Abstract
To investigate the demographic history of human populations from the Caucasus and surrounding regions, we used high-throughput sequencing to generate 147 complete mtDNA genome sequences from random samples of individuals from three groups from the Caucasus (Armenians, Azeri and Georgians), and one group each from Iran and Turkey. Overall diversity is very high, with 144 different sequences that fall into 97 different haplogroups found among the 147 individuals. Bayesian skyline plots (BSPs) of population size change through time show a population expansion around 40–50 kya, followed by a constant population size, and then another expansion around 15–18 kya for the groups from the Caucasus and Iran. The BSP for Turkey differs the most from the others, with an increase from 35 to 50 kya followed by a prolonged period of constant population size, and no indication of a second period of growth. An approximate Bayesian computation approach was used to estimate divergence times between each pair of populations; the oldest divergence times were between Turkey and the other four groups from the South Caucasus and Iran (∼400–600 generations), while the divergence time of the three Caucasus groups from each other was comparable to their divergence time from Iran (average of ∼360 generations). These results illustrate the value of random sampling of complete mtDNA genome sequences that can be obtained with high-throughput sequencing platforms.
Keywords: complete mtDNA genomes, Caucasus, West Asia
Introduction
The Caucasus and neighboring Iran and Turkey are situated between the Levant and Europe, and can be considered as one of the potential pathways during the colonization of Europe by modern humans about 40 000 kya ago.1 A number of Early Upper Paleolithic sites, dating back to about 30–40 kya, have been excavated in this region suggesting an early presence of modern humans in the region.1, 2, 3, 4, 5 Importantly, the Upper Palaeolithic sites were found only in the mountain ridges along the passage between the Caucasus Mountains and the Black Sea that connects the South Caucasus with the southern part of East Europe.1 Thus, genetic study of this region can provide some insights into ancient migrations and can facilitate a reconstruction of modern human migration routes from the Levant to Europe.
Several genetic studies of a number of different groups from this region have been carried out in the last decade.6, 7, 8, 9, 10, 11, 12, 13, 14, 15 These studies were mostly based on sequences of the first hypervariable segment of the noncoding mtDNA control region (HV1) and Y chromosome binary marker variation in various groups from this region. mtDNA studies reveal a high level of diversity, exceeding that within all of Europe and only slightly lower than West Asian mtDNA diversity, which might indicate an old age of human populations from this region.10 Overall, the Caucasus groups showed greater similarity with West Asian than with European groups for both genetic systems, although this similarity was much more pronounced for the Y chromosome than for mtDNA, suggesting that recent male-mediated migrations from West Asia have influenced the genetic structure of Caucasus populations.11
Previous studies of complete mtDNA genome sequences have in general first obtained HV1 sequences (and sometimes, genotyped some haplogroup-defining SNPs), and then selected particular individuals of interest for complete mtDNA genome sequencing. However, such biased sampling renders the resulting sequences unsuitable for many demographic analyses. We have therefore used a high-throughput sequencing approach to generate 147 complete mtDNA genome sequences from random samples of individuals from three groups from the Caucasus (Armenians, Azeri and Georgians), and one group each from Iran and Turkey. We used these sequences to investigate the genetic structure of these groups and more accurately infer their demographic history. In particular, we have examined population size changes through time via Bayesian Skyline Plots (BSPs), and we have used an approximate Bayesian computation (ABC) approach to estimate divergence times between groups; neither of these analyses can be applied to HV1 sequences alone because the HV1 sequences lack sufficient information for such analyses. Our results amply show the value of random sampling of complete mtDNA genome sequences that is afforded by this high-throughput sequencing approach.
Materials and methods
Samples and DNA extraction
Samples were obtained from unrelated individuals, representing three populations from the Caucasus: Armenians (30 cheek cell swabs from Erevan), Azeri (30 cheek cell swabs from Baku) and Georgians (28 saliva samples from Batumi). Samples were also obtained from Turks (29 saliva samples from Ankara) and Iranians (30 blood samples from Tehran). A map of the sampling locations is shown in Figure 1. Genomic DNA from cheek cell swabs was extracted using a standard salting-out procedure.16 DNA from saliva samples was extracted using a previously described method.17 Blood samples were processed using the QIAamp DNA Blood Mini Kit (Qiagen, GmbH, Germany), following the instructions of the manufacturer. Informed consent and information about birthplace, parents and grandparents was obtained from all donors.
Figure 1.
Geographic location of sampling sites.
