Abstract
DFNA5 was first identified as a gene causing autosomal dominant hearing loss (HL). Different mutations have been found, all exerting a highly specific gain-of-function effect, in which skipping of exon 8 causes the HL. Later reports revealed the involvement of the gene in different types of cancer. Epigenetic silencing of DFNA5 in a large percentage of gastric, colorectal and breast tumors and p53-dependent transcriptional activity have been reported, concluding that DFNA5 acts as a tumor suppressor gene in different frequent types of cancer. Despite these data, the molecular function of DFNA5 has not been investigated properly. Previous transfection studies with mutant DFNA5 in yeast and in mammalian cells showed a toxic effect of the mutant protein, which was not seen after transfection of the wild-type protein. Here, we demonstrate that DFNA5 is composed of two domains, separated by a hinge region. The first region induces apoptosis when transfected in HEK293T cells, the second region masks and probably regulates this apoptosis inducing capability. Moreover, the involvement of DFNA5 in apoptosis-related pathways in a physiological setting was demonstrated through gene expression microarray analysis using Dfna5 knockout mice. In view of its important role in carcinogenesis, this finding is expected to lead to new insights on the role of apoptosis in many types of cancer. In addition, it provides a new line of evidence supporting an important role for apoptosis in monogenic and complex forms of HL.
Keywords: tumor suppressor, hearing loss, apoptosis, dfna5, cancer, GSEA
Introduction
DFNA5 first was discovered in a Dutch family with autosomal dominant hearing loss (HL).1 Not much was known concerning its cellular function and how its function was related to HL. Recently, a novel mutation in DFNA5 has been identified in a Korean family, totaling five families with DFNA5 HL.2 These families all have different genomic DFNA5 mutations, but in each case the DFNA5 mRNA transcript skips exon 8, resulting in a frameshift and a premature truncation of the protein.1, 2, 3, 4, 5 These findings have led to the hypothesis that DFNA5-associated HL is attributable to a highly specific gain-of-function mutation, in which skipping of one exon causes disease while mutations in other parts of this gene may not result in HL at all. Further experimental evidence for this hypothesis was provided by the finding that transfection of mutant DFNA5 causes cell death in both yeast6 and mammalian7 cells and by the discovery of a new DFNA5 mutation.8 The latter mutation truncated the protein in the fifth exon, but did not segregate with HL and was present in family members with normal hearing. The hypothesis was further corroborated by a mouse that lacked the Dfna5 protein. This knockout (KO) mouse did not display any HL and, as a consequence, was not a suitable animal model to study DFNA5-associated HL.9
To date, little information is available on the physiological function of DFNA5. However, since its identification, the small number of papers published on DFNA5 almost all point to a possible involvement in cancer biology.10, 11, 12, 13, 14, 15, 16 Especially during the last 3 years, DFNA5 emerged from several genomic screens identifying new tumor suppressor genes. Further analysis showed that the DFNA5 gene is epigenetically inactivated in several types of cancer, including gastric, colorectal and breast cancer.13, 14, 15, 16 Epigenetic silencing through methylation was found in 52–65% of primary tumors. In addition, in vitro studies showed that cell invasion, colony numbers, colony size and cell growth increased in cell lines after DFNA5 knockdown. Forced expression of DFNA5 in colorectal carcinoma cell lines, on the other hand, decreased cell growth and colony-forming ability. In breast carcinoma, the methylation status of DFNA5 was correlated with lymph node metastasis.15 These recent data also explain earlier findings that were not well understood at the time, such as the increased resistance to the chemotherapeutic reagent etoposide in melanoma cells with decreased transcription of DFNA5,11 or the finding that DFNA5 is induced by p53 through a p53-binding site located in intron 1.12 Webb et al17 demonstrated that DFNA5 mRNA expression increased after treatment of human acute lymphoblastic leukemia cell lines with the glucocorticoid dexamethasone. This increase was only found in cells that underwent apoptosis in response to glucocorticoid treatment.17 The conclusion of these data is that DFNA5 is a tumor suppressor gene with an important role in several frequent forms of cancer.
Here we report that DFNA5 contains two domains separated by a hinge region and that the first domain induces apoptosis when transfected into cell lines. The second domain may shield the apoptotic-inducing sequences residing in the first domain. Furthermore, using gene expression microarray experiments on Dfna5 KO mice, we provide evidence that the apoptosis-inducing properties of DFNA5 also occur in a physiological setting and that DFNA5 is involved in cell survival pathways.
