Abstract
The dominant and sometimes competing theories for the aetiology of complex human disease have been the common disease, common variant (CDCV) hypothesis, and the multiple rare variant (MRV) hypothesis. With the advent of genome wide association studies and of second-generation sequencing, we are fortunate in being able to test these ideas. The results to date suggest that these hypotheses are not mutually exclusive. Further, initial evidence suggests that both MRV and CDCV can be true at the same loci, and that other disease-related genetic mechanisms also exist at some of these loci. We propose calling these, pleomorphic risk loci, and discuss here how such loci not only offer understanding of the genetic basis of disease, but also provide mechanistic biological insight into disease processes.
The common disease, common variant (CDCV) hypothesis centres on the notion that the genetic risk for complex, common diseases is mediated by numerous common variants (1). Because such variants by definition have become common, a corollary of this hypothesis is that such risk variants are likely to exert little net negative selective pressure, either because they have quite small biological effect, there is balancing selection, or because the variants are associated with post-reproductive diseases. Genome wide association (GWA) studies have made it possible to explicitly test the CDCV hypothesis and have shown that there is substantive truth to this idea (http://www.genome.gov/26525384) (2). In contrast to the CDCV hypothesis, the multiple rare variant (MRV) hypothesis ascribes the genetic risk component of common, complex diseases to MRVs (3). Unlike common variants, low-frequency alleles are not fixed in the population, and thus, there has been little opportunity for selective pressure to limit the effect size of such variants.
CONSOLIDATION AND BEYOND: PLEOMORPHIC RISK LOCI
Critically, both the CDCV and MRV hypotheses are ideas that have been generalized to exclusively encapsulate the total genetic risk in disease. However, that only one of these ideas can be true is a fallacy, these concepts are not mutually exclusive and a more mature view brings these two ideas together as a general hypothesis for disease susceptibility. We believe, however, that it is not only sensible to consolidate these hypotheses generally but that within a single disease, multiple common and rare risk alleles are likely to co-exist at the same locus. Further, we hypothesize that genetic risk at individual loci will comprise a spectrum of both allele frequency and effect size, encompassing low-risk and high-risk variants; we nominate these as pleomorphic risk loci (PRL).
We have previously discussed the concept of graded haplotype risk in the context of common risk alleles (4,5): briefly this argues that GWA studies initially identify only the most significant disease-associated common variants at any one locus, but that it is likely there are several other common risk variants at the same locus, within or out with the same haplotype block. In this model, there is no single haplotype for disease risk, but rather a collection of haplotypes at or across the same locus that are associated with graded risk of disease (Fig. 1A). As we have discussed, there already exists biological support for this idea (4,6–9). The PRL hypothesis is a natural extension of this idea, incorporating other types of genetic variation of varying risk classes within the same loci. In this model, we suggest that at a single locus, there will likely be: (i) common non-coding variants that effect gene expression and/or splicing; (ii) rare coding and highly penetrant mutations; (iii) common coding and low-to-moderate risk alleles; (iv) rare non-coding variants that exert small effects on risk; and (v) rare whole gene copy number mutations that increase risk for disease substantially and in a dose-dependent manner.