Sequencing complete mtDNA genomes
The entire mtDNA genome was amplified in two overlapping products of about 9.7 and 7.3 kb, using primer pairs L12279/H2986 and L2603/H12314.18 Long-range PCR was carried out using the Expand Long-Range dNTP pack (Roche, Berlin, Germany) and 3 ng of template DNA in a 50 μl volume, using the protocol provided by the manufacturer. The annealing temperature was 57 °C. PCR products were purified with SPRI beads (Agencourt, Denver, MA, USA) using the manufactureŕs instructions. The two PCR products for each individual were mixed in equimolar ratios and nebulized using a Bioruptor Sonicator UCD-200 (Diagenode, Liege, Belgium). The size range of the sonicated DNA fragments was between 150 and 450 bp. MinElute spin columns (Qiagen) were used to remove small DNA fragments, and the purified, nebulized DNA was eluted in 20 μl of elution buffer. About 400 ng of DNA was used for tagging the nebulized PCR product for each individual with a specific tag sequence, as described elsewhere,19 and Illumina Genome Analyzer II (Illumina Inc., San Diego, CA, USA) libraries were prepared according to the protocol described elsewhere.20 Fifty individuals with unique tags were pooled in each library in equimolar ratios. Subsequently, libraries were sequenced with 36 and/or 76-bp reads in one lane of an Illumina flow cell (Cluster Generation kit V2, FC-103–300 × sequencing chemistry, San Diego, CA, USA) according to the manufacturer's instructions.
mtDNA sequence assembly
Each run was processed with RTA 1.5 (Illumina Inc.). Afterward, the PhiX174 control reads of a dedicated control lane were aligned to the corresponding reference sequence to obtain a training data set for the alternative base caller Ibis.21 Raw sequences called from Ibis were separated by sample using their index read, allowing one mismatch and the loss of the first base.20 Reads were then filtered for sequence quality and complexity. In this step, reads having >5 bases with a quality score below 10 (PHRED scale) and reads with sequence entropy (calculated by summing –p*log2(p) for each of the four nucleotides) below 1.0 were removed. Sequence reads lacking an expected tag or with more than one tag were removed. After untagging, reads with identical start and end positions were also removed, since they may represent clones of a single sequence. Sequence reads that passed these filters were sorted according to unique sequence tags. Consensus mtDNA sequences were assembled using the software MIA22 by mapping reads to the revised Cambridge reference sequence (rCRS).23 A multiple alignment of the consensus sequences was carried out using the software MAFFT v6.708b.24 The mtDNA genome sequences have been deposited into Genbank (accession numbers HM852756–HM852902). The list of polymorphic sites and undetermined two positions is shown in the Supplementary Table 1.
Haplogroup assignment
Sequences were assigned to haplogroups according to Phylotree.org Build 6,25 using a custom PERL script. Sequences were assigned to the closest matching haplogroup. As in Phylotree, positions 309.1C(C), 16182C, 16183C, 16193.1C(C) and 16519 were not used to assign haplogroups, because these are highly mutable positions.
Data analysis
DnaSP 4.026 was used to calculate basic parameters of genetic diversity. Analyses of molecular variance (AMOVA) were carried out using Arlequin.27 The statistical significance of Fst values was estimated by permutation analysis, using 10 000 permutations. The software package GENESYN28 was used to calculate the number of polymorphic sites, substitutions, and nonsynonymous and synonymous substitutions. The STATISTICA package (StatSoft Inc., Tulsa, OK, USA) was used for multi-dimensional scaling (MDS) analysis.29
BSPs were produced from the coding region sequences (positions 577–16023) using MCMC sampling in version 5.1 of the program BEAST.30 The plots were obtained with a piecewise linear model and ancestral gene trees were based on a general time-reversible substitution model with invariant sites (GTR+I). A Bayes factor computed via importance sampling indicated that the strict molecular clock could not be rejected and was therefore used for the analysis. We allowed 20 discrete changes in the population history, using a coalescent-based tree prior with a linear model in which population size grows and declines between changing points. Each MCMC sample was based on a run of 40 000 000 generations sampled every 4000 steps, with the first 4 000 000 generations regarded as burn-in. Three independent runs were made for each population, and a mutation rate of 1.691 × 10−8 was used.31
To estimate the divergence time between each pair of populations, we implemented the ABC approach described previously,32 with the exception that all data were simulated with the coalescent simulator ms. Details of the rejection-regression algorithm are as described previously.32 This basically involves fitting a local-linear regression of all simulated parameter values to simulated summary statistics. Furthermore, the observed summary statistics are then substituted into a regression equation. All parameters were transformed with logtan before the actual regression analysis.33 Distances between observed and simulated summary statistics were calculated as Euclidean distances. We simulated 1 million sequence data sets for each estimation procedure. Each data set was simulated as a 15 kb region with a mutation rate of 1.69 × 10−8 substitutions per site per year.31 The recombination rate was set to 0. The demographic history underlying each population was a consensus history from all five populations, obtained from the BSP results: an ancestral population of an effective size of 400 experienced a sudden growth 45 000 years ago to an Ne of 4000, and after a long period of constant size a second sudden growth 15 000 years ago increased the size of the population to 40 000. A simple 1– parameter model of a single population split between two populations with no migration was assumed, and the time of divergence TDIV was the parameter to be estimated. The prior for this parameter was TDIV∼U[1002000] in generations. Each estimation was done as a pairwise population comparison, resulting in 10 ABC estimations in total. Out of the 1 million simulations for each ABC estimation, the 10 000 simulations with the smallest Euclidean distances were kept and used to estimate the posterior distribution. The Euclidean distances for the retained best simulation ranged from 0.2 to 1.2. We used six summary statistics in total: S (the number of segregating sites for each population in every pairwise comparison); π (the average number of pairwise differences for each population in every pairwise comparison); the percentage of shared polymorphisms per pairwise population comparison; and the pairwise Fst value.
