an important aspect of the protein biochemistry of wild-type parkin. Exposure to oxidative stress can rapidly induce the transition of wild-type parkin from a detergent soluble fraction into an insoluble state (Winklhofer et al. 2003; LaVoie et al. 2007), a property of the protein likely linked to its high number of cysteine residues. Not only does direct oxidant exposure (hydrogen peroxide) foster a disruption in parkin solubility, but a variety of cellular stressors can promote the insoluble aggregation of parkin in propagated cell lines (LaVoie et al. 2005, 2007; Wang et al. 2005) and primary cultured neurons (LaVoie et al. 2007). This effect is not limited to chemical-induced stress but rather includes the overexpression of α-synuclein (Kawahara et al. 2008). Our data suggested that the aggregation and insolubility of parkin was associated with intermolecular cross-linking and oligomerization of parkin via disulphide bonds (LaVoie et al. 2007), again consistent with a role for cysteine in the sensitivity of parkin. Evidence from one of our groups and others indicates that parkin is uniquely sensitive to the conditions of the intracellular environment, as other E3 ligases appear far more resistant to stress-induced misfolding (Wang et al. 2005; LaVoie et al. 2007; Schlehe et al. 2008), as are other gene products implicated in PD (LaVoie et al. 2005, 2007). Consistent with these observations of stress-induced aggregation, it has also been reported that parkin solubility decreases with age in the human brain (Pawlyk et al. 2003), which may be particularly relevant to this age-related neurodegenerative disorder.
Posttranslational Modifications that Inactivate Parkin
The reports summarized above raise the intriguing possibility that idiopathic PD pathogenesis or disease risk may arise by processes whereby neuronal stress might inactivate parkin via insolubility and aggregation. As a biochemical phenocopy of PARK2-associated familial PD, a severe loss of active, soluble parkin could compromise the viability of nigral neurons in the sporadic disease.
The one problem with this model is that Lewy bodies have generally not been observed in PARK2-associated familial PD brains, whereas they are almost always present to some extent in postmortem analyses of sporadic PD patients. No biochemical or genetic evidence has yet conclusively tied parkin into α-synuclein aggregation, raising the possibility that neither protein is the direct cause of any form of PD, but that each may act independently through either the same or different downstream initiators of cell death. Of course, the other possibility is that macroscopic α-synuclein aggregates are not involved in pathogenesis, and that loss of parkin function causes disease by producing whatever the actual toxic α-synuclein species is.
In addition to a dramatic shift in the solubility of cellular parkin, other studies have pointed to more direct attacks on the parkin protein that might contribute to disease. Two independent groups showed that not only was parkin vulnerable to S-nitrosylation and that this biochemical modification inactivated the E3 ligase function of parkin, but both studies reported a disease-specific increase in nitrosylated parkin in PD brain (Chung et al. 2004; Yao et al. 2004). The sources of nitric oxide that mediate this effect are not yet known, but may be part of an early pathogenic mechanism or part of a secondary series of events associated with neuroinflammation. Nonetheless, these valuable findings from PD brain tissue provided novel insight into the protein biochemistry that occurs in the course of PD and support the pathophysiological relevance of wild-type parkin dysfunction in idiopathic disease.
As discussed earlier, the cells susceptible to the selective degeneration in PD include the dopaminergic neurons of the substantia nigra (black substance), thusly named because of the age-dependent accumulation of oxidized dopamine and other molecules that make up the dark-colored deposits of neuromelanin. Dopamine is used throughout the animal kingdom as a neurotransmitter. However, the chemical nature of dopamine imparts it with a unique propensity to auto-oxidize and, in the process, both generate free radicals and produce the highly reactive dopamine quinone (Graham 1978). Dopamine quinone is capable of covalently modifying cysteine residues of proteins in an irreversible manner and has been associated with selective neurotoxicity and protein modification in animal models of PD pathology (Hastings et al. 1996; LaVoie and Hastings 1999). Because the neurons that synthesize dopamine require parkin activity for their survival, whether the neurotransmitter dopamine itself might inactivate parkin’s catalytic activity, and thus explain part of the intrinsic vulnerability of these cells, has been investigated. The results showed that exposure to elevated cytoplasmic dopamine induced the aggregation and insolubility of parkin in cell culture (LaVoie et al. 2005). The dopamine quinone, well known for its reactivity toward the free thiols in cysteine, was found to covalently attack the parkin protein, and this novel posttranslational modification resulted in the inactivation of parkin E3 ligase activity (LaVoie et al. 2005). The same workers then explored the intact substantia nigra from normal adult human brain to determine whether parkin was covalently modified in vivo. Data confirmed the selective covalent attack on parkin by dopamine in the substantia nigra, but not other brain regions (LaVoie et al. 2005). These data suggest that factors that influence the synthesis, storage, release, and/or uptake of the neurotransmitter dopamine may influence the risk for idiopathic PD via the modification and inactivation of parkin (Cookson 2005), as well as other potential targets of dopamine-induced modification (van Laar et al. 2008, 2009). Likewise, another posttranslational modification of parkin has been recently reported in a related neurodegenerative disease model (Moszczynska and Yamamoto 2011), and others may soon be discovered as well.
Postmortem Evaluation of Parkin Solubility
Several lines of evidence suggest that the stability of parkin is fragile. Very minor fluctuations within the parkin sequence or the intracellular environment where parkin is expressed can have profound effects on the solubility and activity of the protein. However, many of the studies reviewed here have made exclusive use of cell culture conditions to explore the stability and solubility of parkin, raising the question of whether parkin solubility is an issue within the normal or diseased human brain. Notably, three studies have explored postmortem human brain, and all three reports have found evidence of pathologic changes in parkin solubility in the neurologic disease samples. Two of these studies reported increased levels of insoluble parkin in PD brain tissue, compared to controls (LaVoie et al. 2005; Wang et al. 2005), whereas the third found that parkin was elevated in the insoluble fraction of brains taken from patients with diffuse Lewy body disease (Kawahara et al. 2008), indicating that parkin solubility is affected in human neurologic disease and providing the most tantalizing (albeit indirect) evidence for some link between parkin and α-synuclein. Interestingly, the translocation of parkin from a soluble to insoluble state has also been reported in a rodent model of stroke (Mengesdorf et al. 2002), as well, and is therefore not limited to human neurodegenerative disorders.
In summary, autosomal recessive mutations in parkin are responsible for a subset of the familial cases of PD. These data inform us that parkin serves a critical neuronal function and that its proteostasis is absolutely essential to the dopaminergic neurons of the ventral midbrain. However, there is substantial biochemical and histopathological evidence that the solubility of parkin is fragile, and that subtle perturbations within the brain may alter levels of parkin available to the neuron for support. These challenges may include mitochondrial damage, ischemic insult, external oxidative stress, or combinations of these and others (Fig. 2). Therefore, a deeper understanding of what function is served by parkin within the brain, and precisely why this protein is uniquely essential for the survival of the neurons within the substantia nigra, might point to alternative therapeutics to replace or augment parkin function in the aging or compromised brain, or in idiopathic PD. Discovery of a direct tie-in between parkin and α-synuclein, if one exists, would also help unify models for PD initiation and progression, and possibly lead to more general PD therapeutic strategies.
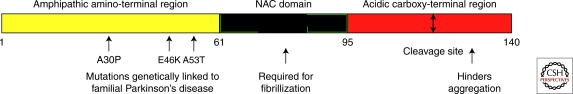
Figure 1.
Schematic of the protein sequence of human α-synuclein, showing various domains. Human α-synuclein is a small protein whose amino acid sequence is often thought of as comprising three domains: an amino-terminal region that is the site of the three known point mutations that cause autosomal dominant, familial PD (yellow); the so-called nonamyloid component (black), which, paradoxically, is required for the formation of α-synuclein amyloid fibrils; and the acidic carboxy-terminal domain (red), which is thought to be disordered in all forms of α-synuclein. Cleavage of approximately the last 20 residues of this region produces a truncated protein that aggregates much more readily than the full-length molecule, suggesting that at least one function of the disordered third of the sequence is to hinder aggregation. Although α-synuclein is widely believed to be a natively unfolded protein except when bound to membranes (where the first 95 residues are believed to become helical), analysis of the sequence by methods designed to detect intrinsically unfolded proteins predicts that only the last 40 residues of α-synuclein have no propensity to fold (J Sussman, pers. comm.). Interestingly, the first ∼100 residues of the sequence contain a mixture of both helix-favoring and β-sheet-forming residues, which may explain the ability of this protein to adopt the cross-β structure of an amyloid when perturbed.