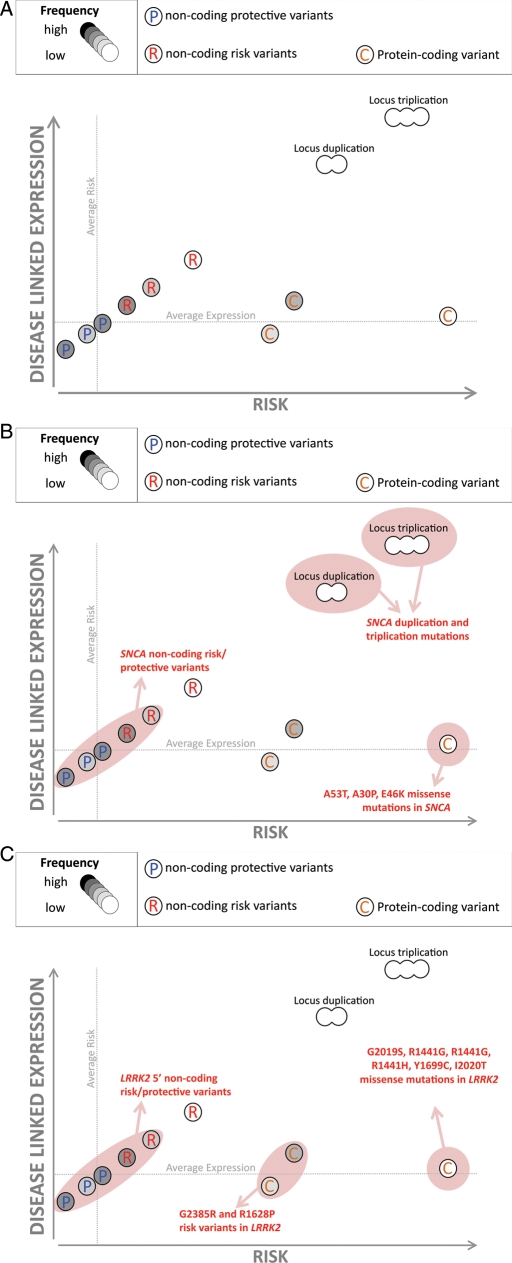
Figure 1.
Schematic of PRL. (A) This schematic illustrates that at the same locus several disease-related genetic mechanisms may co-exist, each influencing disease through different biological effects on a single gene. In this particular model, expression of a gene is positively correlated with risk. Notably, we predict that common alleles affecting expression are also likely to modulate risk conferred by rare mutations and common coding variants. (B) The LPR model as it applies to genetic variability at SNCA in PD. Notably common non-coding variants that alter SNCA expression are linked to increased risk for disease; whole gene duplication and triplication mutations of SNCA are a known cause of disease (and copy number is correlated with severity) and rare protein-coding mutations have been identified. (C) The LPR model as it applies to LRRK2 in PD. Rare protein-coding disease-causing mutations have been identified, common protein-coding risk variants have been found and, recently, common non-coding variability 5′ to LRRK2 has been implicated as a risk factor for PD.
In summarizing this model: at each locus identified as contributing towards the pathogenesis of disease, we predict that there will be many alleles that contribute to the risk profile for that disease. If these are coding changes, their effects will be dependent on the precise details of their biochemistry. If they affect expression or splicing, we would expect their effects on disease risk to be predictable by the size and nature of their effects on this expression and splicing. Disease risk encoded by gene duplications will also be increased by high expressing alleles: disease risk encoded by gene deletions will also be increased by low expressing alleles and nonsense mutations and are likely to be potentiated by low expressing trans alleles.
CONSEQUENCES AND PREDICTIONS OF THE PRL MODEL
We can begin to predict which types of disease-related genes are more likely to be PRL and what predictions can be made about PRL.
We suspect that these loci are more likely to be associated with later onset diseases. One major form of disease risk for late onset disease, we predict, will be heterozygous loss of function mutations at loci where homozygous loss is either lethal or associated with a related recessive disease, and evidence for such phenomena already exist: first, glucosecerebrosidase mutations that when homozygous cause Gaucher's disease and when heterozygous are risk loci for Parkinson's disease (PD) (10); second, loss-of-function mutations as the cause of icthyosis vulgaris when homozygous and as risk loci for atopic dermatitis and asthma when heterozygous (11,12).
