Abstract
As JV Neel put forward the ‘thrifty genotype' hypothesis, many researches tend to support this hypothesis involved in the regulation of energy balance. However, the phrase could equally well encapsulate broader traits and the forms of thrift should be multiple. In particular, genes involved in the regulation of water and sodium balance may also be excellent candidates as thrifty genes. In the present study, we selected the ancestral D allele of angiotensin-converting enzyme (ACE) gene, a key gene involved in water and sodium balance regulation, as a candidate to confirm the ‘thrifty genotype' hypothesis in the framework of evolutionary ecology. On the basis of our compiled worldwide spatial genetics database of I/D frequency of ACE gene and spatial climate database, using techniques of spatial statistics, we found (1) an obvious decreasing geographic genetic cline following the route of out-of-Africa expansion from East Africa, (2) a positive association between D allele and synthetic temperature factor, (3) and a negative relationship between D allele and synthetic humidity factor that covered most regions of the world, and obvious spatial dependence between D allele and these two climate factors followed the route of out-of-Africa expansion from Africa. This suggested that D allele of ACE gene is not only plastic in response to its environmental circumstance but also presents a striking geographic distribution showing the evidence of ‘signatures of selection' by climate factors. Thus, it can be identified as a thrifty allele and could provide a new evolutionary ecological evidence for the ‘thrifty genotype' hypothesis.
Keywords: ‘thrifty genotype' hypothesis, water and sodium regulation, angiotensin-converting enzyme gene, evolutionary ecology, spatial statistics, geographic information system
Introduction
In 1962, JV Neel put forth the ‘thrifty genotype' hypothesis in his famous paper entitled ‘Diabetes mellitus: a ‘thrifty' genotype rendered detrimental by ‘progress' to explain the epidemiology of diabetes. His general thesis, refined in later papers,1 remains a significant influence on our understanding of the evolutionary history of type 2 diabetes and other metabolic syndromes.2, 3, 4 This hypothesis suggested that, in the early years of life, the diabetic genotype was thrifty in the sense of being exceptionally efficient in utilization of food. It would thereby confer a survival advantage in the times of food shortages. However, in contemporary societies, as food is usually available in unlimited amounts, the ‘thrifty genotype' no longer provides a survival advantage, but renders its owners more susceptible to obesity and diabetes.5 Although variants of this hypothesis have formed the topic of vigorous debate,6, 7, 8, 9, 10 there are many researches that tend to support this hypothesis, not only in epidemiology11, 12, 13 but also in population genetics or evolutionary ecology.4, 14, 15
One of the problems of using the ‘thrifty gene' concept is that most individuals associate it with a metabolic trait related to a frugal utilization of fuel, which indeed was Neel's (1962) first intention.3 However, the phrase could equally well encapsulate broader traits and the forms of thrift should be multiple, including metabolic, adipogenic, physiological or behavioral mechanisms.3, 16, 17 A broader range of traits may merit within this typology, including those relating to growth, reproduction and immune function.18 Consistent with the multiple possible forms of thrifty gene, we propose that the thrifty gene should not be limited to genes involved in the regulation of energy balance; genes involved in the regulation of water and sodium balance may also be good candidates as thrifty genes.
In particular, some key genes in the renin–angiotensin–aldosterone system (RAAS), which has a pivotal role in the regulation of blood pressure, volume homeostasis, vascular function and cell growth, may be good candidates to provide evidence for the ‘thrifty genotype' hypothesis. The most impressive case is the angiotensin-converting enzyme (ACE) gene with a common polymorphism represented by the insertion (I) or deletion (D) of a 287-bp AluYa5 element inside intron 16 on chromosome 17q23,19 which is associated with many cardiocerebral vascular diseases (CVDs).20, 21 Moreover, studies on ACE I/D polymorphisms have indicated that the Alu insertions are shown to be absent from the genomes of a number of non-human primates, consistent with the rise in human genetic polymorphisms sometime after the human/African anthropoid divergence,22 which suggests that D allele is the ancestral allele23, 24 and its evolutionary history could be used to provide evidence for the ‘thrifty genotype' hypothesis. On the other hand, accumulating evidence has suggested that genetic susceptibility to CVD as well as salt avidity and cardiovascular reactivity are ancestral and were likely magnified during the early human evolution before the out-of-Africa expansion of anatomically modern humans25, 26, 27 and it is due to diverse selection pressure during the out-of-Africa expansion that occurred 30 000–100 000 years ago, among which, the most important selection pressure was climate.28
A good candidate for a ‘thrifty gene' should exhibit two important features. The first is plasticity in response to environmental circumstance, and the second feature that would be expected is the evidence of ‘signatures of selection', such as striking geographical distribution.3 As an ancestral hypertension susceptibility allele in RAAS, the evolutionary ecology of D allele in ACE gene should be expected to provide some evidence for the ‘thrifty genotype' hypothesis. We therefore propose the following assumption: before the out-of-Africa expansion of anatomically modern humans (Africa was hot and wet during the early Miocene, whereas its climate had become hot and dry since mid-Miocene28), as a salt-sensitivity allele with the propensity to retain salt and water, D allele may have some advantages for humans to adapt to the ancestral environment. Yet, after expanding out of Africa to inhabit the rest of world, where the climate gradually becomes colder or wetter than equatorial Africa, the propensity to retain salt and water becomes less important accordingly. The thrifty D became deleterious, as it increases susceptibility to hypertension and other hypertension-related diseases.20, 21 Consequently, some individuals who carried D allele were eliminated from the population because of hypertension or other hypertension-related diseases. If the above assumption is true, the frequency of D allele in Africans should be higher, and from Africa, following the route of out-of-Africa expansion, an obvious decreasing geographic genetic cline should be detected. Furthermore, a geographic dependence (or association) between spatial genetic cline of D allele and spatial ecologic cline of climate should be discovered because of the selection pressure to hypertension susceptibility.