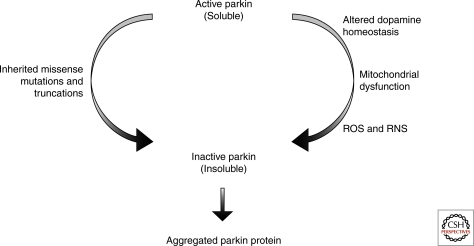
Figure 2.
Parkin proteostasis. The solubility and activity of parkin can be influenced by inherited mutations within the coding sequence of the protein but also the tissue environment and prevalence of cell stressors. Changes in the cytoplasmic concentrations of dopamine, mitochondrial stress and both reactive oxygen species (ROS), and reactive nitrogen species (RNS) can influence the function of parkin and be risk factors in idiopathic PD via modulation of wild-type parkin function.
LRRK2 in Sporadic and Familial PD
Mutations in leucine-rich repeat kinase-2 (LRRK2) are most common genetic cause of PD and are extremely important because they are found in both familial forms of the disease as well as in patients without a recognized family history of the disorder (Dachsel et al. 2010). LRRK2 is an enormous protein kinase comprising a small GTPase domain and several different kinds of putative protein–protein interaction domains in addition to a canonical protein kinase domain. Similar to parkin, the pathogenic mutations in LRRK2 can be found within multiple domains of the protein, but the most commonly studied mutations are either found within the ROC/COR module (containing the GTPase domain) or within the kinase domain. These mutations are autosomal dominant, leading many researchers to suggest that mutant LRRK2 acquires a toxic gain of function. It is uncertain at present whether this gain of function would express its toxicity through downstream misfolding and aggregation of α-synuclein, although a recent paper shows that loss of LRRK2 causes accumulation of α-synuclein, and increases autophagy and cell death in kidneys of aged mice (Tong et al. 2010). These data may indicate LRRK2 mutations could cause loss of function through a dominant negative mechanism, with downstream toxicity mediated via α-synuclein. However, because the effect was seen specifically in the kidneys and not in the brain, the relevance of these findings for PD are uncertain. Even more intriguing, perhaps is the recent observation that LRRK2 knockout mice are somewhat resistant to MPTP toxicity, a result that phenocopies the knockout of synuclein (Andres-Mateos et al. 2009), increasing the likelihood of a connection between these two PD-associated genes.
If the toxic gain of function of LRRK2 is related to increases in either its GTPase or kinase activities, then inhibition of the enzyme may represent a straightforward therapeutic approach for forms of PD whose etiology are dependent on this protein. Early biochemical studies suggested that the pathogenic mutations within LRRK2 might be more prone to aggregation and that the aggregation and toxicity of mutant LRRK2 were dependent on its kinase activity (Greggio et al. 2006). When LRRK2 was examined to determine whether cellular stress would have influences on LRRK2 solubility similar to those found for parkin, no evidence of stress-induced changes in LRRK2 was found (M LaVoie, unpubl.). However, ongoing studies have focused more attention on the importance of localization (Biskup et al. 2006; Higashi et al. 2007), dimerization (Greggio et al. 2008; Sen et al. 2009), and the possibility that these properties are intertwined (Berger et al. 2010). Other work has also focused on the potential association of LRRK2 with other proteins (Ho et al. 2009; Nichols et al. 2010), and these studies may ultimately speak to its neuronal function (still unknown) or how its function is altered by its pathogenic mutations. The biochemistry and cell biology of the LRRK2 protein is still in its infancy as substrates, activators, and the regulation of LRRK2 kinase function are all challenges that need attention. Despite current efforts aimed at these properties, the determinants of LRRK2 stability may turn out to be critical to its function and role in disease as well. LRRK2 is a very large protein (2527 amino acids) with a complex organization. The very fact that LRRK2 protein levels are quite low in the brain alone might suggest that changes in LRRK2 localization, stability, and solubility may be burdensome to the neuron. Given the growing interest in the pathological implications of neuronal proteostasis, this is an issue that will likely be addressed in a major way in the near future.
Other PD-Associated Genes
Three other genes have been linked to familial PD by human genetics: PINK1, UCH-L1, and DJ-1. We do not attempt to review them exhaustively here, but rather consider them briefly from the framework of a general model for PD as a disease of α-synuclein proteostasis (Fig. 3). PINK1 is a putative protein kinase that appears to reside predominantly in the mitochondria (Deas et al. 2009). Autosomal recessive mutations presumably leading to loss of function in PINK1 lead to familial PD. PINK1 has proven remarkably refractory to biochemical studies, and its physiological substrates are not known definitively. There is emerging evidence that it plays a role in mitochondrial quality control, possibly by targeting defective mitochondria for autophagy (see Chu 2010). It is unclear whether PINK1 exerts its Parkinson’s effects through the promotion of α-synuclein aggregation, but given its mitochondrial localization and the fact that many mitochondrial poisons do induce α-synuclein aggregation in cell culture and animal models of PD, a synuclein connection would not be surprising.
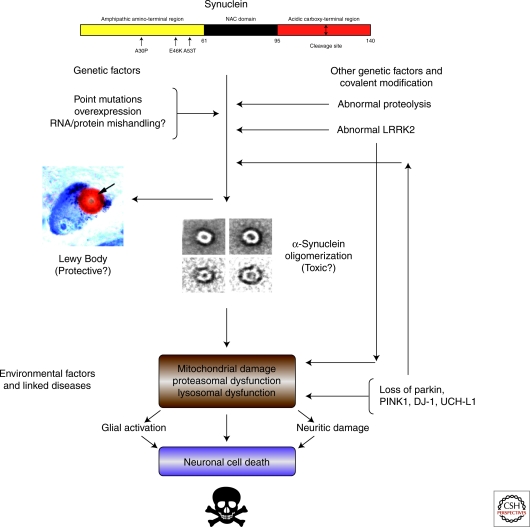
Figure 3.
A pathway for Parkinson’s. A possibly fictitious attempt to fit the known environmental and genetic factors associated with sporadic and familial PD into a common pathological pathway involving α-synuclein proteostasis. Many parts of this diagram are hypothetical, and it is predicated on the assumption that there is a nonpathological, folded form of synuclein that must be unfolded before aggregation can commence. Further, it is assumed that Lewy body formation is off the pathway of toxicity, but this is far from certain. The direct connection between organelle dysfunction/damage and cell death is purely speculative. The pathway is intended to provide a framework for thinking about therapeutic strategies based on proteostasis, but it should be taken with more than just a pinch of salt.
UCH-L1 might be expected to be involved in α-synuclein proteostasis, as it is a ubiquitin carboxy-terminal hydrolase and so in principle it could directly affect α-synuclein levels in the cell, but there are a number of problems with this scenario. First, PD associated with mutations in UCH-L1 has only been reported in a single German family, and the possibility that a different polymorphism in the UCH-L1 gene might confer protection against PD has frequently been disputed (Wang et al. 2011). Only one of the two PD-associated variants has a significant effect on the protein (Andersson et al. 2011). Second, α-synuclein does not appear to be a substrate for UCH-L1. Third, the biochemical function of UCH-L1 is uncertain. Under some conditions, it can be shown to have E2-independent ubiquitin-E3-ligase activity, the opposite of what its sequence similarity to other ubiquitin carboxy-terminal hydrolases might lead one to expect (Liu et al. 2002). An interesting possibility is that UCH-L1 might actually be a ubiquitin-editing enzyme, removing ubiquitin from some sites and then attaching ubiquitin at others, thereby changing the subcellular location and/or half-life of its target proteins (Bheda et al. 2010). However, UCH-L1 has rather low catalytic activity in vitro, and it makes up about 1% of the protein in a neuron, so there is some doubt as to whether it even functions as an enzyme in vivo. The only certainty is that UCH-L1 does seem to have a generally neuroprotective effect, as its overexpression partially restores learning and memory in animal models of Alzheimer disease, suggesting that efforts to increase its steady-state levels in PD brain may be worthwhile (Gong and Lesnik 2007).