Materials and methods
Plasmid construction
Using previously described human full-length DFNA5 cDNA clones as a template,6 we amplified full-length human wild type (WT) and mutant DFNA5 sequences in addition to several specific parts of the DFNA5 cDNA sequence using the iProof High Fidelity PCR kit (Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer's instructions. A kozak consensus sequence (GCCACCATG) was introduced in the specific parts that lacked the regular DFNA5 start codon, either via PCR or via site-directed mutagenesis (Stratagene, La Jolla, CA, USA). PCR primers are listed in Supplementary Table I. All PCR fragments were ligated in pEGFP-N1. To clone the part of Pejvakin (PJVK) that showed high homology to DFNA5, human testis Quick-clone cDNA (BD Biosciences, San Jose, CA, USA) was used as a template. All cloned inserts were bidirectionally sequenced on an ABI 3730xl genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Transfection experiments
HEK293T, HELA and MCF7 cells were subcultured in 60 mm dishes at a density of 2 × 105 cells in Dulbecco's modified Eagle's medium (DME) containing 4500 mg/l glucose supplemented with 10% (volume/volume) fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 m-glutamine (all products from Invitrogen, San Diego, CA, USA). The cells were incubated overnight at 37°C in a 5% CO2 humidified environment. Transfections were performed with a lipofectamine-plasmid emulsion as described previously.7 Twenty-one hour post-transfection (16 h for apoptosis assays; only for HEK293T cells), cells were harvested using TrypLE Express (Invitrogen) and diluted in 2 ml DME. Following centrifugation, the resulting cell pellet was resuspended in 500 μl D-PBS (Dulbecco's Phosphate-buffered Saline; Invitrogen) and was subsequently used for flow cytometric evaluation.
Cell viability measurements
Before measurements, 1 μl propidium iodide (PI) (1 mg/ml) (Invitrogen) was added to the cell suspensions to enable discrimination between viable and dead cells. Cells were then evaluated on a FACScan flow cytometer (BD Biosciences). The gating strategy and cell counting was as described previously.7 All measurements were independently replicated at least three times, unless stated otherwise. Cell viability was determined as the ratio of cells showing no PI fluorescence to the total cell population.
Confocal microscopy
Cells for confocal microscopy were prepared as described previously.7 Subcellular localization of selected constructs was performed using a Zeiss CLSM 510 confocal laser scanning microscope (Zeiss, Göttingen, Germany) equipped with an argon laser (excitation at 488 nm) and two helium/neon lasers (excitation at 543 and 630 nm, respectively). To test whether the mutant DFNA5 exon 9–exon 10 construct was localized in the endoplasmic reticulum (ER), double transfection with this construct and with pDsRed2-ER (Clontech, Mountain View, CA, USA) was performed in HEK293T cells. All images were taken at room temperature (RT) using a Zeiss C-Appochromat × 63/1.2 W Korr lens with Zeiss LSM 510 acquisition software.
Western blotting
HEK293T cells were subcultured at a density of 4 × 105 cells/well and lipofected as described previously.7 After 21 h of incubation, cells were lysed on ice using RIPA buffer (150 m NaCl, 50 m Tris-HCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS), containing Complete Mini protease inhibitors (Roche, Basel, Switzerland). The cell lysate was diluted in sample buffer (60 m Tris-HCl, 50% glycerol, 2% SDS, 14.4 m 2-mercaptoethanol, 0.1% Bromophenol Blue), heated for 5 min at 95°C, loaded on a 12% Tris-Glycine gel (Anamed, Gross-Bieberau, Germany) and electrophoretically separated. Next, the membrane was incubated with a primary rabbit anti-GFP antibody (1/2500; Sigma, St Louis, Missouri, USA) and a secondary goat anti-rabbit-horse radish peroxidase-conjugated antibody (1/10 000; Sigma) afterwards. Subsequently, EGFP-fusion proteins were detected using Pierce ECL western blotting substrate (Thermo Scientific, Rockford, IL, USA).
Annexin V staining
HEK293T cells were lipofected as described above. The standard harvesting time for the annexin V experiment was 16 h post-transfection. For the kinetic experiment, harvesting times were 3, 6, 9, 12, 16 and 24 h post-transfection. After harvesting, the cells were stained for annexin V using annexin V-biotin (Roche), according to the manufacturer's protocol. Before measurements, cells were stained with 1.3 μg/ml PI. Using a CyFlow ML cytometer (Partec, Mürlitz, Germany), viable, early apoptotic, late apoptotic and necrotic cells were analyzed. Transfected annexin V-stained cells were also visualized using confocal microscopy.
TUNEL assay
HEK293T cells were transfected as described above, harvested after 16 h, and a TUNEL assay (‘in situ cell death detection kit', tetramethylrhodamine red; Roche) was performed according to the manufacturer's instructions. Cells were then resuspended in D-PBS (Invitrogen), cytospinned and enclosed in Citifluor (Ted Pella, Redding, CA, USA). Cytospins were studied using confocal microscopy. Random images were taken at RT with Zeiss LSM 510 acquisition software on a Zeiss CLSM510 Meta microscope using a Zeiss C-Appochromat × 40/1.2 W Korr lens. For each experiment, the total number of TUNEL-positive cells and the total number of cells were counted from 10 random images, using the AnalySIS software.