To confirm this hypothesis, in the present study, we compiled a worldwide spatial genetic database of I/D frequency of ACE gene based on the published data, and explored the spatial genetic structure of D allele, as well as the spatial dependence between D allele frequency and climate factors. To characterize the spatial genetic structure of D allele, the Kriging interpolation method was used to visualize geographic genetic clines of D allele, as well as climate clines.29 In addition, the confirmatory factor analysis (CFA),30 in combination with Geographically Weighted Regression (GWR),31 was used to detect spatial dependence between D allele frequency and climate factors. All the maps were created by ArcGIS 9.0 (http://www.esri.com).
Materials and methods
Samples and their spatial databases
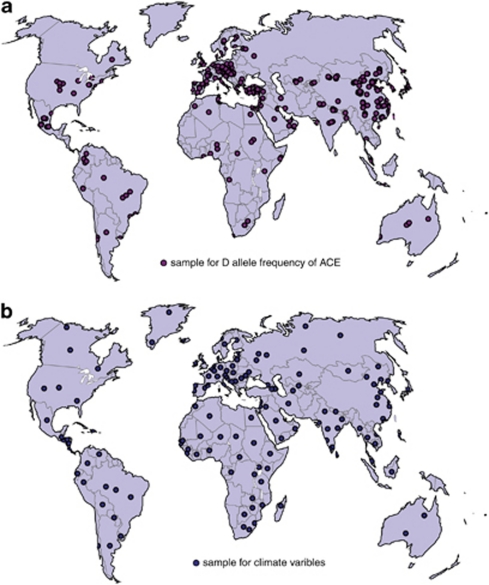
We compiled a worldwide spatial database on I/D frequency distribution using data derived from 299 published sources, with a total of 183 555 individuals from 422 sampling populations. Majority of the records are published in English, searched from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov) using key words such as ‘angiotensin converting enzyme', ‘polymorphism' and country names (eg, ‘Japan'). Small proportion of the data are collected from the literature published in Chinese core journals, which were downloaded from the National Knowledge Infrastructure website (http://www.cnki.net). Studies with a small sample size (<50) and those that do not conform to Hardy–Weinberg equilibrium law were excluded. Gene frequencies from populations with the same geographic coordinates were averaged weighted by sample. The geographic information system (GIS) (http://www.esri.com) was used to build spatial databases. Figure 1a shows the locations of the samples, and a list describing the sources of the data is provided as supplement (see Supplementary Materials 1). The geographic climate data (including Annual precipitation (AP), Annual atmospheric temperature (ATT) and Temperature climate zone (TCZ) and so on) were collected from the World Climate website (http://www.climate-charts.com), and Table 1 shows the value assignment. Data from 151 weather stations were collected with the same or approximate geographic coordinates to genetic data samples. Figure 1b shows the location of the weather stations, and a list describing the sources of the data, together with their value assignment, is provided as supplement (see Supplementary Materials 2)
Figure 1.
Locations of (a) 299 sampled populations for ACE gene D allele and (b) 151 sampled weather stations for climate variables.
Table 1. Value assignment of the geographic climate variables.