DJ-1 was originally discovered simultaneously as a possible oncoprotein in humans and a stress-response protein in yeast. Its crystal structure shows a conserved potential catalytic site that resembles those of the cysteine proteases, but no proteolytic activity has ever been convincingly shown (Wilson et al. 2003). The “active site” cysteine is readily oxidized in vitro and in vivo (Wilson 2011), and oxidation appears to change the subcellular localization of the protein, sending it from the cytoplasm to the mitochondria, and the modification is required for its one established biochemical function: mitochondrial DJ-1 protects a variety of cells from oxidative stress (Canet-Avilés et al. 2001). DJ-1-associated familial cases of PD all present as autosomal recessive, implying that the disease is caused by a loss of this protective function, and structural and biochemical studies of the mutants by Wilson and associates are consistent with this hypothesis (Lakshminarasimhan et al. 2008). Because oxidation appears to increase the tendency of α-synuclein to misfold and aggregate (Fink 2006), a direct connection between DJ-1 and α-synuclein is not hard to envision. However, Lewy bodies have not been observed in DJ-1 knockout mice. Of course, if the actual neurotoxic species of α-synuclein is some small oligomer rather than the macroscopic aggregates, as increasingly seems likely, Lewy body formation may not be a good indication of a link between any other gene and α-synuclein pathology in PD. One report that DJ-1 can act as a chaperone to suppress the formation of α-synuclein amyloid fibrils in vitro is intriguing (Zhou et al. 2006), but the relevance of these observations to the in vivo function of DJ-1 is unclear.
Figure 3 shows an attempt to link all these various genetic and environmental factors into a single model of PD etiology that ultimately funnels through the formation of some cytotoxic species of α-synuclein. Although we cheerfully acknowledge that much of this model is so speculative as to border on the imaginary, it is meant to provide a framework for thinking about the disease from a proteostasis perspective. If it is correct even in part, then it suggests that two strategies not yet widely pursued should be added to the current approaches to treating this disorder: stabilization of the putative native state of α-synuclein, and inhibition of the covalent changes to that protein that promote its destabilization. The model also lends support to efforts to bypass synuclein toxicity by activating protective pathways, especially those aimed at mitochondrial stress and disruption of proteasome and lysosome function.
AMYOTROPHIC LATERAL SCLEROSIS (LOU GEHRIG’S DISEASE)
As mentioned above, although only 1/40th as many people are believed to suffer from ALS as from Parkinson’s disease, their incidence actually only differs about 10-fold because of the rapid progression of the former to mortality. The aggressiveness of ALS actually makes this disease a better candidate for proteostasis approaches in principle, because clinical trials are much easier to design and the outcomes are easier to measure. Thus, it is important to establish whether ALS is in fact a disease that can be treated by strategies based on proteostasis. As we shall see, there are suggestions that it may be, but the issue is far from settled and any such approaches may require thinking about proteostasis in new ways.
SOD1 in Familial ALS
Mutations in the gene encoding Cu,Zn superoxide dismutase-1 (SOD1) were the first genetic link to familial, or inherited, forms of amyotrophic lateral sclerosis (FALS) (Rosen et al. 1993). Since then, more than 150 autosomal dominant mutations have been identified in this gene (http://alsod.iop.kcl.ac.uk/), which collectively account for 20%–25% of FALS cases and represent the most common factor associated with FALS (Bosco and Landers 2010). SOD1 is a 32 kDa homodimeric protein (Fig. 4). The normal posttranslational modifications of each SOD1 subunit include amino-terminal acetylation; coordination of a copper atom that confers the enzymatic antioxidizing activity; and coordination of a Zn atom and formation of an intrasubunit disulfide bond between Cys57 and Cys146, both of which are important for maintaining structural integrity of the protein (Chattopadhyay and Valentine 2009).
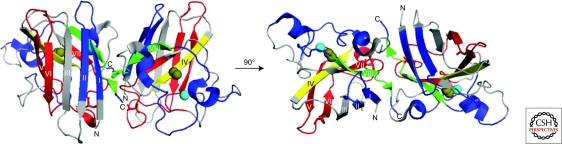
Figure 4.
Probing for misfolded WT-SOD1 in SALS patient tissues with conformation specific antibodies. The epitopes for a series of six anti-SOD1 conformation-specific antibodies designed to detect misfolded SOD1 are mapped onto the structure of the WT-SOD1 dimer (Protein database, accession #2C9V) (Strange et al. 2006), where each β-strand is labeled (I–VIII), copper atoms are colored brown, and zinc atoms cyan. The left view of SOD1 has undergone a 90° rotation about the x-axis to generate the right view. Forsberg et al. generated three polyclonal antibodies that epitope mapped to either SOD1 sequences 4–20, 57–72, or 131–153 (highlighted in blue, except for residues 143–151 that are highlighted in green); all three antibodies detected aggregated WT-SOD1 in SALS spinal cord tissues (Forsberg et al. 2010). The monoclonal C4F6 antibody that detected misfolded, soluble WT-SOD1 in SALS spinal cord tissues was epitope mapped to exon 4 (highlighted in red) (Bosco and Landers 2010). Anti-SOD1 antibodies that failed to detect misfolded SOD1 in SALS tissues include the USOD antibody, which epitope mapped to residues 42–48 (highlighted in yellow) (Kerman et al. 2010), and the SEDI antibody, which epitope mapped to 143–151 (highlighted in green) (Liu et al. 2009). Note that the SOD1 sequence recognized by SEDI (143–151; green) is encoded within the epitope of the Forsberg et al. antibody (131–153; blue and green) that detected WT-SOD1 aggregates.
ALS-linked mutations in SOD1 typically do not induce a loss of normal antioxidizing activity. In fact, many SOD1 mutants retain significant levels of enzymatic activity in cell and animal models (Borchelt et al. 1994; Gurney et al. 1994). Most evidence indicates that SOD1 mutants misfold and subsequently acquire a gain of toxic function. However, the biochemical nature of the misfolded species (e.g., soluble dimers, oligomers, or insoluble aggregates) is controversial (Chattopadhyay and Valentine 2009). Moreover, the aggregation of mutant-SOD1 in vitro has not consistently correlated with ALS pathogenesis in vivo (Wang et al. 2008a,b; Prudencio et al. 2009; Vassall et al. 2011), either because mutant-SOD1 aggregation propensity is not a significant factor in ALS pathogenesis or because the in vitro assays employed do not adequately reflect the aggregation process in vivo. Because multiple cellular processes including axonal transport (Collard et al. 1995; Williamson and Cleveland 1999; Morfini et al. 2009; Bilsland et al. 2010), mitochondrial function (Magrané et al. 2009; Li et al. 2010; Pedrini et al. 2010), ER-stress (Nishitoh et al. 2008; Hetz et al. 2009), and response to oxidative stress (Ferrante et al. 1997; Barber and Shaw 2010) are adversely affected by mutant-SOD1, it is difficult to ascertain which pathways and/or targets of mutant-SOD1 are most significant to the onset of FALS pathogenesis (Rothstein 2009).
Despite the fact that the pathomechanism of mutant-SOD1 in FALS remains elusive, mutant-SOD1 remains the primary target of therapeutic strategies against FALS (Bosco and Landers 2010). Antisense oligonucleotides effectively reduced the load of intracellular SOD1 protein in vivo, delaying disease onset and/or extending survival in ALS-transgenic rodent models (Ralph et al. 2005; Raoul et al. 2005; Smith et al. 2006). The observation that intracellular mutant-SOD1 proteins are actively secreted by chromogranins (Urushitani et al. 2006) opened the possibility for immunotherapeutics against extracellular SOD1. Subsequently, both active and passive immunization trials were shown to have a neuroprotective effect in ALS-transgenic mice (Urushitani et al. 2007; Gros-Louis et al. 2010; Takeuchi et al. 2010). Both antisense oligonucleotide and immunotherapy strategies against mutant-SOD1 are currently under development for human FALS patients.
