Abstract
The successes of genome-wide association (GWA) studies have mainly come from studies performed in populations of European descent. Since complex traits are characterized by marked genetic heterogeneity, the findings so far may provide an incomplete picture of the genetic architecture of complex traits. However, the recent GWA studies performed on East Asian populations now allow us to globally assess the heterogeneity of association signals between populations of European ancestry and East Asians, and the possible obstacles for multi-ethnic GWA studies. We focused on four different traits that represent a broad range of complex phenotypes, which have been studied in both Europeans and East Asians: type 2 diabetes, systemic lupus erythematosus, ulcerative colitis and height. For each trait, we observed that most of the risk loci identified in East Asians were shared with Europeans. However, we also observed that a significant part of the association signals at these shared loci seems to be independent between populations. This suggests that disease aetiology is common between populations, but that risk variants are often population specific. These variants could be truly population specific and result from natural selection, genetic drift and recent mutations, or they could be spurious, caused by the limitations of the method of analysis employed in the GWA studies. We therefore propose a three-stage framework for multi-ethnic GWA analyses, starting with the commonly used single-nucleotide polymorphism-based analysis, and followed by a gene-based approach and a pathway-based analysis, which will take into account the heterogeneity of association between populations at different levels.
INTRODUCTION
Complex traits refer to the phenotypes that are classically believed to result from the interplay of multiple genetic variants and environmental factors. Genome-wide association (GWA) studies, in which phenotypes are compared for differences in genetic variation, have revolutionized the search for genetic risk variants underlying these complex traits. During the past few years, GWA studies have identified robust associations between >3000 single-nucleotide polymorphisms (SNPs) and >700 complex human traits (1). The majority of the GWA studies have been centred on populations of European descent (henceforth referred to as ‘Europeans’). Because complex traits are caused by an interplay of genetic variation and environmental factors, their genetic basis probably reflects the evolution of the human genome and human populations. Genomic surveys have already revealed a substantial divergence of genetic variation across populations in terms of allele frequency, linkage disequilibrium (LD) and haplotype structure (2–4). These inter-population differences in genetic architecture reflect multiple factors such as genetic drift, recent mutations, environmental factors and other evolutionary forces (5). Consequently, complex traits are anticipated to be genetically heterogeneous (6,7). This inter-population heterogeneity of complex traits raises the question as to how far GWA findings can be translated across different ethnic groups. For targeted disease therapy and genetic risk prediction, knowing how much genetic risk loci can be translated between different ethnicities is vital; heterogeneity of genetic risk between populations could considerably limit the applicability of such therapies and risk models across populations. For cross-ethnicity mapping, on the other hand, inter-population heterogeneity can be advantageous; cross-ethnicity mapping combines the association signals across multiple different ethnicities, increasing the power for finding new risk loci and identifying causal variants. Still, it remains unclear to what extent complex traits are heterogeneous between populations; a study by Water et al., for example, assessed the association of 19 loci with type 2 diabetes (T2D) in five ethnic populations and observed consistent association for most of the association signals (8). However, a recent study by Sim et al. (9) concluded that there is considerable locus and allelic heterogeneity in T2D association between populations. Sim et al. performed genome-wide scans for T2D risk loci on three Asian populations and compared the association signals to those in Europeans. This example suggests that the assessment of transferability of risk variants across populations needs to be based on unbiased GWA findings from each population. Such unbiased assessment is currently impossible for several reasons: GWA studies employ tag-SNPs that are more likely to be proxies of the causal variants than true causal variants; hence, any perceived heterogeneity could be due to heterogeneity of the tag-SNP rather than of the true causal variant; GWA platforms are designed for optimal use in European populations and are hence less sensitive in non-European populations; for most complex traits, results from European studies have already been published, colouring the interpretation of results in non-European populations; and finally, there are few non-European GWA studies and they are generally underpowered. Nonetheless, the recent progress of GWA studies in East Asians allows us to make a preliminary empirical comparison of the association signals between Europeans and East Asians as a proxy of the genetic heterogeneity of complex traits between populations, and provides an opportunity to explore the implications of the heterogeneity of association signals in multi-ethnic GWA studies.