| Geographic climate index | Variable | Assignment |
|---|---|---|
| Annual precipitation, AP | 0 mm≤AP<200 mm | 1 |
| 200 mm≤AP<500 mm | 2 | |
| 500 mm≤AP<1000 mm | 3 | |
| 1000 mm≤AP<2000 mm | 4 | |
| 2000 mm≤AP | 5 | |
| Annual atmospheric temperature, AAT | 30°C≤AAT | 6 |
| 20°C≤AAT<30°C | 5 | |
| 10°C≤AAT<20°C | 4 | |
| 0°C≤AAT<10°C | 3 | |
| −10°C≤AAT<0°C | 2 | |
| −20°C≤AAT<−10°C | 1 | |
| Temperature climate zone, TCZ | Frigid zone | 1 |
| Subfrigid zone | 2 | |
| Temperate zone | 3 | |
| Subtropical zone | 4 | |
| Tropical zone | 5 | |
| Geography climatic zone, GCZ | Continental climate | 1 |
| Transitional climate | 2 | |
| Oceanic climate | 3 | |
| Geography latitude, GL | 0°≤GL<25° | 3 |
| 25°≤GL<50° | 2 | |
| 50°≤GL<75° | 1 |
Detection of geographic genetic clines of ACE gene
To quantify the spatial variation of D allele in ACE gene, the ordinary Kriging interpolation32 was used to visualize spatial genetic patterns and geographic genetic clines. Kriging is a spatial interpolation method that predicts unknown values from data observed at known locations. This method uses variogram to express the spatial variation, and it minimizes the error of predicted values,32 which are estimated by spatial distribution of the predicted values. As a virtually unbiased method in a interpolation situation, Kriging model has several advantages over other interpolation and smoothing methods, and has been used to create maps of geographic genetic clines in human population genetics and biological anthropology.29 In the present study, after the Kriging maps were created by ArcGIS9.0, the Natural Breaks (Jenks) method33 was used to classify the geographic genetic clines.
Detection of spatial dependence relationship between D allele and climate
To detect the spatial dependence relationship between D allele and climate, the CFA,30 in combination with Ordinary Least Squares (OLS) regression as well as GWR,31 were used to build the global and local regression model between D allele frequency and climatic factors. CFA was first used to obtain synthetic climate factors and their scores for each climate data point were obtained by the software of lisrel 8.7 (http://www.ssicentral.com/lisrel). Further, the above Kriging algorithm was used to obtain the predicated values of the synthetic climate factors, which were corresponding to the locations of each genetic sample data (D allele frequency). Finally, by using the software of Spatial Analysis in Macroecology (SAM) v4.0 (http://www.ecoevol.ufg.br/sam), OLS regression and GWR were used to set up the global and local regression model between D allele frequency and synthetic climate factors, respectively.
Statistically, CFA30 is a special form of factor analysis. It is used to test whether measures of a construct are consistent with a researcher's understanding of the nature of that latent construct (or latent factor). On the basis of the exploratory factor analysis result of the observed geographic climate variables (AP, ATT, TCZ, geography climatic zone (GCZ) and geography latitude (GL)), CFA was used to extract the latent synthetic climate factors that are expected to be associated with D allele frequency. The P-value of χ2 >0.05, GFI, AGFI and NNFI >0.90 and RMSEA <0.08 are considered as thresholds for a suitable model.34 GWR31 is a local version of spatial regression that generates parameters disaggregated by the spatial units of the analysis. It allows assessment of the spatial heterogeneity in the estimated relationships between independent variables and the dependent variable. Unlike conventional OLS regression, which may only produce a single regression equation to summarize global relationships between D allele and synthetic climate factors, GWR used in this paper can generate spatial dependence that express the local spatial variation between them dynamically, because the regression coefficients of GWR are allowed to vary spatially. As a powerful spatial statistical method to examine geographical variation between dependent and independent variables, GWR has been successfully used in spatial epidemiology35 and similarly in spatial ecology.36
Results
Geographic genetic clines of D allele of ACE gene
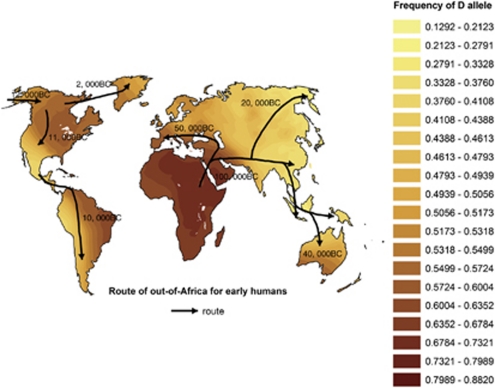
Figure 2 shows the geographic genetic clines interpolated by D allele frequency of ACE gene and the route of out-of-Africa expansion by GIS superimposition analysis. It, generally, reveals an obvious decreasing geographic genetic cline following the route of out-of-Africa expansion from East Africa. The highest frequency of D allele appears in Africa and Arab regions, the lowest frequency locates in East Asia and the medium value emerges in Europe, Australia and America.
Figure 2.
Worldwide geographic genetic clines of D allele frequency of ACE gene interpolated by 299 sampled populations.