Results
After quality filtering and removal of potential duplicate reads, there were an average of 33 219 reads per individual. The average coverage was 69.3 for 90 samples sequenced with 36-bp reads (Supplementary Figure 2A); 211.1 for 21 samples sequenced with 76-bp reads (Supplementary Figure 2B) and 102.5 for 36 samples sequenced twice with 36-bp reads, because of poor coverage in the first sequencing (Supplementary Figure 2C). Overall, the average coverage of each position was 87-fold (Supplementary Table 2).
Summary statistics describing genetic diversity are shown in Table 1. The number of mean pairwise differences (MPDs) ranged from 31.8 to 33.5 for all groups except the Azeri, for which the MPD is 39.3. All groups exhibit similar nucleotide diversity values, as well as an excess of low-frequency variants that is characteristic of a recent population expansion, as shown by significantly negative values for Tajima's D (Table 1).
Table 1. Summary statistics for Armenians, Azeri, Georgians, Iranians and Turks based on complete mtDNA genome sequences.
| Population | No. of samples | No. of haplotypes | Haplotype diversity | Nucleotide diversity | MPD | Tajima's D |
|---|---|---|---|---|---|---|
| Georgians | 28 | 28 (20) | 1±0.01 (0.976±0.015) | 0.002 (0.007) | 33.47 (3.98) | −2.23 (−1.77) |
| Armenians | 30 | 30 (22) | 1±0.009 (0.966±0.021) | 0.002 (0.007) | 31.78 (3.74) | −2.11 (−1.36) |
| Azeri | 30 | 29 (22) | 0.998±0.009 (0.977±0.014) | 0.002 (0.006) | 39.29 (3.11) | −2.08 (−2.01) |
| Iranians | 30 | 30 (24) | 1±0.009 (0.979±0.015) | 0.002 (0.008) | 33.40 (4.79) | −2.12 (−1.79) |
| Turks | 29 | 27 (21) | 0.993±0.009 (0.941±0.001) | 0.002 (0.007) | 32.48 (4.06) | −2.41 (−2.10) |
The same statistics based on HV1 sequences only are in parentheses.
A total of 855 polymorphic sites were detected (Table 2). The highest number of variable sites was found in the control region (192 sites), which is more than three times higher than expected (57 sites) based on the length of the control region. The number of transitions significantly exceeds the number of transversions for all parts of the mtDNA genome (Table 2). The ratio of transitions vs transversions was higher for protein-coding genes (16.71) than for the control region (4.49), but not significantly so (P=0.387). The number of synonymous substitutions was higher than nonsynonymous substitutions for all protein-coding genes except ATP6 and ATP8 (Table 2). Among the 13 protein-coding genes, there were 164 sites with nonsynonymous changes and 385 sites with synonymous changes (Table 2). The ratio of the number of polymorphic nonsynonymous sites per total nonsynonymous sites to the number of polymorphic synonymous sites per total synonymous sites (pn/ps values), is less than one in all cases, but elevated for ATP6 and ATP8 (Table 2).