Microarray expression analysis of Dfna5 KO versus WT mice and subsequent real-time PCR verification
Inner ear samples were removed from nine WT (+/+) postnatal day 0 (P0) and nine Dfna5 KO (−/−) (P0) C57Bl/6J mice and mechanically homogenized, after which total RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA quality was verified using the Experion system (Bio-Rad Laboratories). Subsequently, nine RNA samples from WT mice were pooled in three WT RNA pools and nine RNA samples from KO mice were pooled in three KO RNA pools. RNA pools were labeled with either Cy3 or Cy5 dyes, and hybridized on a Whole Mouse Genome Array (41 K array; Agilent, Santa Clara, CA, USA). Each experiment was followed by a colorflip, resulting in a total of six microarray experiments. The resulting intensity data were analyzed using the R-package ‘limma' (v3.2.1) and the Gene Set Enrichment Analysis (GSEA) program (v2.0, Broad Institute).18 Gene sets for GSEA were downloaded from the Gene Ontology website (http://www.geneontology.org/). These microarray results were validated with real-time PCR using the RNA pools from the microarray experiment and a one-step Power SYBR green PCR master mix (Applied Biosystems) according to the manufacturer's instructions on a Lightcycler 480 instrument (Roche). The resulting gene expression data were analyzed using Qbase plus (Biogazelle, Gent, Belgium) software.
Statistical analysis
All statistical analyses were performed using one-way ANOVA. P-values below 0.05 were considered statistically significant. Statistics were calculated using SPPS 15.0 (SPSS Inc., Chicago, IL, USA).
Results
Gross mapping of the toxic region
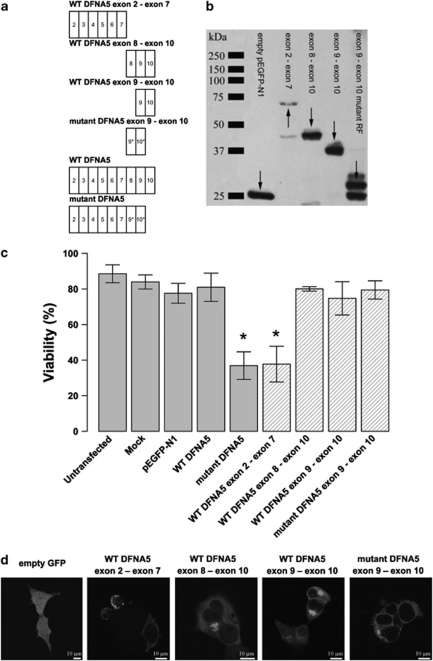
All DFNA5 mutations that cause HL lead to exon 8 skipping. Therefore, we tested whether the toxicity of the mutant protein was due to the aberrant tail of 41 amino acids that is present in the mutant protein, as the first seven exons are identical in WT and mutant DFNA5.7 We cloned the first part of DFNA5 (exon 2–exon 7) and generated three constructs based on the second part of DFNA5 (exon 8–exon 10 and exon 9–exon 10 in the WT as well as in the mutant reading frame; Figure 1a) in a pEGFP-N1 vector. The expression of the fusion proteins was confirmed by western blotting (Figure 1b). Figure 1c clearly demonstrates that transfection of the first part of DFNA5 in HEK293T cells causes a significant decrease in cell viability when compared with transfection of WT DFNA5 (P<0.001). This was an interesting finding because this part of the protein is shared by WT and mutant protein. None of the constructs of the second part of DFNA5 induced cytotoxicity.
Figure 1.
(a) Graphical representation of the insert content in relation to the construct's name. An exon is indicated by a rectangle containing the relevant exon number. The alternative reading frame used in the aberrant tail of mutant DFNA5 is indicated with an asterisk. (b) Western blot analysis of selected DFNA5 constructs demonstrating that all constructs are expressed when transfected in HEK293T cells (using a pEGFP-N1 vector). Arrows point to the relevant position of the fusion proteins. Some degradation products are visible in lanes 2, 3 and 5. (c) Cell viability measurements 21 h post-transfection. Graphical bars represent the percentage of viable cells. An asterisk denotes statistically significant differences (P<0.05) between transfections with the construct of interest and the WT construct. Control measurements are depicted in light gray colored bars, while constructs specific for this experiment are indicated with hatched bars. This series of transfections demonstrates that the toxic part of DFNA5 is located in the first part of DFNA5. Interestingly, this indicates that the motif that is responsible for toxicity is shared by WT and mutant DFNA5. (d) Subcellular localization of EGFP fusion proteins in HEK293T cells. Transfection of empty pEGFP-N1 vector results in an evenly distributed expression pattern throughout the cytosol. The first part of DFNA5 (exon 2–exon 7) is localized mainly at the plasma membrane. Cell blebbing is seen in cells transfected with this construct. A similar pattern was observed when mutant DFNA5 was transfected in HEK293T cells.7 Both WT DFNA5 exon 8–exon 10 and WT DFNA5 exon 9–exon 10 are localized in the cytoplasm, near the nucleus. Cells transfected with the mutant DFNA5 exon 9–exon 10 construct show expression in the endoplasmic reticulum (see also Supplementary Figure 1). A full color version of this figure can be found in the html version of this paper.