Genes that have been previously linked to monogenic disease through a quantitative pathobiological mechanism are likely to be PRL. One would suspect that in such cases, where monogenic disease is driven by large relative effects on splicing or expression, risk will be affected by more subtle effects on splicing expression mediated by rare or common non-coding variation. An example of this is the microtubule associated protein tau (MAPT) locus in the related syndromes of frontotemporal dementia and progressive supranuclear palsy in which dominant mutations cause a dramatic effect on alternate splicing of a single exon cause the former (13), whereas the MAPT haplotype which has a similar but more subtle effect, predisposes to the latter (14). Clear examples of this come from the comparatively well understood study of variability in blood lipids where ∼20% of the loci identified as being important in normal variability had previously been identified as mendelian loci for lipid disorders (15).
Variants at PRL associated with the same disease will likely have a unifying biological mechanism/substrate. Linking the immediate biological consequences of these divergent types of risk variant will be a unique opportunity toward understanding the underlying disease process.
Within PRL, the risk associated with a rare mutation will be modulated by genetic variants in cis to the mutation. Two examples of this are the association of complement factor H with macular degeneration in which coding changes have a major effect on disease risk, but the haplotype also contributes to the risk (6–9) and apolipoprotein E and Alzheimer's disease (16) where the same phenomenon operates and the different risk haplotypes (17) are associated with different levels of expression (18). Furthermore, risk variability at the same locus but in trans to the mutation should exert a major effect on expressivity or penetrance of the disease particularly in cases of where loss of function is critical to pathogenicity. There are already a few examples where variability, both cis and trans to pathogenic changes contribute to the variability in disease phenotypes (19,20). As more high-risk variants are identified, we would expect that this type of effect is likely to be almost a general phenomenon where high expressing trans alleles can partially compensate for loss of function alleles or low expressing cis alleles can mitigate the phenotypes of gain of function alleles.
EXISTING PROOF FOR THE PRL HYPOTHESIS
The data surrounding common risk variability have matured considerably over the previous 4 years, and now there are hundreds of replicated risk loci (2). With even more history, linkage and positional cloning efforts have revealed mutations that underlie >1500 monogenic diseases (http://www.ncbi.nlm.nih.gov/Omim/mimstats.html). It is readily apparent that several loci contain both common risk variants and rare, highly penetrant mutations, associated with the same or related diseases. From our own work, common genetic variability at the SNCA, MAPT and LRRK2 loci confers mild risk for PD (21–23); these three genes have previously been shown to contain highly penetrant point mutations that cause familial forms of parkinsonism.(13,24–26). We use two of these loci to illustrate the concept of PRL.
First, common and rare variants at the SNCA locus: missense mutations in SNCA were the first-described genetic cause of PD (26): subsequently both whole locus triplication and duplication were shown as a cause of rare familial forms of PD, mediating disease through increased expression of the protein product, α-synuclein. Notably, in these cases, disease severity and age at onset were related to SNCA copy number, and implicitly α-synuclein levels (27–29) in a dose-dependent manner (30). Given our model, one would predict that common non-coding variability, which alters SNCA expression, would be a risk factor for PD. This indeed seems to be the case with the identification of multiple risk haplotypes at SNCA. Furthermore, these risk haplotypes underpin increased SNCA expression (21,22). Thus, in the case of SNCA, rare coding mutations, rare whole gene copy number variants and common expression-related variants, all occur and contribute in different ways to disease risk (Fig. 1B).
Secondly, genetic variability at LRRK2: missense mutations in LRRK2 were identified as a relatively common cause of PD in 2004 (24,25). Subsequent work has shown that common missense variants within the Asian population act as risk factors for PD, increasing risk for disease ∼2-fold (31). Further work using GWA testing suggests that in addition to these protein-coding risk variants, there are low-risk, non-coding variants immediately 5′ to LRRK2 (22). Thus, in the case of LRRK2, there exists three known categories of variant associated with PD; high penetrance missense mutations, moderate risk missense variants and low-risk non-coding risk variants (Fig. 1C). Parsimony would suggest that at this locus common, non-coding variability that presumably mediates risk effects by splicing/expression and apparently qualitative changes to the protein-coding sequence, ultimately exert their effects through similar pathobiological mechanisms.