Table 2. Number of variable sites, transitions, transversions, synonymous and nonsynonymous differences, and pa/ps ratio.
| Region/ Gene | No. of variable sites | Transitions | Transversions | Synonymous | Nonsynonymous | pa/ps |
|---|---|---|---|---|---|---|
| Control region | 192 | 157 | 35 | NA | NA | NA |
| Other noncoding | 8 | 6 | 2 | NA | NA | NA |
| 12S rRNA | 23 | 22 | 1 | NA | NA | NA |
| 16S rRNA | 44 | 35 | 9 | NA | NA | NA |
| tRNAs | 39 | 38 | 1 | NA | NA | NA |
| ATP6 | 41 | 40 | 1 | 16 | 25 | 0.756 |
| ATP8 | 14 | 14 | 0 | 6 | 8 | 0.570 |
| COX1 | 66 | 63 | 3 | 56 | 10 | 0.079 |
| COX2 | 32 | 31 | 1 | 24 | 8 | 0.145 |
| COX3 | 39 | 35 | 4 | 26 | 13 | 0.218 |
| CYTB | 66 | 61 | 5 | 46 | 20 | 0.193 |
| ND1 | 44 | 41 | 3 | 31 | 13 | 0.198 |
| ND2 | 46 | 42 | 4 | 30 | 16 | 0.241 |
| ND3 | 15 | 15 | 0 | 11 | 4 | 0.174 |
| ND4L | 11 | 9 | 2 | 8 | 3 | 0.184 |
| ND4 | 60 | 59 | 1 | 49 | 11 | 0.103 |
| ND5 | 87 | 81 | 6 | 57 | 30 | 0.236 |
| ND6 | 28 | 27 | 1 | 25 | 3 | 0.045 |
| Total | 855 | 776 | 79 | 385 | 164 |
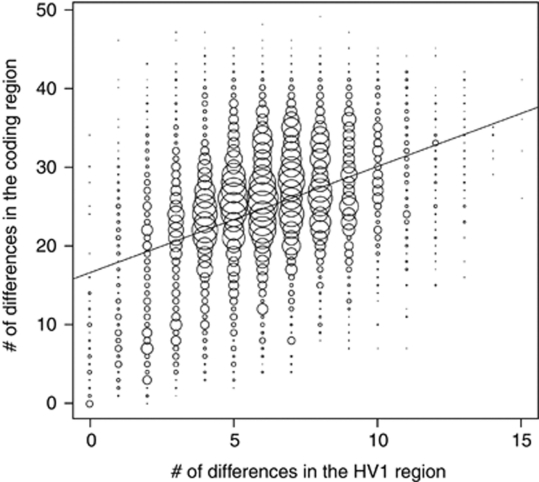
A plot of the number of differences in the HV1 sequences vs the number of differences in the coding region sequences for each pair of individuals showed that the pairwise comparisons with no differences in the HV1 sequences goes up to a maximum of 34 differences in the coding region (Figure 2). Thus, there can be appreciable coding region variation among individuals with identical HV1 sequences.
Figure 2.
Plot of the number of differences in the HV1 sequences vs the number of differences in the coding region sequences for each pair of individuals. The best-fit regression line is indicated.
The haplogroup assignment for each individual, according to the nomenclature of Phylotree.org,23 is given in Supplementary Table 1. As none of the sequences in this study matched any previous haplogroup exactly, a sequence was assigned to a haplogroup if it contained all the mutations that define that haplogroup. All of our sequences therefore contain all of the mutations that define the haplogroup plus additional mutations.
A total of 97 different haplogroups were identified (Supplementary Table 1), which fall into 16 major haplogroups (Table 3). Haplogroups H, HV, J, T and U were found in all groups, and are generally among the most frequent West Eurasian mtDNA haplogroups. Several haplogroups typical for Central and East Asia (A, C, D and F) were found only in Azeri and Turks (Table 3). One haplogroup (X) was restricted to the three Caucasus groups.
Table 3. Haplogroup frequencies.