The C-terminal part of DFNA5 does not have an inhibitory effect on the cytotoxicity displayed by the N-terminal part of DFNA5, as the viability after double transfections with the WT DFNA5 exon 2–exon 7 and the WT DFNA5 exon 9–exon 10 construct or with the WT DFNA5 exon 2–exon 6 and the WT DFNA5 exon 8–exon 10 construct was comparable with that observed when transfecting the WT DFNA5 exon 2–exon 7 construct alone (results not shown).
The subcellular localization of these EGFP-tagged proteins was determined (Figure 1d). Transfection of the WT DFNA5 exon 2–exon 7 construct resulted in a fusion protein that was expressed mainly at the plasma membrane. In addition, it was observed in the cytoplasm in a granulated manner. The majority of cells transfected with this construct showed an unhealthy appearance (ie, cell blebbing). Overall, the expression pattern was very similar to that of mutant DFNA5.7 WT DFNA5 exon 8–exon 10 and WT DFNA5 exon 9–exon 10 exhibited a highly similar expression pattern. Both fusion proteins were distributed along the cytoplasm and cluster near the nucleus. Finally, cells transfected with mutant DFNA5 exon 9–exon 10 showed EGFP expression primarily in the ER. This was confirmed by co-transfection experiments with pDsRed2-ER, in which the EGFP signal of mutant DFNA5 exon 9–exon 10 colocalized with the signal of DsRed (Supplementary Figure 1).
Hydrophobic cluster analysis
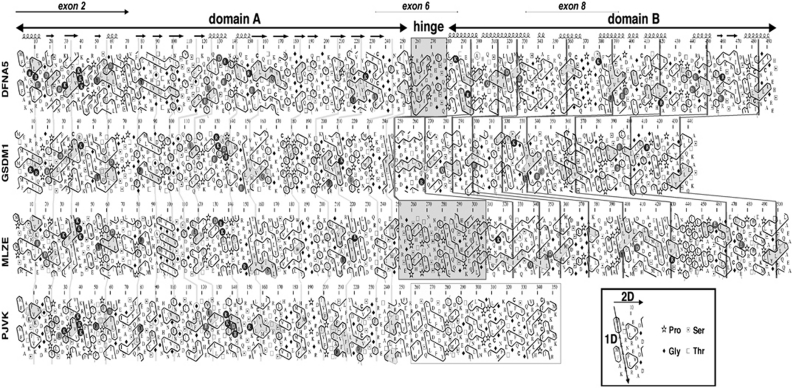
Simultaneously with the experiments described above, the sequence of DFNA5 was analyzed by hydrophobic cluster analysis (HCA) and compared with those of the DFNA5-gasdermin family, to which DFNA5 belongs (Figure 2). This technique of protein sequence analysis allows the simultaneous comparison of primary and secondary structures.19, 20, 21
Figure 2.
HCA analysis of DFNA5, GSDM1, MLZE and PJVK. The sequences are shown on a duplicated α-helical net, in which hydrophobic residues (VILFMYW) are contoured, forming clusters, which mainly correspond to the internal faces of regular secondary structures.27 The way to read the sequences (1D) and secondary structures (2D) as well as special symbols are indicated in the inset. Conserved hydrophobic residues are shaded in gray, the vertical lines indicating the correspondences between the different sequences. Noticeable identities/similarities outside the hydrophobic clusters are encircled in gray. The predicted globular domains (A and B) are depicted with arrows. A hinge of variable length (gray box) separates domain A from domain B. Predicted secondary structures of these globular domains are indicated at the top. Domain B does not exist in PJVK, in which it is replaced by a shorter domain (contoured in gray), likely containing a zinc-finger domain. A full color version of this figure can be found in the html version of this paper.
From the comparison of DFNA5-gasdermin family members, DFNA5 can be divided into three distinct domains: a globular domain A (exon 2–exon 6), a hinge region (exon 6–exon 7), which is variable in length and sequence among the different DFNA5-gasdermin members, and a globular domain B (exon 7–exon 10). Domain A, which is common to all members of the DFNA5-gasdermin family, had an α/β-fold, whereas domain B, which is present in GSDM1 and MLZE but not in PJVK (in this case, it is replaced by a zinc-finger domain), had an all α-fold (mainly α-helical structures). The lengths and shapes of the hydrophobic clusters within domain B suggested the presence of long helical structures, which might form coiled-coils.
Based on the division proposed by HCA, we constructed two plasmids, the first one containing HCA part A and the second HCA part B. When transfected in HEK293T cells, domain A but not domain B was able to induce a toxic effect when compared with WT DFNA5 (see Supplementary Table II). This finding enabled us to exclude exon 7 as part of the toxic region, as this exon was not present in domain A of DFNA5.