A Role for Wild-Type SOD1 in Sporadic ALS
Structural analyses of SOD1 mutants compared to wild-type (WT)-SOD1 have failed to reveal significant mutation-induced alterations to SOD1 tertiary and quaternary structure (Rakhit and Chakrabartty 2006). These observations suggest that relatively subtle perturbations to the SOD1 protein are sufficient to induce a gain of toxic function related to ALS pathogenesis, or that the pathological species is some minor conformation in equilibrium with the native-like one. Importantly, nongenetic perturbations to the normal posttranslational modifications (described above) of WT-SOD1 also cause the protein to misfold, which has important implications for the sporadic forms of ALS that account for 90% of disease cases. For example, Cu and Zn depletion (Estevez et al. 1999; Durazo et al. 2009), Cys57-Cys146 disulfide reduction (Lindberg et al. 2004), and defective subunit dimerization (Rakhit et al. 2004) have all been shown to destabilize and enhance the aggregation propensity of WT-SOD1. In addition, aberrant covalent modifications of WT-SOD1, such as oxidation of amino acid side chains (Rakhit et al. 2002; Ezzi et al. 2007; Bosco et al. 2010a), also induce the protein to misfold.
Although these studies provide compelling evidence that the WT-SOD protein can misfold in vitro, what are the implications of misfolded WT-SOD1 in SALS in vivo (Bredesen et al. 1997; Kabashi et al. 2007)? A chemical modification approach applied to extracts of SALS and FALS spinal cord tissues identified a 32kDa species detected by an anti-SOD1 antibody that was present only in SALS and FALS, but not in healthy or other disease controls (Gruzman et al. 2007). Although these results initially indicated a misfolded SOD1 species that was common to both SALS and FALS, subsequent studies revealed this 32kDa species was in fact carbonic anhydrase I and not SOD1 (Liu et al. 2010).
An alternative approach to chemical labeling for the characterization of misfolded SOD1 species in vivo has been the use of SOD1 conformation-specific antibodies. Misfolded WT-SOD1 was detected in ∼40% of SALS spinal tissues by immunohistochemistry with the monoclonal C4F6 antibody (Bosco et al. 2010a), which was generated against the FALS-linked SOD1 G93A mutant (Urushitani et al. 2007). These data suggest that modified forms of WT-SOD1 associated with SALS in vivo may adopt a similar, misfolded conformation to mutant-SOD1 proteins associated with FALS. This study also shed light onto the functional consequences of WT-SOD1 misfolding; both misfolded WT-SOD1 from SALS tissues and FALS-linked mutant-SOD1 proteins inhibited axonal transport in a squid axoplasm model through activation of the p38 MAPK pathway (Morfini et al. 2009; Bosco et al. 2010a).
The detection of misfolded WT-SOD1 in SALS tissues was also reported in an independent study that employed different SOD1 conformation-specific antibodies (Forsberg et al. 2010). Intriguingly, the antibodies reported in this study detected granular inclusions of SOD1, whereas the C4F6 antibody detects soluble misfolded forms of WT-SOD1 (Bosco et al. 2010a). Two conformation-specific antibodies that failed to detect misfolded SOD1 in SALS tissues include the USOD (unfolded SOD1) antibody and the SEDI (SOD1 exposed dimer interface) antibody (Rakhit et al. 2007; Liu et al. 2009). The difference in the immunohistochemistry staining between these studies is likely because of the different antibodies used. Although there is some overlap in the epitopes recognized by the antibodies employed by Forsberg et al. (2010) and the SEDI antibody (see sequence highlighted in green in Fig. 4), none of the epitopes are identical across these studies. Although further investigations are expected to reveal the extent to which WT-SOD1 is involved in SALS, these initial studies show that aberrant SOD1 proteostasis may provide a common link between FALS and SALS. They also raise the possibility of employing the therapeutic strategies highlighted above for FALS to also treat SALS (Bosco et al. 2010a).
This raises the proteostasis question. How should one stabilize the native, benign fold of SOD1? Strategies involving cross-linking (Ray et al. 2004; Auclair et al. 2010) and ligand binding (Ray et al. 2005) have all showed promise of stabilizing the dimer in vitro. None has been tried in vivo yet.
ALS as a Disease of RNA Quality Control
Although ALS research was dominated for many years by the apparent connection between SOD1 misfolding and FALS, the recent discovery of two additional genes, mutations which appear to be associated with both familial disease and increased risk for sporadic ALS, has caused a rethinking of the molecular etiology of most forms of the disorder. In contrast to the antioxidant enzymatic activity of SOD1 and its tendency to form pathological aggregates, these two new genes, TDP-43 and FUS/TLS, encode primarily nuclear proteins that function in RNA biology. As we shall see, they not only raise the possibility that ALS is an RNA-based disease, they also suggest that a broader view of the concept of proteostasis may be valuable for developing new approaches to its treatment.
TDP-43
ALS research was turned on its head in 2006 with the report by Trojanowski and Lee and coworkers that inclusions found in sporadic ALS patients contained a single-strand RNA/DNA-binding protein, TDP-43 (Neumann et al. 2006). Ubiquitin-positive, tau-, and α-synuclein-negative inclusions are histopathological hallmarks of ALS. Pathologic TDP-43 was hyperphosphorylated, ubiquitinated, and cleaved to generate carboxy-terminal fragments and was recovered only from affected central nervous system regions, including hippocampus, neocortex, and spinal cord. Subsequently, mutations in the TDP-43 gene were shown to cause a significant percentage of FALS cases (reviewed in Chen-Plotkin et al. 2010).
TDP-43 is a large protein comprising a number of domains, including RNA-binding and putative protein–protein interaction modules (Fig. 5). TDP-43 is normally localized to the nucleus and its diverse functions appear to include regulation of gene expression and inhibition of mRNA splicing. There is some evidence that it can shuttle to the cytoplasm, at least in small amounts, where it associates with stress granules, punctate bodies that sequester mRNA molecules and ribosomal components, during oxidative stress (Columbrita et al. 2009; Dewey et al. 2011). Recent data even suggest that one of its functions is to regulate stress granule dynamics via differential regulation of the G3BP and TIA-1 proteins (McDonald et al. 2011).
Figure 5.
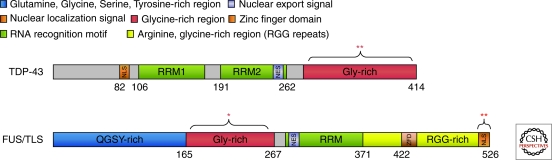
Domain structures of TDP-43 and FUS/TLS. TDP-43 and FUS have somewhat similar domain structures and each contain multiple, functional domains (see color coded key). More than 35 ALS-linked mutations have been identified in TDP-43, the vast majority of which are located within the Gly-rich domain (denoted by **) (Lagier-Tourenne et al. 2010). A similar number (∼30) of ALS-linked mutations have been reported in FUS; however, most FUS mutations are within the nuclear localization signal (NLS; denoted by **) with additional mutations in the Gly-rich region (denoted by *) (Lagier-Tourenne et al. 2010).
In both TDP-43-dependent FALS patients and many SALS patients, TDP-43 appears to be mislocalized to the cytoplasm, where it forms punctate inclusions (Giordana et al. 2010). Mislocalized TDP-43 is also toxic to neuronal cells in culture (Barmada et al. 2010). Transgenic mice expressing human TDP-43 with a defective nuclear localization signal have been generated and compared with mice expressing WT-hTDP-43 to determine the effects of mislocalized cytoplasmic TDP-43 on neuronal viability (Igaz et al. 2011). Expression of either hTDP-43-ΔNLS or WT-hTDP-43 led to neuron loss in selectively vulnerable forebrain regions, corticospinal tract degeneration, and motor spasticity that recapitulated many aspects of TDP-43-associated motor neuron disease in people. Importantly, only rare cytoplasmic phosphorylated and ubiquinated TDP-43 inclusions were seen in hTDP-43-ΔNLS mice, in contrast to humans, suggesting that cytoplasmic inclusions may not be required to induce neuronal death. Instead, neurodegeneration in h-TDP-43- and hTDP-43-ΔNLS-expressing neurons was accompanied by a dramatic down-regulating and increased expression/mislocalization to the cytoplasm of the endogenous mouse TDP-43. Moreover, mice expressing hTDP-43-ΔNLS showed profound changes in gene expression in cortical neurons, implying that mislocalization of endogenous nuclear THP-43 exerts at least some of its toxic effects through a loss of normal TDP-43 function(s). If this is indeed the case, then a novel proteostasis approach aimed at preserving the proper localization of this protein may represent a viable therapeutic strategy for at least some FALS and SALS cases. It may also be possible to identify and activate bypass pathways that restore at least some of the functions lost when TDP-43 is mislocalized.