Latent geographic climate factors and their spatial ecological clines
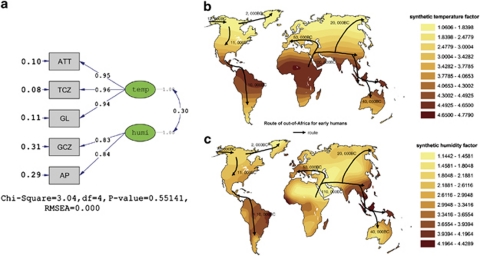
Figure 3a shows geographic climate structure model created by the CFA method, two latent geographic climate factors are extracted from this structure model. The first one, which is reflected by ATT, TCZ and GL, was named as synthetic temperature factor (STF; Figure 3a). The second one, which is composed of AP and GCZ, was named as synthetic humidity factor (SHF; Figure 3a). According to the value assignment of the geographical climate factors (Table 1), the larger the STF, the hotter the climate, and the larger the SHF, the wetter the climate. Figures 3b and c show the spatial ecological clines interpolated by STF and SHT, respectively. Conforming to the real world of climate distribution, STF reaches the maximum around the equatorial zone by decreasing gradually toward Antarctic Pole and Arctic Pole symmetrically and reaching the minimum in north Russia, north Canada and Greenland (Figure 3b), whereas SHF shows the highest values in South-East Asia, central Africa and south America, and lower values in north Africa, Arab region, central Asia, Greenland and south-west Australia (Figure 3c).
Figure 3.
Worldwide geographic clines of two synthetic factors. The P-value,GFI, AGFI, NNFI and RMSEA of the confirmatory factor analysis model were 0.55141, 0.99, 0.97, 1.00 and <0.01,respectively, in (a), which suggest the model was fitted well.34 (b) and (c) Show interpolated maps of synthetic temperature factor and synthetic humidity factor interpolated by 151 sampled weather stations, respectively.
Global spatial dependence between D allele and synthetic climate factors
Table 2 shows the result of OLS regression between D allele and synthetic climate factors. D allele frequency was significantly associated with STF and SHT (F=180.75, P<0.001), and 27.47% of the total variance of D allele was explained by the two synthetic climate factors (Radj2=0.2747). Moreover, the regression coefficients (Table 2) showed that the higher the STF and the lower the SHF, the higher the D allele frequency. With STF, SHF and D allele all revealing obvious geographic clines (Figures 2, 3b and c), the OLS regression result (Table 2) indicated a significant global relationship between D allele and climate.
Table 2. Parameter estimates comparison of OLS model and GWR model.
| β1 (synthetic temperature factor) | β2 (synthetic humidity factor) | Adjusted R2 | |
|---|---|---|---|
| OLS model | 0.275 | ||
| Estimates | 0.04075 | −0.06242 | |
| Standard error, SE | 0.00242 | 0.00397 | |
| β±1 SE | 0.03833–0.04317 | −0.06639 to −0.05845 | |
| t-Value | 16.85 | −15.72 | |
| Pr>∣t∣ | <0.0001 | <0.0001 | |
| GWR model | 0.838 | ||
| 25% Quartile | 0.01583 | −0.06781 | |
| 50% Quartile | 0.0358 | −0.02855 | |
| 75% Quartile | 0.05168 | −0.00716 |
Local spatial dependence between D allele and synthetic climate factors
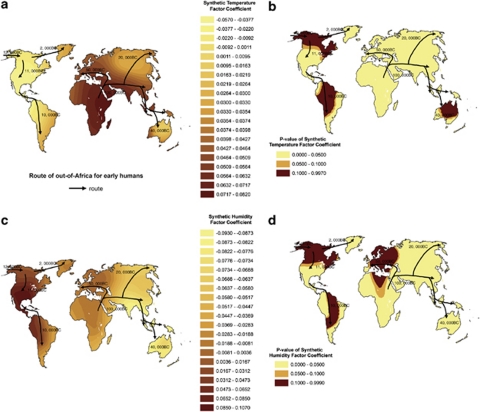
Table 2 summarizes the results of GWR between D allele and synthetic climate factors. In the GWR model, the adjusted R2 increased from 0.275 (OLS) to 0.838 (GWR), demonstrating that GWR had a much better explanatory ability than OLS model. The F-test rejected the null hypothesis (F=316.25, P<0.001), suggesting that the improvement to model fit using GWR was statistically significant. Furthermore, as the interquartile range of the local estimates ([0.01583–0.05168] for STF, and [−0.06781 to −0.00716] for SHF were greater than that of the global OLS parameter estimation±1 SE ([0.03833–0.04317] for STF, and [−0.06639 to −0.05845] for SHF), the spatial dependence relationship between D allele and the two synthetic climate factors could be considered as non-stationary,31 indicating that the spatial dependence between them (reflected by the regression coefficients of GWR) should vary spatially. Figures 4a and b show a contour map of the regression coefficients of STF and its P-values, respectively, interpolated by the Kriging method. This demonstrated that the regression coefficients are significant in most areas of the world, and the local spatial dependency between D allele and STF exhibits a non-constant mean and variance across the whole study area. The standardized regression coefficients estimates of STF from the GWR were mostly positive except in Australia, North America and South America, which indicates a positive association between D allele and STF in most regions of the world. Furthermore, it reveals an obvious decreasing spatial dependency between D allele and STF following the route of out-of-Africa expansion from Africa, and the strongest relationship between them was concentrated at the tropical areas, especially in most of Africa, Arab and India (Figure 4a), whereas the negative standardized regression coefficients estimates of STF were mostly in North America, South America and Australia (some were not statistically significant (Figures 4a and b)), which are predominantly occupied by the immigrants from Europe recently (during sixteenth to eighteenth century).