Domain A of DFNA5 is highly similar to the first part of PJVK. To investigate whether the mechanism of toxicity was a conserved feature among the two members of the DFNA5 family, we cloned the first part of PJVK (as indicated by HCA) in the pEGFP-N1 vector. After overexpression of the first part of PJVK in HEK293T cells, we measured no viability drop (see Supplementary Table II).
Further delineation of the toxic region
The toxic region of DFNA5 was further delineated through the generation of different constructs. In a first series of constructs, DFNA5 was truncated exon per exon starting from exon 7 toward the N-terminal region (Supplementary Figure 2). In another series of constructs, DFNA5 was truncated exon per exon from the N-terminal region toward exon 7 (Supplementary Figure 3). The conclusion of these series of experiments was that an essential part of the toxic DFNA5 region was located somewhere in exon 6 and that the presence of exon 2 was essential for the toxic effect.
The toxic motif did not consist of a single exon as was demonstrated by the transfection of DFNA5 exons 2–7 separately (see Supplementary Table II). In addition, transfection of constructs similar to WT DFNA5 HCA domain A, but lacking specific exons (either lacking exon 3, exon 4 or exon 5), did not lead to a detectable viability loss (see Supplementary Table II), indicating that the complete region between exon 2 and 6 is required for correct folding of the toxic domain.
Cell death evaluation
The fate of cells transfected with mutant DFNA5 was investigated using two independent assays: annexin V staining and TUNEL labeling.
Annexin V staining
To distinguish between apoptotic and necrotic cells, we performed a double labeling, using annexin V and PI. Early apoptotic cells were positive for annexin V, but not for PI. Late apoptotic cells were positive for both annexin V and PI (due to secondary necrosis), while necrotic cells emitted fluorescent signals only from PI.
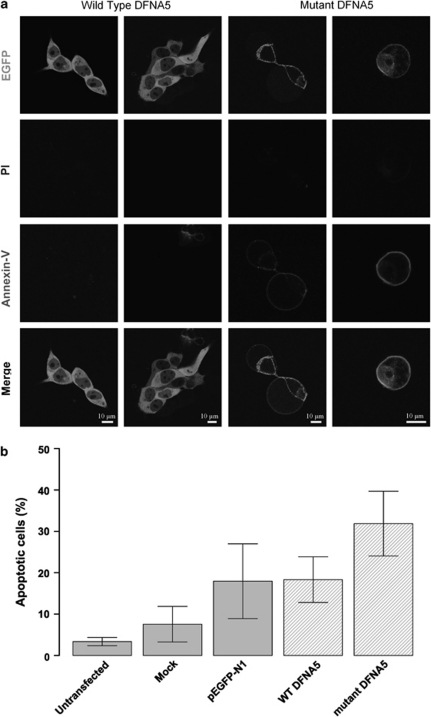
Cells were transfected with an empty EGFP vector and with WT and mutant DFNA5–EGFP constructs and were visualized using confocal laser microscopy (Figure 3a). The vast majority of cells transfected with WT DFNA5 showed no signs of PI or annexin V staining, while cells transfected with mutant DFNA5 exhibited clear annexin V-Cy5 staining at their plasma membranes. As no PI staining could be detected, this finding clearly indicated that cell death caused by mutant DFNA5 was due to an apoptotic process.
Figure 3.
(a) Confocal images of annexin V-Cy5-stained cell transfections. Cells transfected with WT DFNA5 are negative for PI and annexin V-Cy5. Cells transfected with mutant DFNA5 are negative for PI, but show clear annexin V at the plasma membrane, which is suggestive for apoptotic cell death. All cells were harvested 16 h post-transfection. A full color version of this figure can be found in the html version of this paper. (b) Flow cytometric quantification of annexin V-Cy5-stained transfected cells. Cells were harvested 16 h post-transfection. Percentages of apoptotic cells within the total cell population are shown. The total apoptotic fraction was calculated as the sum of the early (annexin V+ PI−) and late (annexin V+ PI+) apoptotic fractions.
To obtain quantitative results, we stained transfected HEK293T cell suspensions with annexin V-Cy5. Prior to flow cytometric analysis we also added PI to the cell suspensions. This allowed to discriminate between the viable, early apoptotic, late apoptotic and necrotic fractions. A typical flow cytometric result is shown in Supplementary Figure 4. To obtain the total apoptotic fraction in each cell suspension, we added the early apoptotic to the late apoptotic fraction. The bar charts in Figure 3b summarize the average total apoptotic fraction (three independent measurements) for each transfected cell suspension. Cells transfected with mutant DFNA5 had a higher total apoptotic fraction (31.87%±7.82) than cells transfected with WT DFNA5 (18.32%±5.51). Although the difference between the two groups was not statistically significant (P=0.07), the trend was clearly visible.