Recent advances in GWA studies in East Asians
Since 2009, the focus of genetic studies in East Asians has clearly switched from the replication of small sets of risk variants reported in Europeans to genome-wide analyses to discover new risk loci. The total number of GWA studies in East Asians, including Chinese, Japanese and Korean populations, has increased greatly in the last 30 months from 5 at the beginning of 2009 to 84 by May 2011 (1). Although many of the hypothesis-free GWA studies in East Asians resemble those in the early stages of the GWA era in Europeans, with relatively small sample sizes, these studies have already successfully reported risk loci not previously detected in Europeans, thereby yielding new insights into the aetiology of complex traits (10). In addition, the GWA studies in East Asians may provide unique information, especially for those complex diseases that have a much higher prevalence in East Asians than in Europeans. This is, for example, the case with hepatocellular carcinoma, the incidence of which is <2 in 100 000 males in the Western world but 40–60 in 100 000 males in Africa and East Asia (11,12). Three GWA studies on hepatocellular carcinoma in East Asians report a total of six risk loci, whereas no such study has so far been performed in Europeans (13–15). GWA studies that include non-European populations thus hold the promise of being able to provide a broader spectrum of complex trait loci and a higher resolution for exploring the genetic architecture of such traits.
Heterogeneity of association signals between Europeans and East Asians
To gain maximal power from multi-ethnic GWA studies, it is important to know the extent of heterogeneity of association signals between populations and the implications for genetic studies. Here, we confined ourselves to four different complex traits or diseases that have been studied in both Europeans and East Asians, which represent a broad range of complex phenotypes: (i) a metabolic disease: T2D, with a similar prevalence in both populations (Europeans 5.7–7.8% and East Asians 5.5–11.7%); (ii) two inflammatory diseases: systemic lupus erythematosus (SLE), with a higher prevalence in East Asians (161 in 100 000) than in Europeans (91 in 100 000), and ulcerative colitis (UC), with a higher prevalence in Europeans (100 in 100 000) than in East Asians (6–16.1 in 100 000) and (iii) an anthropomorphic trait, height, with an average difference of 7 cM between East Asians and Europeans. For each trait we derived all the reported SNPs associated to the traits from the Catalogue of Published Genome-Wide Association Studies (GWAS Catalogue, available at: www.genome.gov/gwastudies; accessed on 15 May 2011) (1). The sample sizes of GWA studies in East Asians are generally much smaller than those in Europeans. This relative underpowering of GWA studies in East Asians has resulted in a lower number of susceptibility loci detected in East Asians. We therefore based further analysis exclusively on associations reported in East Asians at a genome-wide significant level (P < 5 × 10−8). We first clustered all reported SNPs into LD blocks (r2> 0.8) and assigned an index SNP, the most associated SNP, to each LD block. Then we took a window size of 1 Mb around the index SNPs (0.5 Mb on each side) and clustered all reported SNPs into genomic loci. In this manner, we selected 43 risk loci (47 index SNPs): 13 for T2D, 19 for SLE, 3 for UC and 8 for height (Table 1). For these risk loci, we assessed whether they were associated in Europeans, where the power of GWA studies should be much greater. We extracted all SNPs reported in European studies from the GWAS Catalogue and then manually checked the supplementary data of each of the papers to include, where available, all the SNPs reported at 1 × 10−5 in the original papers. We considered the association loci to be either: (i) specific to East Asians, when the power of the study in Europeans was >80% but no association was reported in Europeans at P < 1 × 10−5 at this locus (Supplementary Material, Notes); or (ii) shared with Europeans, when either the same SNP was associated at P < 1 × 10−5 or different SNPs within the same region were associated at P < 5 × 10−8 in Europeans. For the shared loci, we further assessed whether the association was: (i) to the same SNPs, (ii) to highly linked SNPs or (iii) to independent SNPs (Table 1 and Fig. 1).