| Population | |||||
|---|---|---|---|---|---|
| Haplogroup | Armenia | Azerbaijan | Georgia | Iran | Turkey |
| A | — | — | — | — | 0.069 |
| C | — | 0.033 | — | — | — |
| D | — | 0.033 | — | — | 0.069 |
| F | — | 0.067 | — | — | 0.034 |
| H | 0.200 | 0.067 | 0.179 | 0.233 | 0.138 |
| HV | 0.067 | 0.067 | 0.071 | 0.067 | 0.241 |
| I | — | — | — | 0.100 | 0.034 |
| J | 0.167 | 0.033 | 0.036 | 0.133 | 0.034 |
| K | 0.067 | 0.067 | 0.071 | 0.034 | |
| M | — | 0.067 | — | 0.133 | — |
| N | 0.033 | — | 0.071 | 0.033 | — |
| R | — | 0.033 | 0.107 | 0.067 | 0.069 |
| T | 0.100 | 0.200 | 0.107 | 0.067 | 0.034 |
| U | 0.267 | 0.267 | 0.286 | 0.133 | 0.241 |
| X | 0.100 | 0.067 | 0.071 | — | — |
| Z | — | — | — | 0.033 | — |
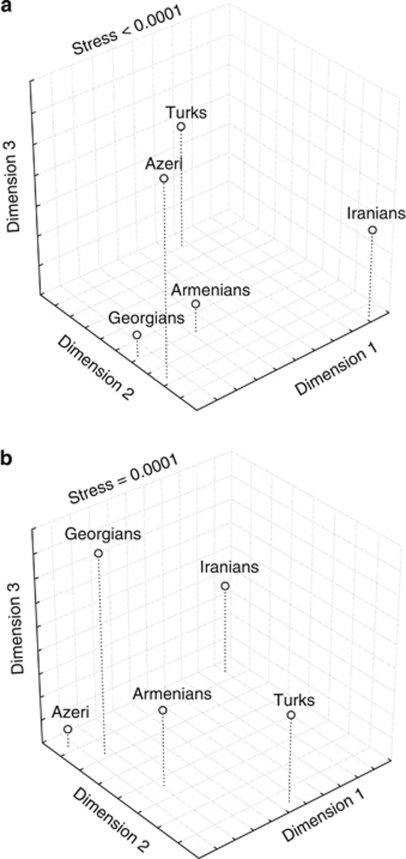
In order to visualize the relationships of these five groups based on complete mtDNA genome sequences, an MDS plot was constructed from the pairwise Fst values (Supplementary Table 3). The results (Figure 3a) showed a cluster of groups from the Caucasus in the first two dimensions, but a similar placement of Azeri and Turks in the third dimension For comparison, we constructed an MDS plot for just the HV1 region (Figure 3b), which showed a similar cluster of the Caucasian groups in the first two dimensions, but did not associate Azeri and Turks in the third dimension. Thus, the complete mtDNA sequences reveal an association between the two Turkic-speaking groups that is not seen in the HV1 sequences.
Figure 3.
MDS plot for five groups from the South Caucasus, Iran and Turkey. (a) Based on complete mtDNA genome sequences. (b) Based on HV1 sequences only.
The geographic and linguistic structure of the Caucasian, Iranian and Turkic groups was investigated by an AMOVA (Supplementary Table 4). Approximately 99% of the variance was due to the within-population component. A geographic classification of populations gave a slightly better fit to the genetic data than did a linguistic classification, although permutation tests showed that the higher among-group components are not significantly different from zero.
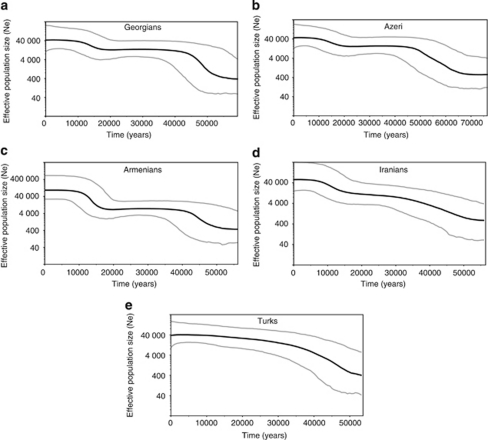
Many of the above descriptive analyses can be carried out on HV1 sequences as well as complete mtDNA genome sequences. However, HV1 sequences are not amenable to demographic inference, and we utilized our random samples of complete mtDNA genome sequences to estimate population size change over time and pairwise population divergence times. To investigate changes in population size over time, BSPs were constructed using the coding region (positions 577–16022). The BSPs for the three Caucasus groups as well as the Iranian group (Figure 4) are generally similar, showing a population expansion around 45–50 kya, followed by a constant population size, and then another expansion around 15–18 kya. However, the BSP for the Turkic group differs, with an increase from 35 to 50 kya followed by a prolonged period of constant population size, and no indication of a second growth period.
Figure 4.
Bayesian skyline plots. The Y axis for each plot is the effective population size and the X axis is time in years. (a) Georgians; (b) Azeri; (c) Armenians; (d) Iranians and (e) Turks. The gray lines represent the 95% posterior density intervals.
An ABC approach was used to estimate the divergence time between each pair of populations. For each estimation, 1 million simulations were carried out and compared with empirical data based on six summary statistics. The posterior distributions based on the best-fitting 10 000 simulations show pronounced peaks (Supplementary Figure 1), and the Euclidean distances between the simulated and observed parameters vary between 0 and 1.2, indicating good support for the divergence time estimates (Table 4). In general, the oldest divergence times are between the group from Turkey and other groups from the South Caucasus and Iran (∼400–600 generations). The divergence time for the South Caucasus groups from each other is about the same as their divergence from Iran (average Iran-Caucasus is 361 generations, average within Caucasus is 365 generations).