TUNEL assay
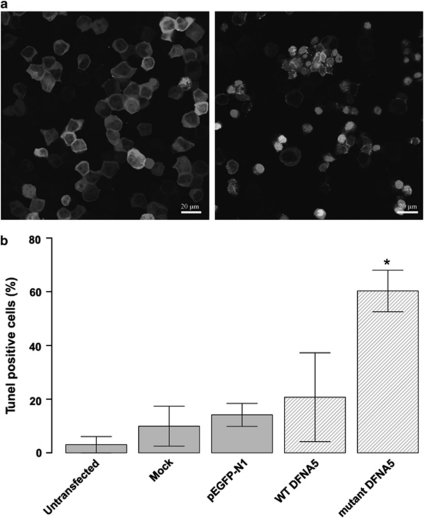
We transfected HEK293T cells with WT DFNA5 and mutant DFNA5–EGFP constructs and an empty EGFP vector as negative control. Further controls included an untransfected and a mock-transfected cell population. After staining, we counted the average TUNEL-positive cells in 10 randomly captured images and made a ratio of these positive cells to the total number of cells present in these images (Figure 4). A statistically significant difference (P=0.02) was observed between mutant DFNA5-transfected cells and cells transfected with WT DFNA5. The combined results of TUNEL and annexin V staining clearly indicate that toxicity induced by mutant DFNA5 was due to apoptosis.
Figure 4.
(a) Confocal images of WT DFNA5 (left pane of image)- versus mutant DFNA5 (right pane of image)-transfected HEK293T cells, stained with the in situ cell death detection kit, TM Red. TUNEL-positive cells show bright staining of the nucleus. A full color version of this figure can be found in the html version of this paper. (b) Quantitative measurement of TUNEL-positive cells relative to the total cell population. Cells transfected with mutant DFNA5 show significantly more apoptosis compared to controls. All cells were harvested 16 h post-transfection.
Microarray experiments using WT and Dfna5 KO mice
To evaluate whether the apoptosis-inducing features of DFNA5 could also be demonstrated in a physiological setting, we performed a microarray gene expression analysis on WT and Dfna5 KO mice using the Agilent Whole Mouse Genome Array (41 K). Differential gene expression was estimated and 92 genes were statistically significantly upregulated (adjusted P-value <0.05) in KO mice, while 88 genes were statistically significantly downregulated in KO mice when compared with WT mice. From this list, four genes were selected and their expression patterns were evaluated using real-time PCR and compared with those obtained from the microarray study. The differential expression of these genes was very similar between the two techniques (Supplementary Figure 5 and Supplementary Table III), indicating that the microarray results were reliable. Next, a GSEA was performed to check whether specific biological pathways were up- or downregulated in these mice to extract biologically meaningful conclusions. The analysis revealed 73 upregulated gene sets and 225 downregulated gene sets in KO mice (pathways with a False Discovery Rate <25% see Supplementary Tables IV and V for a list of these pathways). Particularly, gene sets involved in cartilage maintenance (indicated in red in Supplementary Table IV) and DNA repair (indicated in blue in Supplementary Table IV) are upregulated in Dfna5 KO mice, while gene sets involved in energy metabolism (indicated in orange in Supplementary Table V) and apoptosis (indicated in purple in Supplementary Table V) are downregulated in Dfna5 KO mice.
Discussion
In this study, we showed that induction of apoptosis is an intrinsic feature of the physiological function of DFNA5. We previously assumed that transfection of mutant DFNA5 causes necrotic cell death.7 As increasing evidence of tumor suppressor gene characteristics of DFNA5 has been provided,11, 12, 14 we re-evaluated the nature of mutant DFNA5-induced cell death. In this report, we used annexin V as well as TUNEL staining. Both techniques showed that transfection of mutant DFNA5 resulted in cell death due to apoptosis. A likely explanation for the misinterpretation7 may be that the time-lapse between transfection and assaying for cell death was too long in our previous report. We previously waited 24 h before harvesting the cells,7 versus 16 h in this study. We further investigated this concept with an experiment to establish the kinetics of mutant DFNA5-induced cell death. In a kinetic experiment, we harvested cells transfected with WT and mutant DFNA5 after 3, 6, 9, 12, 16 and 24 h (results not shown). Apoptotic events, identified as a positive signal for annexin V staining, became evident as soon as 6 h after transfection with mutant DFNA5, and remained evident up to the time point of 16 h, our standard harvesting time. Cells that were harvested after 24 h had already shifted from late apoptotic cell death to secondary necrotic cell death.
In this study, we localized two essential sections of the apoptosis-inducing domain. Our results demonstrate that the apoptosis-inducing domain is not merely a linear domain located in a single or a few exons, but a three-dimensional structure consisting of different non-contiguous parts of the protein, and that the complete region between exon 2 and exon 6 may be critical for the correct formation of the apoptosis-inducing motif. This implicates that proper folding of the globular domain formed by the region including exon 2 to exon 6 is essential.