Figure 4.
Worldwide geographic clines of two synthetic factor coefficients. (a) and (c) Show the distribution of synthetic temperature factor and synthetic humidity factor coefficients derived from the Geographically Weighted Regression model, respectively. (b) and (d) Show P-value distribution of synthetic temperature factor and synthetic humidity factor coefficients derived from the Geographically Weighted Regression model, respectively.
Figures 4c and d show contour map of the regression coefficients of SHF and its P-values, respectively. Similar to the regression coefficients of STF, it also demonstrated that the regression coefficients are significant in most areas, and the local spatial dependency between D allele and SHF varies across the world. A negative association between D allele and SHF was revealed in most regions of the world, except North America and South America. Generally, this negative association is consistent with the spatial distribution of SHF, and the drier the area, the stronger the negative association (Figure 3b). In arid areas, such as in Africa and Arab regions, Greenland, parts of Central Asia and Australia, negative association between D allele and SHF are stronger than that of wet areas, such as South-East Asia. However, the positive or nonsignificant regression coefficients of SHF are mostly in North America and South America, which are occupied by immigrants from Europe recently. In addition, another region with a nonsignificant regression coefficient reveals in Europe.
Discussion
The ‘thrifty genotype' hypothesis posits that there exist genes in human populations that facilitate the efficient utilization of food and its conversion into stored fat. Populations in early stages of human evolution, who were exposed to unpredictable periods of starvation with such genes, as a result are more likely to survive during periods of famine. Yet, in contemporary societies, as food is usually available in unlimited amounts and life is sedentary, thrifty genes become deleterious, as they increase susceptibility to diabetes mellitus.1, 2, 3, 4, 37 This hypothesis has been widely supported by epidemiological,11, 12, 13 genetic and evolutionary researches.4, 14, 15 In particular, some specific genes have been identified or considered as ‘thrifty genes' (or ‘thrifty SNP'), including calpain-10 gene and G-protein-coupled receptor 35 gene,4 APOE*4 allele,14 SNPs in FABP2, MTP, β3AR (the Trp64Arg SNP), apo-E, UCP2, UCP3-p, PPARγ2, LEPR, adiponectin and so on.15 However, current studies on ‘thrifty gene' are mainly limited in genes involved in the regulation of energy balance. Although the concept of ‘multiple thrifty gene form' has been put forward, the research and discussion scope is still constrained in genes involved in regulation of energy balance.3, 16, 17, 18 In the present study, we further extended the concept of thrifty gene, and proposed that genes involved in regulation of water and sodium balance could also be good candidates as thrifty genes. As an impressive case, we selected the ACE gene, a key gene in RAAS, to explore the new evolutionary ecological evidence for the ‘thrifty genotype' hypothesis.
RAAS has a pivotal role in the regulation of blood pressure, volume homeostasis, vascular function and cell growth. The RAAS pathway and the functions of different parts are summarized in Supplementary Materials 3. Some ancestral key genes in RAAS may be good candidates to provide evidence for the ‘thrifty genotype' hypothesis. In fact, as an ancestral gene28 in RAAS, the angiotensinogen (AGT) gene has been considered as a thrifty gene.28, 38, 39, 40 In the present study, we chose the ACE gene, another ancestral hypertension susceptibility gene in RAAS, to explore the evolutionary ecological evidence for the ‘thrifty genotype' hypothesis within the framework of spatial genetics and spatial ecology.
Heat and drought are cardinal environmental factors responsible for natural selection since the beginning of life on Earth.41 Effective heat dissipation is essential in hot environments and is achieved most efficiently through evaporative heat loss.42 As a result, along with the arid environment, the high temperature contributes to the consequence that the African ancestors developed a markedly enhanced capacity to sweat.43 Sweating, however, can lead to loss of large amounts of salt and water.42 Large salt losses due to sweating made sodium and water conservation essential for survival.44 For example, humans and non-human primates from tropical climates have enhanced salt and water avidity.25, 26, 27 Another consequence of this reliance on sweating is decreasing blood volume.28 To maintain blood pressure and ensure organ perfusion, the vessel contraction is important. Because heat dissipation through sweating results in large volume losses, as well as the water scarcity due to the drought, further enhancement of salt and water avidity and cardiovascular reactivity were likely part of human adaptation to hot, dry African environments since the Miocene.28 Therefore, under the ancient African environmental condition, which was arid and hot, the sodium and water retaining capacity was crucial, carrying D allele frequency could be favored, given that the higher the D allele frequency, the higher the activity of serum ACE level.19, 20 When each population reached their new location and settled down, they were subject to new different evolutionary forces, such as cold temperatures and humid surroundings, which likely became important selective forces after human out-of-Africa expansion.