Table 4. Divergence time (Tdiv) (in generations) between pairs of populatiuons.
| Pair of populations | Tdiv (generations) | 95% CI |
|---|---|---|
| Armenians-Azeri | 492 | 32–1132 |
| Armenians-Iranians | 360 | 21–997 |
| Armenians-Turks | 493 | 37–1069 |
| Azeri-Iranians | 333 | 16–990 |
| Azeri-Turks | 514 | 24–1181 |
| Georgians-Azeri | 405 | 20–1071 |
| Georgians-Iranians | 317 | 17–955 |
| Georgians-Turks | 418 | 21–1046 |
| Georgians-Armenians | 197 | 12–738 |
| Iranians-Turks | 600 | 35–1185 |
Discussion
Overall, the randomly sampled complete mtDNA genome sequences indicated extraordinarily high genetic diversity in the groups from the South Caucasus, Iran and Turkey. For the Georgians, Armenians and Iranians, all individuals had different sequences, while for the Azeri two individuals shared the same sequence, and for the Turks there were 27 haplotypes among 29 individuals (Table 1). Overall, there were 144 different sequences among the 147 individuals. By contrast, when only the HV1 sequences are considered, the level of haplotype sharing is much higher (Table 1), indicating that individuals with identical HV1 sequences often had different complete mtDNA genome sequences.
The assignment of sequences to haplogroups further reinforces the extraordinary diversity in the complete mtDNA genome sequences, as none of the 147 sequences in this study was identical to a sequence in the Phylotree.org database.23 This is all the more remarkable when one considers that the complete mtDNA genome sequences present in the public databases are heavily biased toward Eurasia; for example, nearly 40% of >5000 sequences analyzed by Pereira et al34 are from Europe and the Middle East. Thus, even in well-studied areas of the world, there is much mtDNA diversity yet to be discovered.
The high level of diversity extends to the level of major haplogroups, as the individual haplogroups fall into 16 major haplogroups (Table 3). Most of these haplogroups (H, HV, I, J, K, M, N, R, T, U and Z) are typical for West Eurasia,34 while others (A, C, D and F) occur mostly in Central and East Asia.34 Haplogroup X is widespread, albeit at low frequencies, across Eurasia.34 Among the West Eurasian haplogroups, the Caucasian groups are characterized by relatively high frequencies of haplogroups U and X. The Central/East Asian haplogroups were notable in that they were found only in a few individuals from the Azeri and Turkish groups (the two Turkish-speaking groups in the study), suggesting some Central Asian influence specifically on these groups. Indeed, there is historical documentation of such contact via the Oguz migrations from Central Asia to Anatolia and the South Caucasus in the eleventh century.35 These groups brought the Turkic language into the Azeri and Turkish populations, and presumably left some genetic footprint along with their language. Although the specific contact(s) between Azeri and Turks with Central Asian groups that brought in these Central Asian mtDNA lineages is unknown, overall the low frequency of these mtDNA lineages is in good agreement with previous estimates of low levels of gene flow from Asia into Anatolia.36
Overall, the complete mtDNA genome sequences indicate that the three South Caucasus groups are genetically similar, although they represent three different language families. This can be seen in the MDS plots (Figures 3a and b), where Indo European-speaking Armenians and Turkic-speaking Azeri cluster with the Caucasian-speaking Georgians. The clustering of groups on the basis of geographic rather than linguistic relationships is in keeping with previous studies of genetic diversity among Caucasian populations.10, 11 However, the complete mtDNA genome sequences do reveal some additional genetic similarity between the two Turkic-speaking groups (Azeri and Turks) that was not evident in previous studies. Presumably, the sharing of some Central Asian mtDNA haplogroups by Azeri and Turks (Table 3) accounts for this signal of genetic similarity between the Azeri and the Turks. Thus, the greater genetic resolution afforded by the complete mtDNA genome sequences both confirms and extends previous studies.
We also investigated patterns of variation in the mtDNA genome itself (Table 2). Of the 855 variable positions, about 22% occurred in the control region, which is significantly more than expected if variable positions occur randomly across the mtDNA genome (P<0.0001). The excess in polymorphisms and transversions for the control region most likely reflects weaker functional constraints on this major noncoding region of the mtDNA genome. Among the 13 protein-coding genes, there was a significant excess of nonsynonymous polymorphisms in the overlapping ATP6 and ATP8 genes, relative to the other mtDNA protein-coding genes (Table 2). Previously, an excess of nonsynonymous polymorphisms in the ATP6 gene was found in Siberian populations and hypothesized to reflect positive selection for cold adaptation.37 However, subsequent studies suggested that relaxation of functional constraints, rather than positive selection, can explain the higher pa/ps ratio for ATP6.38 The finding of a significantly elevated pa/ps ratio for ATP6 in this study, as well as in a previous study of Filipino groups,39 argue against the cold adaptation hypothesis.