The apoptosis-inducing mechanism of DFNA5 is not limited to transfections of HEK293T cells. Previously, we showed that transfection of mutant DFNA5 results in a viability loss in COS cells.7 Here, we extended our analysis and transfected additional cell lines (MCF7 and HELA) with mutant and WT DFNA5 (see Supplementary Figure 6). It appeared that transfection efficiencies in these cell lines were much lower than those obtained in HEK293T. Hence, transfection of mutant DFNA5 did not impact viability of the overall cell population. However, when only GFP-positive sub-populations were taken into account, it was clear that mutant DFNA5 causes a significant drop in viability, which is not seen after transfection of WT DFNA5. In conclusion, the apoptosis-inducing mechanism of DFNA5 is not limited to transfections in HEK293T, but is present in all investigated cell lines so far.
Gene expression microarray experiments on WT and Dfna5 KO mice, followed by pathway analysis, revealed that apoptosis inducing pathways are downregulated in Dfna5 KO mice. As such, we provide the first experimental evidence that the apoptotic features of DFNA5 are not only limited to overexpression experiments, but are also present in a physiological setting. Additionally, we have observed an upregulation of the DNA repair mechanisms in Dfna5 KO mice. The latter could be explained by the fact that when apoptotic pathways are downregulated, DNA repair mechanisms need to be upregulated to maintain viable conditions.
Interestingly, genes involved in cartilage maintenance were also upregulated in Dfna5 KO mice, linking our findings to those obtained by Busch-Nentwich et al.22 They found that zebrafish injected with Dfna5 morpholino's displayed an absent expression of Ugdh (UDP-glucose dehydrogenase), leading to reduced production of hyaluronic acid, resulting in impaired facial cartilage differentiation. As Dfna5 KO mice do not display this phenotype, it is possible that the gene sets involved in cartilage development provide compensatory mechanisms resulting in the lack of phenotype in these mice.
DFNA5 and PJVK clearly form a separate group within the DFNA5-gasdermin family,23 while the other group includes GSDM1, MLZE and homologs. Although the two families evolved from a common ancestor, phylogenetic analysis has indicated that the two clades are distinct,24 leaving DFNA5 with only a single closely related protein: PJVK. The first part of PJVK shows high similarity with DFNA5, but does not induce apoptosis. This is a first indication that the apoptosis-inducing mechanism may be specific for DFNA5 and that it may not be a general feature of the DFNA5 protein family.
Still, some questions remain to be clarified. Given the finding that the apoptosis-inducing domain is present in both WT and mutant DFNA5, the question arises as to why transfections with WT DFNA5 did not result in cell death. A plausible explanation could be that the last part of the protein may quench or shield the part in which the apoptosis-inducing domain resides, so that apoptosis is not expressed in normal cellular conditions. Because the mutation changes and shortens the last part of the protein (starting from exon 8), the shielding function of this part of the protein may be neutralized and, as a consequence, apoptosis may be unleashed. This remains to be further investigated, for example by the resolution of the crystal three-dimensional structure of DFNA5.
Methylation of the 5′-flanking region of DFNA5, as frequently observed in gastric, colorectal and breast carcinoma,13, 14, 15, 16 may be a way to prevent the apoptotic actions of DFNA5. We believe that our description of the apoptosis-inducing mechanism of DFNA5 contributes to the growing evidence that DFNA5 may be a tumor suppressor gene. As apoptosis induction seems to be an intrinsic feature of DFNA5's function, it is likely that the WT DFNA5 protein can be activated to induce apoptosis in the cell under certain conditions. The nature of this regulating mechanism, however, remains to be elucidated.
DFNA5-associated HL is nonsyndromic and has so far not been associated with other symptoms. Still, DFNA5 is present in every tissue investigated so far, albeit at low levels of expression. In our opinion, it is unlikely that massive apoptotic cell death takes place in cochleae of DFNA5 mutation carriers, or in any of the other tissues where DFNA5 is expressed. A more plausible scenario for the in vivo situation is that expression of mutant DFNA5 is a proapoptotic factor in the cell and that especially terminally differentiated cells, such as cochlear hair cells, are vulnerable to mutant DFNA5-induced programmed cell death. This cochlear hair cell loss may then be manifested as HL. The fact that DFNA5-associated HL is progressive in nature seems compatible with this hypothesis. The role of apoptosis in cochlear cells resulting in auditory pathology has been described previously. For example, it has been demonstrated that apoptosis occurs in outer hair cells in mice displaying age-related HL.25 Recently, a report was published implicating apoptosis in monogenic HL. Overexpression of TJP2 leads to altered expression of apoptosis-related genes, ultimately causing HL.26 We believe that our study provides a new line of evidence supporting an important role of apoptosis in HL.
In conclusion, we have demonstrated that DFNA5 is an apoptosis-inducing protein. In addition, it becomes increasingly clear that DFNA5 not only functions in the auditory pathway, but is probably also implicated in a more fundamental role involving cell survival and apoptosis.