Table 1.
The association of East Asian-associated loci in individuals of European ancestry
| T2D | SLE | UC | Height | |
|---|---|---|---|---|
| Number of unique risk loci detected in East Asians (P < 5 × 10−8) | 13 (15 SNPs) | 19 (19 SNPs) | 3 (3 SNPs) | 8 (10 SNPs) |
| Number of loci specific to East Asiansa | 5 | 1 (8) | 1 | 0 |
| Number of loci shared in Europeansb | 8 | 6 | 2 | 8 |
| Same SNPs | 3 | 1 | 2 | 1 |
| Highly linked SNPs | 3 | 1 | 0 | 2 |
| Independent SNPs | 2 | 3 | 0 | 1 |
| Weakly linked SNPs | 0 | 1 | 0 | 4 |
T2D, type 2 diabetes; SLE, systemic lupus erythematosus; UC, ulcerative colitis.
aThe number of loci specific to East Asians refers to the loci that are significantly associated in East Asians at a genome-wide significance level P-value <5 × 10−8, but for which no association was detected in Europeans at a significance level P-value < 1 × 10−5 with at least 80% power. The number in brackets refers to the number of spurious East Asian-specific loci where European studies had <80% power to detect the association signals.
bThe LD, in terms of r2, between the SNPs was assessed based on HapMap reference panels. For individuals of European ancestry, we used HapMap CEU reference panel (Utah residents with European ancestry). For the East Asian population, we used HapMap CHB + JPT reference panel (Han Chinese from Beijing and Japanese from Tokyo). The association with highly linked SNPs refers to the loci where r2 is >0.8 in any population. The association with independent SNPs refers to the loci where r2 is <0.2 in both populations. The weakly linked SNPs are the remaining loci with r2 between 0.2 and 0.8.
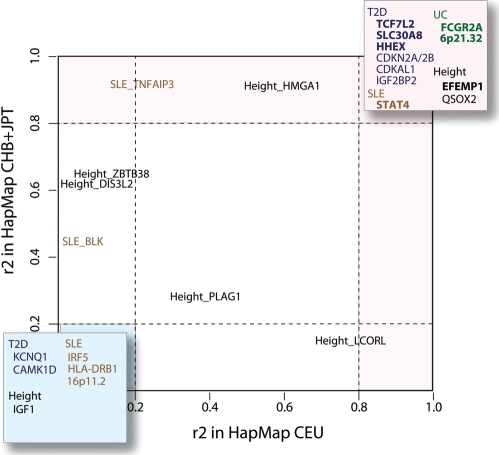
Figure 1.
The linkage disequilibrium (LD) between European-associated SNPs and East Asian-associated SNPs at the shared susceptibility loci. The susceptibility loci shared between the Europeans and East Asians are named after the gene closest to the index SNP in each locus. The colours of the loci refer to the different traits: blue for type 2 diabetes (T2D), brown for SLE, green for UC and black for height. The loci in bold indicate the loci with the same index SNPs reported in both populations. The LD is the pairwise r2 between the European-associated SNP and the East Asian-associated SNP at each locus. The x-axis represents the r2 in the HapMap CEU reference panel (Utah residents with European ancestry) and the y-axis represents the r2 in HapMap CHB + JPT reference panel (Han Chinese from Beijing and Japanese from Tokyo). The light pink region indicates the loci with linked signals (r2> 0.8) in any reference panel. The light blue region indicates the loci with independent signals (r2<0.2 in both reference panels or distance >500 kb).