In the interval of ca 30–40 kya, Europe underwent numerous changes known as the Upper Paleolithic revolution, which involved dramatic changes in technologies, hunting techniques, human burials and an artistic traditions revealed in the archeological records.40 These changes presumably also involved population size increases, and indeed the BSPs for all five groups indicate a strong expansion around this time (Figure 4). Following this initial expansion, the BSPs indicate constant population size during the period of the Last Glacial Maximum (LGM), about 18–30 kya. The dates for the second expansion event evident in the BSPs from the Caucaus and Iranian groups are in good agreement with the start of the continuous deglaciation of the ice shield ∼18 kya ago, when environmental conditions were almost fully glacial.41 Curiously, the BSP for the mtDNA sequences from Turkey do not show any evidence of this second expansion (Figure 4). The different BSP for Turkey cannot be explained by Central Asian mtDNA sequences in this population, as a BSP with the Central Asian sequences removed is identical to the BSP for all of the sequences (data not shown). This suggests that the ancestors of the group from Turkey have a different history than the ancestors of the Caucasian and Iranian groups in this study. Specifically, these results suggest that the ancestors of the group from Turkey did not expand after the LGM. This could be explained by the fact that Turkey was not influenced heavily by glaciation during the LGM. The most extensive LGM glaciers descended only to an altitude of 2150 m above sea level in central Turkey,42 suggesting that the end of the LGM may not have caused as dramatic changes in the lowland environment as occurred in the Caucasus.
In addition, we estimated the divergence time between pairs of populations using an ABC–MCMC approach with uniform priors.32 Assuming a generation time of 28 years for mtDNA,43 the earliest divergence occurred between the group from Turkey and the other four groups about 11.2–16.8 kya. This event coincides with the second expansion event for the South Caucasus and Iranian groups (Figure 4). This was a time of post-glacial recolonization; in the wake of climatic amelioration, temperate species expanded their distribution range to the north, following the expansion of favorable habitats (‘habitat tracking').44 Most likely, modern humans followed the expansion of temperate species and recolonized the South Caucasus, which was mostly glaciated during the LGM.45 Along with the colonization of new lands and territories, human groups settled in different parts of the region, giving rise to new populations. This scenario is supported by divergence time estimates among South Caucasus groups, ranging from 5.6 to 11 kya, as well as divergence time estimates between Iran and South Caucasian groups of 8.9–10 kya. Thus, in this part of West Eurasia there appear to have been two major periods of population expansion and divergence after the LGM.
Admittedly, some caveats are in order. Divergence time estimates were obtained assuming no subsequent migration between groups, and hence should be viewed as minimum estimates of the actual divergence time. We did attempt to incorporate migration into the model, however, we were unable to obtain reliable estimates of both migration rate and divergence time with the ABC–MCMC approach (results not shown). Nevertheless, the pronounced peaks in the posterior distributions (Supplementary Figure 1), small Euclidean distances, and concordance between the BSPs and the divergence time estimates indicate that the demographic inferences are probably reliable. Moreover, the relatively low level of sequence sharing between groups further indicates that recent migration among these groups has been low.
In conclusion, the finding of extraordinarily high mtDNA diversity and resulting demographic inferences for the South Caucasian and West Asian groups studied here was enabled by random sampling of individuals, which in turn was made possible by the high-throughput sequencing approach implemented here. The speed, low cost and reliability of the resulting sequences demonstrated by this and similar studies39 further indicate that this is the approach of choice for generating complete mtDNA genome sequences.