The role of TDP-43 fragmentation and other posttranslational modifications in its trafficking and pathogenesis has yet to be established. In any case, although TDP-43 containing aggregates are detected in a majority of ALS cases, the results to date imply TDP-43 aggregation does not necessarily correlate with toxicity. Moreover, the emerging reports linking TDP-43 pathology to a spectrum of neurodegenerative disorders, including AD and FTLD (Chen-Plotkin et al. 2010), are consistent with a general loss of regulation of this protein under conditions of stress and degeneration. Therefore, it may not be possible to treat TDP-43-dependent disorders by chaperone up-regulation or alteration of other conventional proteostasis pathways as for α-synuclein and SOD1.
FUS/TLS
Fused in sarcoma/translocated in liposarcoma (FUS/TLS or FUS) is an RNA binding, nuclear protein that was originally identified in the context of an oncoprotein comprising the amino terminus of FUS and the transcription factor CHOP (Crozat et al. 1993; Rabbitts et al. 1993). In 2009, mutations at the carboxyl terminus of FUS were linked to FALS and shown to induce cellular FUS mislocalization from the nucleus to the cytoplasm (Kwiatkowski et al. 2009; Vance et al. 2009). This cytoplasmic mislocalization was later explained by studies demonstrating that many of the ALS-associated mutations disrupt a functional, albeit nonclassical, nuclear localization signal (NLS) (Fig. 5) (Dormann et al. 2010; Gal et al. 2010). Subsequent reports have documented additional mutations within the carboxyl terminus and Glycine-rich domains of FUS in association with both FALS (Belzil et al. 2009; Blair et al. 2009; Chiò et al. 2009; Corrado et al. 2009; Damme et al. 2009; Ticozzi et al. 2009; Waibel et al. 2010; Yan et al. 2010) and SALS (Belzil et al. 2009; Corrado et al. 2009; DeJesus-Hernandez et al. 2010; Lai et al. 2010) across various populations. An association of FUS with other neurodegenerative disorders has been suggested by immunological costaining of FUS within the pathological aggregates of frontotemporal lobar degeneration (FTLD)-related (Munoz et al. 2009; Neumann et al. 2009; Lagier-Tourenne et al. 2010) and polyglutamine diseases (Doi et al. 2009; Lagier-Tourenne et al. 2010). Although TDP-43 pathology is also detected across different neurological disorders (Chen-Plotkin et al. 2010), FUS and TDP-43 pathology appear to be mutually exclusive in a majority of disease cases (Lagier-Tourenne et al. 2010), and in yeast models of FUS and TDP-43 proteotoxicity (Ju et al. 2011; Sun et al. 2011).
As in the case of TDP-43, FUS has been implicated in various RNA- and DNA-related processes including transcription, mRNA splicing, nucleocytoplasmic mRNA shuttling, and stress granule assembly (Lagier-Tourenne et al. 2010; Lemmons et al. 2010; van Blitterswijk and Landers 2010). To date, there are no published ALS-FUS animal or mammalian cell-based models wherein exogenous ALS-linked FUS induces cellular toxicity in a manner that is mutant specific; at lower expression levels, neither mutant nor WT-FUS is toxic (Bosco et al. 2010b) and yet in yeast models of FUS proteotoxicity (Ju et al. 2011; Sun et al. 2011) both WT and mutant-FUS are toxic (discussed below). These models are dissimilar to ALS-linked SOD1 models, where exogenous mutant-SOD1 is toxic even in the presence of endogenous WT-SOD1 and overexpression of WT-SOD1 is not overtly toxic (Bruijn et al. 2004). Thus, although ALS-linked mutant SOD1 appears to cause ALS through a gain of toxic function mechanism (discussed above), it is not yet clear whether mislocalized mutant-FUS exerts a gain of toxic function within the cytoplasm, or whether a loss of normal nuclear FUS function contributes to ALS pathogenesis in mammalian cells.
Although a loss of FUS function induced by FALS-linked mutations has not been shown, the silencing of endogenous FUS expression has resulted in a loss of function phenotype in the context of different cellular processes. For example, hippocampal neurons from FUS-null mice show abnormal spine morphology (Fujii et al. 2005), indicating a role for FUS in spine maintenance that may be relevant to processes such as synaptic plasticity. In addition, knocked-down expression of FUS in mouse macrophage cells revealed a loss of function with respect to transcriptional regulation of CCND1/cyclin D1. FUS was shown to bind the CCND1 promoter in response to DNA damaging signals, which in turn represses the expression of cyclin D1 and inhibits the cell cycle (Wang et al. 2008a). Although an analogous role for FUS in CCND1/cyclin D1 regulation in neurons has not yet been shown, a potential misregulation of the cell cycle has been linked to neurodegenerative disorders (Klein and Ackerman 2003), including ALS (Ranganathan and Bowser 2003).
Studies in yeast models of FUS proteotoxicity (Ju et al. 2011; Sun et al. 2011) indicate that a gain of toxic FUS function may result from altered FUS expression levels and/or aberrant cellular localization. In contrast to mammalian cells (Bosco et al. 2010b; Dormann et al. 2010; Gal et al. 2010), expression of WT-FUS is almost as toxic as FALS-linked FUS mutants in yeast. The toxicity of WT-FUS is due in part to the inefficiency of the human-FUS NLS in yeast, which leads to increased cytoplasmic expression levels of WT-FUS. Importantly, the toxicity of WT-FUS is ameliorated when FUS is redirected to the nucleus by an efficient yeast NLS, indicating that, at least in this model organism, mislocalization of FUS is sufficient for a gain of toxic function. In contrast, cytoplasmic FUS aggregates were not sufficient to induce toxicity. Therefore, the correlation of FUS toxicity and cytoplasmic mislocalization, but not necessary cytoplasmic aggregation, is reminiscent of the TDP-43 mouse models discussed above. FUS-toxicity in yeast was also modulated by factors involved in RNA processing, including RNA quality control and the stress granule pathway (Ju et al. 2011; Sun et al. 2011). In fact, FALS-linked mutant FUS proteins that mislocalize to the cytoplasm in mammalian cells were shown to incorporate into stress granules under conditions of cellular stress (e.g., oxidative stress, heat-shock, etc.) (Fig. 6) (Bosco et al. 2010b; Dormann et al. 2010; Gal et al. 2010), and both WT-FUS and mutant-FUS incorporated into what may be stress granules in yeast (Ju et al. 2011; Sun et al. 2011). Stress granules are stalled translational complexes that form during induced cellular stress, and function to regulate the expression of proteins that are required to overcome stress (Anderson et al. 2008). That both FUS and TDP-43 (Columbrita et al. 2009; Liu-Yesucevitz et al. 2010; McDonald et al. 2011) are associated with stress granules has generated the hypothesis that altered stress response mediated by FUS and TDP-43 proteotoxicity may be linked to ALS pathogenesis. This hypothesis is further supported by the detection of stress granule markers in end-stage pathological aggregates in ALS and FTLD (Dormann et al. 2010; Liu-Yesucevitz et al. 2010).
Figure 6.