The next stage of investigation is likely to centre on finding rare variants, with much lower risk effects than disease-causing mutations. Because of the technological challenges associated with identifying such variants, particularly those that are non-coding, there is a paucity of data surrounding this mechanism. However, initial work in this area has begun to provide examples of this phenomenon; perhaps most notable has been described in diabetes. Common variability at the IFIH1 locus was implicated in risk for type 1 diabetes (T1D) by GWA (32). Subsequent resequencing of this locus revealed that MRVs exist within this gene that confer a small protective effect against T1D, and that these variants are independent of the originally identified GWA signal (32,33). This resequencing work centred on exons and immediate regulatory sequences; however, one might expect that as second-generation sequencing affords more ability to perform highly multiplexed sequencing of large genomic regions, the field will begin to identify more non-coding variants associated with disease. The increasing resolution of the 1000 genomes project, and related efforts, coupled with the availability of ever more GWA data for common diseases will afford a complementary approach to this, through the ability to impute even rare variants as significantly increasing disease risk.
HOW TO TEST THE PRL HYPOTHESIS
We predict that as genetic data accumulate for common diseases, the PRL hypothesis will be explicitly proven. The initial testing of this idea will likely result from follow-on experiments to GWA studies, which aim to perform deep resequencing of genomic loci implicated by this method. While we recognize that in terms of our hypothesis, this type of experiment may provide support that is circular in its logic, these types of focused data will be invaluable as we begin to construct methods to understand and delineate benign, risk and protective rare variants. At some point, this type of effort will transition from targeted resequencing, to whole genome resequencing. Should the PRL hypothesis be generalizable, the pre hoc prioritization for analysis of loci previously implicated in the disease in question, either because of a link to monogenic forms of disease or because of identified common risk variants at the locus, may provide considerable statistical benefit.
CONCLUDING REMARKS
The CDCV and MRV hypotheses were proposed as models for disease pathogenesis largely before systematic data were available. Now that data have become available through the application of array and sequencing technologies, we can see that they both have some elements of truth to them, but that a greater and more interesting truth comes from their synthesis: that disease pathogenesis is initiated by a mixture of common and rare variants which interact in a manner which is, to some extent, predictable at each locus. We suggest that this synthesis, which we encapsulate as the pleomorphic risk locus hypothesis, can be used to explain much of the risk for common disease. Of course, we recognize that there will be interactions between disease loci and between, more generally, genes and environment and that stochastic effects may often have a role. Additionally, as large numbers of loci are identified, as has now been possible for human height (34), it will be possible to elucidate the biochemical and the developmental pathways which the genetic findings underpin. Eventually, these too will need to be incorporated into our models as we develop a complete description of disease risk.
Conflict of Interest statement. None declared.
FUNDING
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project number Z01 AG000957-07 (A.S.) and by the Wellcome Trust and MRC (J.H.).