We, therefore, deduce that the D allele of ACE gene, which is an ancestral salt-sensitivity allele with the propensity to retain salt and water and increase hypertension susceptibility,24 is likely to suffer the following selection pressure and evolutionary history: before the out-of-Africa expansion of anatomically modern humans, D allele had some advantages for humans to adapt to the ancestral environment. Yet, after they expanded out of Africa to inhabit the rest of world, where the climate gradually becomes colder or wetter than that in Africa and the propensity to retain salt and water becomes less important accordingly, the thrifty allele D becomes deleterious, as it increases susceptibility to hypertension and other hypertension-related diseases.20, 23 Consequently, some individuals who carried D allele were eliminated from the population because of hypertension or other hypertension-related diseases. By conducting the Kriging spatial interpolation of D allele and its GIS superimposition analysis with the route of out of Africa, in the present study, we have detected an obvious decreasing geographic genetic cline following the route of out-of-Africa expansion from East Africa (Figure 2); these results coincide with the published results of the worldwide D allele distribution by using non-spatial genetic methods.45 Obviously, this striking geographical distribution of D allele offers its ‘signatures of selection' evidence, and supports the above inference.
Accumulating evidence suggests that the most important selection pressure to genetic susceptibility to hypertension is climate.37 Thus, a geographic correlation between genetic cline of D allele and ecological cline of climate should be detected. From the results, we have detected this relationship and trend (Table 2, Figures 3 and 4). For the STF, we found that a positive association between D allele and STF covers most regions of the world, and an obvious decreasing spatial-dependent cline between D allele and STF, which conforms to the spatial distribution of STF, follows the route of out-of-Africa expansion from East Africa (Figures 3b, 4a and b). The strongest relationship between them was concentrated at the tropical areas, especially in most parts of Africa, Arab and India. Clearly, this striking spatial dependence of the D allele on STF supports the above expectation. As an exception, in Australia, North America and South America, the local spatial dependence with negative association or non-statistically significant associations could not conform with the regulations (Figures 4a and b); however, it can be explained by special population genetic structure in these regions, which are predominantly occupied by the immigrants from Europe recently (during sixteenth to eighteenth century). For the SHF, we found a negative association between the D allele and SHF in most regions of the world. This negative association is consistent with the spatial distribution of SHF, and the drier the climate, the stronger the negative association (Figures 3c, 4c and d). Again, this striking spatial dependence of the D allele on SHF also obviously supports the expectation. For the positive association between the D allele and SHF emerging in North America and South America (Figures 4c and d), its non-conformity to the hypothesis can be also explained by special population genetic structure in these regions, as discussed above.
Studies showed that the D allele frequency is closely related to serum ACE levels.19, 20 Therefore, under the ancient African environmental condition, which was arid and hot, the capacity to retain sodium and water was crucial, so the D allele would be favored. As humans spread out of equatorial Africa to regions at higher latitudes, the cold temperatures and humid surroundings likely became important selective pressures. Consequently, traits under stabilizing selection at near-equatorial latitudes may have become deleterious at higher latitudes.44 The prevalence of hypertension in this population should be expected to be higher than that of Africa. This expectation has actually been validated by the investigation of Worldwide prevalence of hypertension,46 which suggested that the African countries had relatively low prevalence of hypertension compared with European and Asian countries.
In summary, the D allele of ACE gene is not only plastic in response to its environmental circumstance but also presents a striking geographical distribution showing the evidence of ‘signatures of selection' by climate factors. Thus, it can be identified as a ‘thrifty allele' and can provide new evolutionary ecological evidence for the ‘thrifty genotype' hypothesis.
Limitation and further work
Because of the limitation of our spatial genetic database, we cannot obtain the climate data at each geographical location of the sampled population. We have to estimate the climate values by Kriging interpolation method at each point of D allele frequency. Further, the observed gene frequency and the interpolated climatic data were used to conduct the OLS and GWR model. Although this may lead to some errors, it is an acceptable method regarding our current data set. On the other hand, because of the multicollinearity between the climate variables (AP, AAT, TCZ, GL, GCZ), it is difficult to build the multiple regression model by these variables directly. We, therefore, extracted the latent variables as the independent variables of the regression model. Using this method, we not only avoid multicollinearity but also obtain the synthetic factors to measure and interpret the climate data.