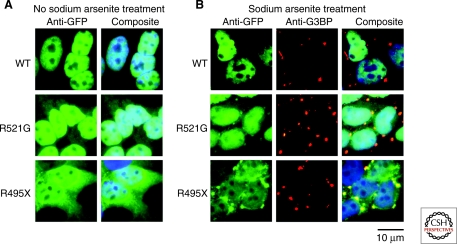
FUS incorporates into stress granules. (A) GFP-FUS WT is predominately localized to the nucleus of stably-transfected HEK-239 cells. Both a point mutation (R521G) within the nuclear localization signal (NLS; Fig. 5) and a truncation mutation (R495X) that eliminates the NLS increase the cytoplamsic expression level of GFP-FUS according to the following trend R495X > R521G > WT (Bosco et al. 2010b). DAPI (blue) is used to stain the nuclei. (B) On exposure to sodium arsenite, which induces oxidative stress, mutant-FUS proteins become incorporated into cytoplasmic foci called stress granules, which are detected with the stress granule marker anti-G3BP (red) (Bosco et al. 2010b). The extent to which FUS incorporates into stress granules correlates with the cytoplamsic levels of the FUS protein (R495X > R521G > WT).
How is one to think about the various protein factors that have been implicated in sporadic and familial forms of ALS? In the end, our view of this disorder as a proteostasis disease, or one in which proteostasis-based approaches are useful therapeutically, will depend on whether commonalities among the different types of ALS can be established. Perhaps SOD1 misfolding via, e.g., oxidation can eventually be linked conclusively to all forms of sporadic ALS; then a proteostasis approach aimed at this protein might be of widespread therapeutic benefit. Or perhaps, as some researchers are now starting to speculate, ALS is not a single disease but a spectrum of disorders with many nonoverlapping causes. If that is the case, proteostasis-based therapies may need to be tailored to each specific form—a daunting challenge in the case of the sporadic, idiopathic disease. Or perhaps SOD1-dependent familial ALS is an outlier and most of the other familial and sporadic forms of ALS have a common, RNA-based etiology. In that case, the SOD1 form of the disease may be the easiest type to treat via a proteostsis-based approach, but other forms of ALS may require approaches directed at RNA quality control pathways. Or it may be that all forms of ALS ultimately arise from derangement of some RNA quality-control pathway, in which case proteostasis-based strategies may need to include ways to control the subcellular location of proteins like TDP-43 and FUS/TLS that are not in any sense misfolded. Or, just maybe, the very notion of proteostasis should be expanded to include RNAstasis or ribonucleostasis, or whatever new term we still need to invent.
REFERENCES
- Abeliovich A, Schmitz Y, Fariñas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, et al. 2000. Mice lacking α-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25: 239–252 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Andersson FI, Werrell EF, McMorran L, Crone WJ, Das C, Hsu ST, Jackson SE 2011. The effect of Parkinson’s-Disease-associated mutations on the deubiquitinating enzyme UCH-L1. J Mol Biol 407: 261–272 [DOI] [PubMed] [Google Scholar]
- Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, et al. 2009. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine). J Neurosci 29: 15846–15850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair JR, Boggio KJ, Petsko GA, Ringe D, Agar JN 2010. Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc Natl Acad Sci 107: 21394–21399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber SC, Shaw PJ 2010. Oxidative stress in ALS: Key role in motor neuron injury and therapeutic target. Free Radic Biol Med 48: 629–641 [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S 2010. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci 30: 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, S2D team, Hince P, Desjarlais A, et al. 2009. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology 73: 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Z, Smith KA, LaVoie MJ 2010. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry 49: 5511–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J 2010. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: Implication in mitosis. Cell Cycle 9: 980–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland LG, Sahai E, Kelly G, Golding M, Greensmith L, Schiavo G 2010. Deficits in axonal transport precede ALS symptoms in vivo. Proc Natl Acad Sci 107: 20523–20528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, et al. 2006. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol 60: 557–569 [DOI] [PubMed] [Google Scholar]
- Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA 2009. FUS mutations in amyotrophic lateral sclerosis: Clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry 81: 539–545 [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH Jr, Price DL, Sisodia SS, Cleveland DW 1994. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci 91: 8292–8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Landers JE 2010. Genetic determinants of amyotrophic lateral sclerosis as therapeutic targets. CNS Neurol Disord Drug Targets 9: 779–790 [DOI] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, et al. 2010a. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci 13: 1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ Jr, Sapp P, McKenna-Yasek D, Brown RH Jr, Hayward LJ 2010b. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19: 4160–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredesen DE, Ellerby LM, Hart PJ, Wiedau-Pazos M, Valentine JS 1997. Do posttranslational modifications of CuZnSOD lead to sporadic amyotrophic lateral sclerosis? Ann Neurol 42: 135–137 [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW 1998. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281: 1851–1854 [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW 2004. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci 27: 723–749 [DOI] [PubMed] [Google Scholar]
- Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC 2010. α-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 329: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet-Avilés RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR 2001. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci 101: 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Lansbury PT 2003. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci 26: 267–298 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M, Valentine JS 2009. Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antioxid Redox Sign 11: 1603–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ 2010. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol 6: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, Sabatelli M, Monsurrò MR, Battistini S, Mandrioli J, et al. 2009. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging 30: 1272–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT 2010. A pivotal role for PINK1 and autophagy in mitochondrial quality control: Implications for Parkinson disease. Hum Mol Genet 19: R28–R37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM 2004. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 304: 1328–1331 [DOI] [PubMed] [Google Scholar]
- Collard JF, Cote F, Julien JP 1995. Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature 375: 61–64 [DOI] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A 2009. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem 111: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Cookson MR 2005. Neurons inflict self-harm. Nat Med 11: 1159–1161 [DOI] [PubMed] [Google Scholar]
- Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Sorarù G, Carlomagno Y, Ghezzi S, Pensato V, et al. 2009. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet 47: 190–194 [DOI] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363: 640–644 [DOI] [PubMed] [Google Scholar]
- Dachsel JC, Farrer MJ 2010. LRRK2 and Parkinson disease. Arch Neurol 67: 542–547 [DOI] [PubMed] [Google Scholar]
- Damme PV, Goris A, Race V, Hersmus N, Dubois B, Bosch LV, Matthijs G, Robberecht W 2009. The occurrence of mutations in FUS in a Belgian cohort of patients with familial ALS. Eur J Neurol 17: 754–756 [DOI] [PubMed] [Google Scholar]
- Dawson TM, Ko HS, Dawson VL 2010. Genetic animal models of Parkinson’s Disease. Neuron 66: 646–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Wood NW 2009. PINK1 function in health and disease. EMBO Mol Med 1: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, et al. 2010. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat 31: E1377–E1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P III, Good SK, Johnson BA, Herz J, Yu G 2011. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol 31: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y 2009. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res 66: 131–133 [DOI] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. 2010. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 29: 2841–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufty BM, Warner LR, Hou ST, Jiang SX, Gomez-Isla T, Leenhouts KM, Oxford JT, Feany MB, Masliah E, Rohn TT 2007. Calpain-cleavage of α-synuclein: Connecting proteolytic processing to disease-linked aggregation. Am J Pathol 170: 1725–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazo A, Shaw BF, Chattopadhyay M, Faull KF, Nersissian AM, Valentine JS, Whitelegge JP 2009. Metal-free superoxide dismutase-1 and three different ALS variants share a similar partially unfolded {β}-barrel at physiological temperature. J Biol Chem 284: 34382–34389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L, Beckman JS 1999. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science 286: 2498–2500 [DOI] [PubMed] [Google Scholar]
- Ezzi SA, Urushitani M, Julien JP 2007. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem 102: 170–178 [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH Jr, Beal MF 1997. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem 69: 2064–2074 [DOI] [PubMed] [Google Scholar]