Acknowledgments
This work was supported by grants of the European Community (Eurohear) and the ‘Fonds voor Wetenschappelijk Onderzoek Vlaanderen' (FWO grant G.0245.10N). KODB holds a predoctoral research position with the ‘Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT)'.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Van Laer L, Huizing EH, Verstreken M, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–197. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- Park HJ, Cho HJ, Baek JI, et al. Evidence for a founder mutation causing DFNA5 hearing loss in East Asians. J Hum Genet. 2009;55:59–62. doi: 10.1038/jhg.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Han DY, Dai P, et al. A novel DFNA5 mutation, IVS8+4 A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a Chinese family. Clin Genet. 2007;72:471–477. doi: 10.1111/j.1399-0004.2007.00889.x. [DOI] [PubMed] [Google Scholar]
- Bischoff AMLC, Luijendijk MWJ, Huygen PLM, et al. A second mutation identified in the DFNA5 gene in a Dutch family. A clinical and genetic evaluation. Audiol Neuro-otol. 2004;9:34–36. doi: 10.1159/000074185. [DOI] [PubMed] [Google Scholar]
- Yu C, Meng X, Zhang S, Zhao G, Hu L, Kong X. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a Chinese family. Genomics. 2003;82:575–579. doi: 10.1016/s0888-7543(03)00175-7. [DOI] [PubMed] [Google Scholar]
- Gregan J, Van Laer L, Lieto LD, Van Camp G, Kearsey SE. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim Biophys Acta. 2003;1638:179–186. doi: 10.1016/s0925-4439(03)00083-8. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Vrijens K, Thys S, et al. DFNA5: hearing impairment exon instead of hearing impairment gene. J Med Genet. 2004;41:401–406. doi: 10.1136/jmg.2003.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Meyer NC, Malekpour M, et al. A novel DFNA5 mutation does not cause hearing loss in an Iranian family. J Hum Genet. 2007;52:549–552. doi: 10.1007/s10038-007-0137-2. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Pfister M, Thys S, et al. Mice lacking Dfna5 show a diverging number of cochlear fourth row outer hair cells. Neurobiol Dis. 2005;19:386–399. doi: 10.1016/j.nbd.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Weigel RJ. Characterization of a gene that is inversely correlated with estrogen receptor expression (ICERE-1) in breast carcinomas. Eur J Biochem. 1998;252:169–177. doi: 10.1046/j.1432-1327.1998.2520169.x. [DOI] [PubMed] [Google Scholar]
- Lage H, Helmbach H, Grottke C, Dietel M, Schadendorf D. DFNA5 (ICERE-1) contributes to acquired etoposide resistance in melanoma cells. FEBS Lett. 2001;494:54–59. doi: 10.1016/s0014-5793(01)02304-3. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Futamura M, Kamino H, et al. The potential role of DFNA5, a hearing impairment gene, in p53-mediated cellular response to DNA damage. J Hum Genet. 2006;51:652–664. doi: 10.1007/s10038-006-0004-6. [DOI] [PubMed] [Google Scholar]
- Akino K, Toyota M, Suzuki H, et al. Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer. Cancer Sci. 2007;98:88–95. doi: 10.1111/j.1349-7006.2006.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chang X, Yamashita K, et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 2008;27:3624–3634. doi: 10.1038/sj.onc.1211021. [DOI] [PubMed] [Google Scholar]
- Kim MS, Lebron C, Nagpal JK, et al. Methylation of the DFNA5 increases risk of lymph node metastasis in human breast cancer. Biochem Biophys Res Commun. 2008;370:38–43. doi: 10.1016/j.bbrc.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikane T, Nishikawa N, Toyota M, et al. Genomic screening for genes upregulated by demethylation revealed novel targets of epigenetic silencing in breast cancer. Breast Cancer Res Treat. 2009;122:699–710. doi: 10.1007/s10549-009-0600-1. [DOI] [PubMed] [Google Scholar]
- Webb MS, Miller AL, Thompson EB. In CEM cells the autosomal deafness gene dfna5 is regulated by glucocorticoids and forskolin. J Steroid Biochem Mol Biol. 2007;107:15–21. doi: 10.1016/j.jsbmb.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Labesse G, Durand P, et al. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes R, Le Tuan K, Delettre J, Mornon JP, Callebaut I. A generalized analysis of hydrophobic and loop clusters within globular protein sequences. BMC Struct Biol. 2007;7:2. doi: 10.1186/1472-6807-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Busch-Nentwich E, Sollner C, Roehl H, Nicolson T. The deafness gene dfna5 is crucial for ugdh expression and HA production in the developing ear in zebrafish. Development (Cambridge, England) 2004;131:943–951. doi: 10.1242/dev.00961. [DOI] [PubMed] [Google Scholar]
- Delmaghani S, del Castillo FJ, Michel V, et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38:770–778. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- Tamura M, Tanaka S, Fujii T, et al. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics. 2007;89:618–629. doi: 10.1016/j.ygeno.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Sha SH, Chen FQ, Schacht J. Activation of cell death pathways in the inner ear of the aging CBA/J mouse. Hearing Res. 2009;254:92–99. doi: 10.1016/j.heares.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Pierce SB, Lenz DR, et al. Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet. 2010;87:101–109. doi: 10.1016/j.ajhg.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock S, Mornon JP, Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992;5:629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.