In RAAS, besides ACE gene, there exists other genes involved in the regulation of water and sodium balance. Further work is needed to explore the evolutionary ecological evidence using the other genes in the whole system, such as AGT gene, Angiotensin II receptor type 1 gene, aldosterone synthase (CYP11B2) gene and so on.
Acknowledgments
We are pleased to acknowledge the support of the national Natural Science Foundation of China (NSFC) through grant 30890034. We are also pleased to acknowledge the World Climate website for providing us the climate data. We appreciate investigators whose data about the ACE I/D loci were included in our database.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Neel JV, Weder AB, Julius S. Type II diabetes, essential hypertension, and obeisty as ‘syndromes of impaired genetic homeostasis': the ‘thrifty genotype' hypothesis enters the 21st century. Perspect Biol Med. 1998;42:44–74. doi: 10.1353/pbm.1998.0060. [DOI] [PubMed] [Google Scholar]
- Prentice AM. Early influences on human energy regulation: thrifty genotypes and thrifty phenotypes. Physiol Behav. 2005;86:640–645. doi: 10.1016/j.physbeh.2005.08.055. [DOI] [PubMed] [Google Scholar]
- Prentice AM, Rayco-Solon P, Moore SE. Insights from the developing world: thrifty genotypes and thrifty phenotypes. Proc Nutr Soc. 2005;64:153–161. doi: 10.1079/pns2005421. [DOI] [PubMed] [Google Scholar]
- Vander Molen J, Frisse LM, Fullerton SM, et al. Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am J Hum Genet. 2005;76:548–560. doi: 10.1086/428784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott R. Ethics, epidemiology and the thrifty gene: biological determinism as a health hazard. Soc Sci Med. 1998;47:1189–1195. doi: 10.1016/s0277-9536(98)00191-9. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Thrifty genes for obesity and the metabolic syndrome--time to call off the search. Diab Vasc Dis Res. 2006;3:7–11. doi: 10.3132/dvdr.2006.010. [DOI] [PubMed] [Google Scholar]
- Prentice AM, Hennig BJ, Fulford AJ. Evolutionary origins of the obesity epidemic: natural selection of thrifty genes or genetic drift following predation release. Int J Obes (Lond) 2008;32:1607–1610. doi: 10.1038/ijo.2008.147. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: the ‘drifty gene' hypothesis. Int J Obes (Lond) 2008;32:1611–1617. doi: 10.1038/ijo.2008.161. [DOI] [PubMed] [Google Scholar]
- Speakman JR. A nonadaptive scenario explaining the genetic predisposition to obesity: the ‘predation release' hypothesis. Cell Metab. 2007;6:5–12. doi: 10.1016/j.cmet.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Neel JV. Update to ‘The study of natural selection in primitive and civilized human populations'. Human Biol. 1989;61:811–823. [PubMed] [Google Scholar]
- Wendorf M, Goldfine ID. Archaeology of NIDDM: excavation of the ‘thrifty' genotype. Diabetes. 1991;40:161–165. doi: 10.2337/diab.40.2.161. [DOI] [PubMed] [Google Scholar]
- Zimmet P, Dowse G, Finch C, Serjeantson S, King H. The epidemiology and natural history of NIDDM. Lessons from the south. Pacific Diabetes Metab Rew. 1990;6:91–124. doi: 10.1002/dmr.5610060203. [DOI] [PubMed] [Google Scholar]
- Dowse GK, Zimmet PZ, Finch CF, Collins VR. Decline in incidence of epidemic glucose intolerance in Nauruans: implications for the ‘thrifty genotype'. Am J Epidemiol. 1991;133:1093–1104. doi: 10.1093/oxfordjournals.aje.a115822. [DOI] [PubMed] [Google Scholar]
- Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty' allele. Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- Kagawa Y, Yanagisawa Y, Hasegawa K, et al. Single nucleotide polymorphisms of thrifty genes for energy metabolism: evolutionary origins and prospects for intervention to prevent obesity-related diseases. Biochem Biophys Res Commun. 2002;295:207–222. doi: 10.1016/s0006-291x(02)00680-0. [DOI] [PubMed] [Google Scholar]
- Bouchard C. The biological predisposition to obesity: beyond the thrifty genotype scenario. Int J Obes (Lond) 2007;31:1337–1339. doi: 10.1038/sj.ijo.0803610. [DOI] [PubMed] [Google Scholar]