REFERENCES
- 1.Lander E.S. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. doi:10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 2.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pritchard J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001;69:124–137. doi: 10.1086/321272. doi:10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy J., Singleton A. Genomewide association studies and human disease. N. Engl. J. Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. doi:10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singleton A., Myers A., Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum. Mol. Genet. 2004;13:R123–R126. doi: 10.1093/hmg/ddh093. Spec No. 1 doi:10.1093/hmg/ddh093. [DOI] [PubMed] [Google Scholar]
- 6.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., San Giovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. doi:10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R., et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. doi:10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 8.Edwards A.O., Ritter R., 3rd, Abel K.J., Manning A., Panhuysen C., Farrer L.A. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. doi:10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 9.Li M., Atmaca-Sonmez P., Othman M., Branham K.E., Khanna R., Wade M.S., Li Y., Liang L., Zareparsi S., Swaroop A., Abecasis G.R. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat. Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. doi:10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sidransky E., Nalls M.A., Aasly J.O., Aharon-Peretz J., Annesi G., Barbosa E.R., Bar-Shira A., Berg D., Bras J., Brice A., et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. doi:10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith F.J., Irvine A.D., Terron-Kwiatkowski A., Sandilands A., Campbell L.E., Zhao Y., Liao H., Evans A.T., Goudie D.R., Lewis-Jones S., et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006;38:337–342. doi: 10.1038/ng1743. doi:10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 12.Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., Goudie D.R., Sandilands A., Campbell L.E., Smith F.J., et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38:441–446. doi: 10.1038/ng1767. doi:10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 13.Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. doi:10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 14.Myers A.J., Pittman A.M., Zhao A.S., Rohrer K., Kaleem M., Marlowe L., Lees A., Leung D., McKeith I.G., Perry R.H., et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. doi:10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. doi:10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 17.Chartier-Harlin M.C., Parfitt M., Legrain S., Perez-Tur J., Brousseau T., Evans A., Berr C., Vidal O., Roques P., Gourlet V., et al. Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum. Mol. Genet. 1994;3:569–574. doi: 10.1093/hmg/3.4.569. doi:10.1093/hmg/3.4.569. [DOI] [PubMed] [Google Scholar]
- 18.Lambert J.C., Perez-Tur J., Dupire M.J., Galasko D., Mann D., Amouyel P., Hardy J., Delacourte A., Chartier-Harlin M.C. Distortion of allelic expression of apolipoprotein E in Alzheimer's disease. Hum. Mol. Genet. 1997;6:2151–2154. doi: 10.1093/hmg/6.12.2151. doi:10.1093/hmg/6.12.2151. [DOI] [PubMed] [Google Scholar]
- 19.Risch N.J., Bressman S.B., Senthil G., Ozelius L.J. Intragenic cis and trans modification of genetic susceptibility in DYT1 torsion dystonia. Am. J. Hum. Genet. 2007;80:1188–1193. doi: 10.1086/518427. doi:10.1086/518427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldfarb L.G., Petersen R.B., Tabaton M., Brown P., LeBlanc A.C., Montagna P., Cortelli P., Julien J., Vital C., Pendelbury W.W., Gajdusek D.C. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: disease phenotype determined by a DNA polymorphism. Science. 1992;258:806–808. doi: 10.1126/science.1439789. doi:10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 21.Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. doi:10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalls M.A., Plagnol V., Hernandez D.G., Sharma M., Sheerin U.M., Saad M., Simón-Sánchez J., Schulte C., Lesage S., Sveinbjörnsdóttir S., et al. International Parkinson Disease Genomics. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2000;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer C.C., Plagnol V., Strange A., Gardner M., Paisan-Ruiz C., Band G., Barker R.A., Bellenguez C., Bhatia K., Blackburn H., et al. UK Parkinson's Disease Consortium; Wellcome Trust Case Control Consortium 2. Wood NW (2011) Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum. Mol. Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. doi:10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. doi:10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. doi:10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 27.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. doi:10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 28.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. doi:10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 29.Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. doi:10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 30.Singleton A., Gwinn-Hardy K. Parkinson's disease and dementia with Lewy bodies: a difference in dose? Lancet. 2004;364:1105–1107. doi: 10.1016/S0140-6736(04)17117-1. doi:10.1016/S0140-6736(04)17117-1. [DOI] [PubMed] [Google Scholar]
- 31.Zabetian C.P., Yamamoto M., Lopez A.N., Ujike H., Mata I.F., Izumi Y., Kaji R., Maruyama H., Morino H., Oda M., et al. LRRK2 mutations and risk variants in Japanese patients with Parkinson's disease. Mov. Disord. 2009;24:1034–1041. doi: 10.1002/mds.22514. doi:10.1002/mds.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth D.J., Cooper J.D., Bailey R., Field S., Burren O., Smink L.J., Guja C., Ionescu-Tirgoviste C., Widmer B., Dunger D.B., et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. doi:10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 33.Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. doi:10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. doi:10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]