Acknowledgments
We are grateful to the original donors for providing DNA samples. We thank Marc Bauchet for writing a script that allows us to assign sequences to mtDNA haplogroups. This research was supported by funding from the Max Planck Society, Germany.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Arslanov KhA, Dolukhanov PM, Gei NA. Climate, Black Sea levels and human settlements in Caucasus Littoral 50 000–9000 BP. Quaternary Int. 2007;167:121–127. [Google Scholar]
- Golovanova LV, Doronichev VB. The middle paleolithic of the Caucasus. J World Prehistory. 2003;17:71–140. [Google Scholar]
- Stiner MC, Gulec E. Initial: upper palaeolithic in south-central Turkey and its regional context: a preliminary report. Antiquity. 1999;73:505–517. [Google Scholar]
- Pinhasi R, Gasparian B, Wilkinson K, et al. Hovk 1 and the middle and upper paleolithic of Armenia: a preliminary framework. J Hum Evol. 2008;55:803. doi: 10.1016/j.jhevol.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Biglari F, Shidrang S. The lower paleolithic occupation of Iran. Near Eastern Archaeol. 2006;69:160–168. [Google Scholar]
- Barbujani G, Nasidze IS, Whitehead GN.Genetic diversity in the Caucasus Hum Biol 1994a66639668. [PubMed] [Google Scholar]
- Barbujani G, Whitehead GN, Bertorelle G, Nasidze I. Testing hypotheses on processes of genetic and linguistic change in the Caucasus. Hum Biol. 1994b;66:843–864. [PubMed] [Google Scholar]
- Comas D, Calafell F, Bendukidze N, et al. Georgian and Kurd mtDNA sequence analysis shows a lack of correlation between languages and female genetic lineages. Am J Phys Anthropol. 2000;112:5–16. doi: 10.1002/(SICI)1096-8644(200005)112:1<5::AID-AJPA2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, et al. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331–1334. doi: 10.1016/s0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Stoneking M. Mitochondrial DNA variation and language replacements in the Caucasus. Proc R Soc Lond B. 2001;268:1197–1206. doi: 10.1098/rspb.2001.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Ling ES, Quinque D, et al. Mitochondrial DNA and Y-chromosome variation in the Caucasus. Ann Hum Genet. 2004;68:205–221. doi: 10.1046/j.1529-8817.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Rahmani M, Alemohamad SA, Stoneking M. Concomitant replacement of language and mtDNA in South Caspian populations of Iran. Current Biol. 2006;16:668–673. doi: 10.1016/j.cub.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Rahmani M, Ali Alemohamad S, Stoneking M. Close genetic relationship between semitic-speaking and Indo-European-speaking groups in Iran. Ann Hum Genet. 2008;72:241–252. doi: 10.1111/j.1469-1809.2007.00413.x. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Rahmani M, Ali Alemohamad S, Asadova P, Zhukova O, Stoneking M. MtDNA and Y-chromosome variation in the Talysh of Iran and Azerbaijan. Am J Phys Anthropol. 2009;138:82–89. doi: 10.1002/ajpa.20903. [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Myles S, Prüfer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucl Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protocols. 2008;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M.Illumina sequencing library preparation for highly multiplexed target capture and sequencing Cold Spring Harbor Protocols 20101doi:pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Kircher M, Stenzel U, Kelso J. Improved base calling for the illumina genome analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Malaspinas AS, Krause J, et al. A complete neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Meth Mo Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin ver 2.000: A Software for Population Genetics Data Analysis. Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Pavesi G, Mauri G, Iannelli F, Gissi C, Pesole G. GeneSyn: a tool fordetecting conserved gene order across genomes. Bioinformatics. 2004;20:1472–1474. doi: 10.1093/bioinformatics/bth102. [DOI] [PubMed] [Google Scholar]
- Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc R Soc B. 2009;276:367–373. doi: 10.1098/rspb.2008.0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Estoup A, Cornuet JM. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics. 2005;169:1727–1738. doi: 10.1534/genetics.104.036236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Currat M, Ray N. Bayesian estimation of recent migration rates after a spatial expansion. Genetics. 2005;170:409–417. doi: 10.1534/genetics.104.034199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Freitas F, Fernandes V, et al. The diversity present in 5140 human mitochondrial genomes. Am J Hum Genet. 2009;84:628–640. doi: 10.1016/j.ajhg.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew C. Before Bibel: speculations on the origins of linguistic diversity. Camb Archaeol J. 1991;1:13–23. [Google Scholar]
- Di Benedetto G, Ergüven A, Stenico M, et al. DNA diversity and population admixture in Anatolia. Am J Phys Anthropol. 2001;115:144–156. doi: 10.1002/ajpa.1064. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. Rate variation between mitochondrial domains and adaptive evolution in humans. Hum Mol Genet. 2007;16:2281–2287. doi: 10.1093/hmg/ddm180. [DOI] [PubMed] [Google Scholar]
- Gunnarsdottir ED, Li M, Bauchet M, Finstermeier K, Stoneking M. High-throughput sequencing of complete human mtDNA genomes. Genome Res. 2011;21:1–11. doi: 10.1101/gr.107615.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yosef O. The upper paleolithic revolution. Annu Rev Anthropol. 2002;31:363–393. [Google Scholar]
- Sommer Zachos FE. Fossil evidence and phylogeography of temperate species: ‘glacial refugia' and post-glacial recolonization. J Biogeogr. 2009;36:2013–2020. [Google Scholar]
- Zreda M, Çiner A. Glaciations and paleoclimate of Mount Erciyes, central Turkey, since the lastglacial maximum, inferred from 36Cl cosmogenic dating and glacier modeling. Quaternary Sci Rev. 2009;28:2326–2341. [Google Scholar]
- Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence Studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linnean Soc. 1999;68:87–112. [Google Scholar]
- Provan J, Bennett K. Phylogeographic insights into cryptic glacial refugia. Trends in Ecol Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.