- Siervo M, Wells JC, Cizza G. The contribution of psychosocial stress to the obesity epidemic: an evolutionary approach. Horm Metab Res. 2009;41:261–270. doi: 10.1055/s-0028-1119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JC. Thrift: a guide to thrifty genes, thrifty phenotypes and thrifty norms. Int J Obes (Lond) 2009;33:1331–1338. doi: 10.1038/ijo.2009.175. [DOI] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Hirata RD, Hirata MH. Five polymorphisms in gene candidates for cardiovascular disease in Afro-Brazilian individuals. J Clin Lab Anal. 2004;18:309–316. doi: 10.1002/jcla.20044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennon B, Petrie JR, Small M, Connell JM. Angiotensin-converting enzyme gene and diabetes mellitus. Diabet Med. 1999;16:448–458. doi: 10.1046/j.1464-5491.1999.00071.x. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Alegria-Hartman M, et al. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci USA. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn CF, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for angiotensin-converting enzyme. Pharmacogenet Genomics. 2010;20:143–146. doi: 10.1097/FPC.0b013e3283339bf3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EH, Ireson NJ, Carney C, Markandu ND, MacGregor GA. Transepithelial sodium absorption is increased in people of African origin. Hypertension. 2001;38:76–80. doi: 10.1161/01.hyp.38.1.76. [DOI] [PubMed] [Google Scholar]
- Vollmer WM, Sacks FM, Ard J, et al. Effects of diet and sodium intake on blood pressure: subgroup analysis of the DASH-sodium trial. Ann Intern Med. 2001;135:1019–1028. doi: 10.7326/0003-4819-135-12-200112180-00005. [DOI] [PubMed] [Google Scholar]
- Sawka MN, Montain SJ, Latzka WA. Hydration effects on thermo-regulation and performance in the heat. Comp Biochem Physiol A MolIntegr Physiol. 2001;128:679–690. doi: 10.1016/s1095-6433(01)00274-4. [DOI] [PubMed] [Google Scholar]
- Young JH. Evolution of blood pressure regulation in humans. Curr Hypertens Rep. 2007;9:13–18. doi: 10.1007/s11906-007-0004-8. [DOI] [PubMed] [Google Scholar]
- Relethford JH. Geostatistics and spatial analysis in biological anthropology. Am J Phys Anthropol. 2008;136:1–10. doi: 10.1002/ajpa.20789. [DOI] [PubMed] [Google Scholar]
- Kline RB. Principles and Practice of Structural Equation Modeling (Second Edition) New York: Guilford Press; 2004. [Google Scholar]
- Fotheringham AS, Brunsdon C, Charlton M. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships. Chichester, UK: Wiley; 2002. [Google Scholar]
- Oliver MA, Webster R. Kriging: a method of interpolation for geographical information system. INT J Geographical Information Systems. 1990;4:313–332. [Google Scholar]
- Jenks GF, George F. The data model concept in statistical mapping. International Yearbook of Cartography. 1967;7:186–190. [Google Scholar]
- Hou J, Wen Z, Cheng Z. Structural Equation Model and Its Applications. Beijing, China: Education Science Publishing House; 2004. [Google Scholar]
- Schuurman N, Peters PA, Oliver LN. Are obesity and physical activity clustered? A spatial analysis linked to residential density. Obesity (Silver Spring) 2009;17:2202–2209. doi: 10.1038/oby.2009.119. [DOI] [PubMed] [Google Scholar]
- Kim SG, Cho SH, Lambert DM, Roberts RK. Measuring the value of air quality: application of the spatial hedonic model. Air Qual Atmos Health. 2010;3:41–51. doi: 10.1007/s11869-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV. Diabetes mellitus: a ‘thrifty' genotype rendered detrimental by ‘progress'. AM J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Katsuya T, Ishikawa K, Sugimoto K, Rakugi H, Ogihara T. Salt sensitivity of Japanese from the viewpoint of gene polymorphism. Hypertens Res. 2003;26:521–525. doi: 10.1291/hypres.26.521. [DOI] [PubMed] [Google Scholar]
- Young JH, Chang YP, Kim JD, et al. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Sakagami T, et al. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet. 2004;74:898–916. doi: 10.1086/420793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcroft A. Body Temperature Control in Anaesthesia, Surgery and Intensive Care. London, UK: Bailliere Tindall; 1980. [Google Scholar]
- Newman RW. Why man is such a sweaty and thirsty naked animal: a speculative review. Hum Biol. 1970;42:12–27. [PubMed] [Google Scholar]
- Ruff C. Climate and body shape in hominid evolution. J Hum Evol. 1991;21:81–105. [Google Scholar]
- Denton D, Weisinger R, Mundy NI, et al. The effect of increased salt intake on blood pressure of chimpanzees. Nat Med. 1995;1:1009–1016. doi: 10.1038/nm1095-1009. [DOI] [PubMed] [Google Scholar]
- Rotimi C, Puras A, Cooper R, et al. Polymorphisms of renin-angiotensin genes among Nigerians, Jamaicans, and African Americans. Hypertension. 1996;27:558–563. doi: 10.1161/01.hyp.27.3.558. [DOI] [PubMed] [Google Scholar]
- Kearney PM, Whelton M, Reynolds K, Whelton PK, He J. Worldwide prevalence of hypertension: a systematic review. J Hypertens. 2004;22:11–19. doi: 10.1097/00004872-200401000-00003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.