- Fink AL 2006. The aggregation and fibrillation of α-synuclein. Acc Chem Res 39: 628–634 [DOI] [PubMed] [Google Scholar]
- Forsberg K, Jonsson PA, Andersen PM, Bergemalm D, Graffmo KS, Hultdin M, Jacobsson J, Rosquist R, Marklund SL, Brännström T 2010. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS One 5: e11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T 2005. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol 15: 587–593 [DOI] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Zhu H 2010. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordana MT, Piccinini M, Grifoni S, De Marco G, Vercellino M, Magistrello M, Pellerino A, Buccinnà B, Lupino E, Rinaudo MT 2010. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol 20: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Leznik E 2007. The role of ubiquitin C-terminal hydrolase L1 in neurodegenerative disorders. Drug News Perspect 20: 365–370 [DOI] [PubMed] [Google Scholar]
- Graham DG 1978. Oxidative pathways for catecholamines in the genesis of neuromelanin and cytotoxic quinones. Mol Pharmacol 14: 633–643 [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. 2006. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23: 329–341 [DOI] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniëls V, Lewis P, Jain S, Ding J, Syed A, et al. 2008. The Parkinson’s disease associated Leucine rich repeat kinase 2 (LRRK2) is a dimer that undergoes intra-molecular autophosphorylation. J Biol Chem 283: 16906–16914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Soucy G, Lariviere R, Julien JP 2010. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J Neurochem 113: 1188–1199 [DOI] [PubMed] [Google Scholar]
- Gruzman A, Wood WL, Alpert E, Prasad MD, Miller RG, Rothstein JD, Bowser R, Hamilton R, Wood TD, Cleveland DW, et al. 2007. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci 104: 12524–12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. 1994. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264: 1772–1775 [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O 2006. Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet 15: 2059–2075 [DOI] [PubMed] [Google Scholar]
- Hastings TG, Lewis DA, Zigmond MJ 1996. Role of oxidation in the neurotoxic effects of intrastriatal dopamine injections. Proc Natl Acad Sci 93: 1956–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Kubo S, Sato S, Hattori N 2009. Pathogenesis of familial Parkinson’s disease: New insights based on monogenic forms of Parkinson’s disease. J Neurochem 111: 1075–1093 [DOI] [PubMed] [Google Scholar]
- Henn IH, Gostner JM, Lackner P, Tatzelt J, Winklhofer KF 2005. Pathogenic mutations inactivate parkin by distinct mechanisms. J Neurochem 92: 114–122 [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH 2009. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev 23: 2294–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S, Biskup S, West AB, Trinkaus D, Dawson VL, Faull RL, Waldvogel HJ, Arai H, Dawson TM, Moore DJ, et al. 2007. Localization of Parkinson’s disease-associated LRRK2 in normal and pathological human brain. Brain Res 1155: 208–219 [DOI] [PubMed] [Google Scholar]
- Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT 2009. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci 29: 1011–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, et al. 2011. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 121: 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW 2009. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol 187: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RH Jr, Lindquist S, et al. 2011. A yeast model of FUS/TLS-dependent cytoxicity. PLoS Biol. 9: e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Rouleau GA 2007. Oxidized/misfolded superoxide dismutase-1: The cause of all amyotrophic lateral sclerosis? Ann Neurol 62: 553–559 [DOI] [PubMed] [Google Scholar]
- Kahle PJ, Neumann M, Ozmen L, Haass C 2000. Physiology and pathophysiology of α-Synuclein: Cell culture and transgenic animal models based on a Parkinson’s Disease-associated protein. Ann NY Acad Sci 920: 33–41 [DOI] [PubMed] [Google Scholar]
- Kawahara K, Hashimoto M, Bar-On P, Ho GJ, Crews L, Mizuno H, Rockenstein E, Imam SZ, Masliah E 2008. α-Synuclein aggregates interfere with Parkin solubility and distribution: Role in the pathogenesis of Parkinson disease. J Biol Chem 283: 6979–6987 [DOI] [PubMed] [Google Scholar]
- Kerman A, Liu HN, Croul S, Bilbao J, Rogaeva E, Zinman L, Robertson J, Chakrabartty A 2010. Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta Neuropathol 119: 335–344 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Klein JA, Ackerman SL 2003. Oxidative stress, cell cycle, and neurodegeneration. J Clin Invest 111: 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW 2010. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19: R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, Dillman A, Cookson M, Calvo A, Battistini S, Giannini F, et al. 2010. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA 2008. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry 47: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel HA, Lansbury PT Jr 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys 39: 167–201 [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Hastings TG 1999. Dopamine quinone formation and protein modification associated with the striatal neurotoxicity of methamphetamine: Evidence against a role for extracellular dopamine. J Neurosci 19: 1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ 2005. Dopamine covalently modifies and functionally inactivates parkin. Nat Med 11: 1214–1221 [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Cortese GP, Ostaszewski BL, Schlossmacher MG 2007. The effects of oxidative stress on parkin and other E3 ligases. J Neurochem 103: 2354–2368 [DOI] [PubMed] [Google Scholar]
- Lemmens R, Moore MJ, Al-Chalabi A, Brown RH Jr, Robberecht W 2010. RNA metabolism and the pathogenesis of motor neuron diseases. Trends Neurosci 33: 249–258 [DOI] [PubMed] [Google Scholar]
- Li W, West N, Colla E, Pletnikova O, Troncoso JC, Marsh L, Dawson TM, Jäkälä P, Hartmann T, Price DL, et al. 2005. Aggregation promoting C-terminal truncation of α-synuclein is a normal cellular process and is enhanced by the familial Parkinson’s disease-linked mutations. Proc Natl Acad Sci 102: 2162–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Vande Velde C, Israelson A, Xie J, Bailey AO, Dong MQ, Chun SJ, Roy T, Winer L, Yates JR, et al. 2010. ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import. Proc Natl Acad Sci 107: 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg MJ, Normark J, Holmgren A, Oliveberg M 2004. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci 101: 15893–15898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr 2002. The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease susceptibility. Cell 111: 209–218 [DOI] [PubMed] [Google Scholar]
- Liu HN, Sanelli T, Horne P, Pioro EP, Strong MJ, Rogaeva E, Bilbao J, Zinman L, Robertson J 2009. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Ann Neurol 66: 75–80 [DOI] [PubMed] [Google Scholar]
- Liu J, Akhavan A, Lu M, Gruzman A, Lingappa VR, An J, Bowser R 2010. Carbonic anhydrase I is recognized by an SOD1 antibody upon biotinylation of human spinal cord extracts. Int J Mol Sci 11: 4051–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sehrman M, et al. 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: Analysis of cultured cells and pathological brain tissue. PLoS One 5: e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G, Sidransky E 2010. Autosomal recessive mutations in the development of Parkinson’s disease. Biomark Med 4: 713–721 [DOI] [PubMed] [Google Scholar]
- Magrané J, Hervias I, Henning MS, Damiano M, Kawamata H, Manfredi G 2009. Mutant SOD1 in neuronal mitochondria causes toxicity and mitochondrial dynamics abnormalities. Hum Mol Genet 18: 4552–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K 2006. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281: 3204–3209 [DOI] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C 2011. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet 20: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Mengesdorf T, Jensen PH, Mies G, Aufenberg C, Paschen W 2002. Down-regulation of parkin protein in transient focal cerebral ischemia: A link between stroke and degenerative disease? Proc Natl Acad Sci 99: 15042–15047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, Brown RH Jr, Brown H, Tiwari A, Hayward L, et al. 2009. Axonal transport defects in neurodegenerative diseases. J Neurosci 29: 12776–12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moszczynska A, Yamamoto BK 2011. Methamphetamine oxidatively damages parkin and decreases the activity of 26S proteasome in vivo. J Neurochem 116: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR 2009. FUS pathology in basophilic inclusion body disease. Acta Neuropathol 118: 617–627 [DOI] [PubMed] [Google Scholar]
- Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, Trojanowski JQ, Lee VM 2003. Role of α-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry 42: 8530–8540 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR 2009. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132: 2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, et al. 2010. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J 430: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H 2008. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev 22: 1451–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati A, Fournier M, Lashuel HA 2010. Role of post-translational modifications in modulating the structure, function and toxicity of α-synuclein: Implications for Parkinson’s disease pathogenesis and therapies. Prog Brain Res 183: 115–145 [DOI] [PubMed] [Google Scholar]
- Outeiro TF, Lindquist S 2003. Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302: 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleologou KE, Oueslati A, Shakked G, Rospigliosi CC, Kim HY, Lamberto GR, Fernandez CO, Schmid A, Chegini F, Gai WP, et al. 2010. Phosphorylation at S87 is enhanced in synucleinopathies, inhibits α-synuclein oligomerization, and influences synuclein-membrane interactions. J Neurosci 30: 3184–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim KL, Dawson VL, Dawson TM, Palmiter RD, Trojanowski JQ, Lee VM 2003. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J Biol Chem 278: 48120–48128 [DOI] [PubMed] [Google Scholar]
- Pedrini S, Sau D, Guareschi S, Bogush M, Brown RH Jr, Naniche N, Kia A, Trotti D, Pasinelli P 2010. ALS-linked mutant SOD1 damages mitochondria by promoting conformational changes in Bcl-2. Hum Mol Genet 19: 2974–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. 1997. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276: 2045–2047 [DOI] [PubMed] [Google Scholar]
- Prudencio M, Hart PJ, Borchelt DR, Andersen PM 2009. Variation in aggregation propensities among ALS-associated variants of SOD1: Correlation to human disease. Hum Mol Genet 18: 3217–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Goldberg AL 2002. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci 99: 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P 1993. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet 4: 175–180 [DOI] [PubMed] [Google Scholar]
- Rakhit R, Chakrabartty A 2006. Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim Biophys Acta 1762: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Rakhit R, Cunningham P, Furtos-Matei A, Dahan S, Qi XF, Crow JP, Cashman NR, Kondejewski LH, Chakrabartty A 2002. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem 277: 47551–47556 [DOI] [PubMed] [Google Scholar]
- Rakhit R, Crow JP, Lepock JR, Kondejewski LH, Cashman NR, Chakrabartty A 2004. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem 279: 15499–15504 [DOI] [PubMed] [Google Scholar]
- Rakhit R, Robertson J, Vande Velde C, Horne P, Ruth DM, Griffin J, Cleveland DW, Cashman NR, Chakrabartty A 2007. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med 13: 754–759 [DOI] [PubMed] [Google Scholar]
- Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, et al. 2005. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat Med 11: 429–433 [DOI] [PubMed] [Google Scholar]
- Ranganathan S, Bowser R 2003. Alterations in G(1) to S phase cell-cycle regulators during amyotrophic lateral sclerosis. Am J Pathol 162: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P 2005. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat Med 11: 423–428 [DOI] [PubMed] [Google Scholar]
- Ray SS, Nowak RJ, Strokovich K, Brown RH Jr, Walz T, Lansbury PT Jr 2004. An intersubunit disulfide bond prevents in vitro aggregation of a superoxide dismutase-1 mutant linked to familial amytrophic lateral sclerosis. Biochemistry 43: 4899–4905 [DOI] [PubMed] [Google Scholar]
- Ray SS, Nowak RJ, Brown RH Jr, Lansbury PT Jr 2005. Small-molecule-mediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proc Natl Acad Sci 102: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. 1993. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362: 59–62 [DOI] [PubMed] [Google Scholar]
- Ross OA, Braithwaite AT, Skipper LM, Kachergus J, Hulihan MM, Middleton FA, Nishioka K, Fuchs J, Gasser T, Maraganore DM, et al. 2008. Genomic investigation of α-synuclein multiplication and parkinsonism. Ann Neurol 63: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein JD 2009. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol 65 (Suppl 1): S3–S9 [DOI] [PubMed] [Google Scholar]
- Schlehe JS, Lutz AK, Pilsl A, Lämmermann K, Grgur K, Henn IH, Tatzelt J, Winklhofer KF 2008. Aberrant folding of pathogenic Parkin mutants: Aggregation versus degradation. J Biol Chem 283: 13771–13779 [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seehus CR, Capowski EE, Aebischer P, Zhang SC, Svendsen CN 2007. Over-expression of α-synuclein in human neural progenitors leads to specific changes in fate and differentiation. Hum Mol Genet 16: 651–656 [DOI] [PubMed] [Google Scholar]
- Schneider B, Zufferey R, Aebischer P 2008. Viral vectors, animal models and new therapies for Parkinson’s disease. Parkinsonism Relat D 14 (Suppl 2): S169–S171 [DOI] [PubMed] [Google Scholar]
- Sen S, Webber PJ, West AB 2009. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J Biol Chem 284: 36346–36356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, et al. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302–305 [DOI] [PubMed] [Google Scholar]
- Shulman JM, De Jager PL, Feany MB 2011. Parkinson’s disease: Genetics and pathogenesis. Annu Rev Pathol 28: 193–222 [DOI] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, Ward CM, McAlonis-Downes M, Wei H, et al. 2006. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest 116: 2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM 2005. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of Parkin. Hum Mol Genet 14: 2571–2586 [DOI] [PubMed] [Google Scholar]
- Steele JC 2005. Parkinsonism-dementia complex of Guam. Mov Disord 20 (Suppl 12): S99–S107 [DOI] [PubMed] [Google Scholar]
- Strange RW, Antonyuk SV, Hough MA, Doucette PA, Valentine JS, Hasnain SS 2006. Variable metallation of human superoxide dismutase: Atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol 356: 1152–1162 [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD 2011. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9: e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surguchov A 2008. Molecular and cellular biology of synucleins. Int Rev Cell Mol Biol 270: 225–317 [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Fujiwara N, Ido A, Oono M, Takeuchi Y, Tateno M, Suzuki K, Takahashi R, Tooyama I, Taniguchi N, et al. 2010. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. J Neuropathol Exp Neurol 69: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ Jr, McKenna-Yasek DM, Sapp PC, et al. 2009. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology 73: 1180–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ III, Shen J 2010. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of α-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci 107: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP 2006. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci 9: 108–118 [DOI] [PubMed] [Google Scholar]
- Urushitani M, Ezzi SA, Julien JP 2007. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc Natl Acad Sci 104: 2495–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Eliezer D 2009. Biophysics of Parkinson’s disease: Structure and aggregation of α-synuclein. Curr Protein Pept Sci 10: 483–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Landers JE 2010. RNA processing pathways in amyotrophic lateral sclerosis. Neurogenetics 11: 275–290 [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Laar VS, Dukes AA, Cascio M, Hastings TG 2008. Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: Implications for Parkinson disease. Neurobiol Dis 29: 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Laar VS, Mishizen AJ, Cascio M, Hastings TG 2009. Proteomic identification of dopamine-conjugated proteins from isolated rat brain mitochondria and SH-SY5Y cells. Neurobiol Dis 34: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassall KA, Stubbs HR, Primmer HA, Tong MS, Sullivan SM, Sobering R, Srinivasan S, Briere LA, Dunn SD, Colón W, et al. 2011. Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS. Proc Natl Acad Sci 108: 2210–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AC, Crum WR, Johansson SM, Modo M 2011. Evolution of extra-nigral damage predicts behavioural deficits in a rat proteasome inhibitor model of Parkinson’s disease. PLoS One 6: e17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel S, Neumann M, Rabe M, Meyer T, Ludolph AC 2010. Novel missense and truncating mutations in FUS/TLS in familial ALS. Neurology 75: 815–817 [DOI] [PubMed] [Google Scholar]
- Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay SP, Ho MW, Lim TM, Soong TW, Pletnikova O, et al. 2005. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet 14: 3885–3897 [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R 2008a. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Johnson JL, Agar NY, Agar JN 2008b. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol 6: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo JF, Nie LL, Luo L, Zuo X, Shen L, Jiang H, Yan XX, Xia K, Pan Q, et al. 2011. Case-control study of the UCH-L1 S18Y variant in sporadic Parkinson’s disease in the Chinese population. J Clin Neurosci 18: 541–544 [DOI] [PubMed] [Google Scholar]
- Williamson TL, Cleveland DW 1999. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci 2: 50–56 [DOI] [PubMed] [Google Scholar]
- Wilson MA 2011. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid Redox Sign [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA 2003. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci 100: 9256–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklhofer KF, Henn IH, Kay-Jackson PC, Heller U, Tatzelt J 2003. Inactivation of parkin by oxidative stress and C-terminal truncations: A protective role of molecular chaperones. J Biol Chem 278: 47199–47208 [DOI] [PubMed] [Google Scholar]
- Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, Liu E, Donkervoort S, Zheng JG, Shi Y, et al. 2010. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology 75: 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, et al. 2004. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci 101: 10810–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL 2006. The oxidation state of DJ-1 regulates its chaperone activity toward α-synuclein. J Mol Biol 356: 1036–1048 [DOI] [PubMed] [Google Scholar]