Population-specific loci
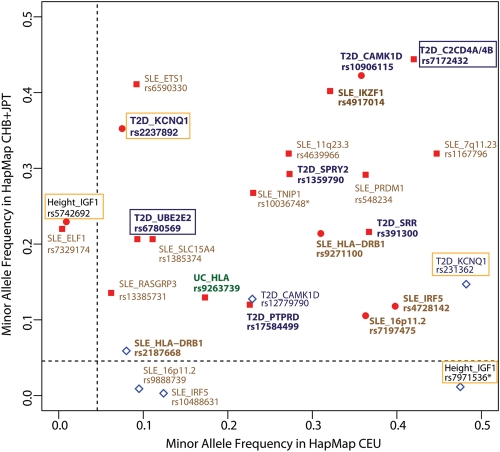
Considering the risk loci detected in East Asians and the power calculations for these loci in the European studies (Supplementary Material, Notes), we observed that there were only a few loci which show population-specific association in East Asians (Table 1). For example, the population-specific T2D risk loci, UBE2E2 and C2CD4A-C2CD4B, were reported by two relatively small East Asian GWA studies for T2D, while these loci did not reach genome-wide significance in European studies performed on cohorts 10 times larger (10,16,17). A possible explanation for the apparent population specificity of association signals can be a very low minor allele frequency (MAF) of a risk SNP, thereby reducing the power for detecting the signal (Fig. 2). For example, the most associated SNP in the UBE2E2 locus, rs6780569, has a relatively low allele frequency in HapMap CEU panel (MAF = 0.093) compared with CHB + JPT panel (MAF = 0.222) (Fig. 2). However, the association in East Asians was clearly genome-wide significant (P = 1.04 × 10−9), whereas in Europeans it was completely absent (P = 0.976) (10), although the European study had sufficient power to detect the association signal (Supplementary Material, Table S1). This renders a false-negative signal in Europeans unlikely. For the C2CD4A-C2CD4B locus, the most associated SNP is equally common in both populations (Fig. 2), so the power for detecting the association signal is high in the European population (Supplementary Material, Table S1). The association to SNP rs7172432 could be replicated in Europeans at P = 6.36 × 10−5 (10), which does not pass our defined threshold (P < 1 × 10−5). This suggests that we need to be cautious in defining risk loci as shared or non-shared between populations. Seemingly population-specific risk loci may well be spurious due to the threshold for significance and the power of the study. Thus, population-specific loci need further replication with larger sample sizes. The presence of population-specific loci, whether spurious or true, is however something that should be taken into account when performing multi-ethnic GWA studies.
Figure 2.
Comparison of the minor allele frequencies of the population-specific SNPs. Each dot represents the population-specifically associated SNPs from each East Asian specific locus or shared risk locus: the red squares indicate East Asian-associated SNPs from the East Asian-specific loci; the red circles indicate the East Asian-associated SNPs from the shared loci; and the diamonds show the European-associated SNPs from the shared loci. The SNP IDs, their associated traits and the gene closest to the signal are indicated. The colours refer to the different traits: blue for type 2 diabetes (T2D), brown for SLE, green for UC and black for height. The x-axis represents the MAFs of the SNPs in the HapMap CEU reference panel (Utah residents with European ancestry); the y-axis represents the MAFs of the SNPs in the HapMap CHB + JPT reference panel (Han Chinese from Beijing and Japanese from Tokyo). The dashed grey lines indicate the MAF = 0.05, which is the cut-off between common SNPs and rare SNPs. The SNPs with different minor allele are highlighted by asterisk. The SNPs highlighted by open boxes are example cases for a population-specific locus (blue box) and for independent SNPs at shared loci (orange box). The SNPs in bold refer to the population-specific SNPs for which studies in Europeans had >80% power to detect the association signal. Other SNPs refer to spurious population-specific SNPs for which studies in Europeans had <80% power to detect the association signal.
Shared risk loci between East Asians and Europeans
Out of 43 loci associated in East Asians, we observed that 32% (6 of 19 SLE loci) to 100% (8 of 8 height loci) of the loci per trait were shared with Europeans (Table 1). For these shared loci, we further assessed whether the same or different SNPs were associated in each population and we compared the LD between the European- and East Asian-associated SNPs.
Association signal to the same SNP
In our analysis, we observed several incidences in which an association signal to the same SNP was found in East Asians and Europeans (Table 1 and Fig. 1). One example of these is the T2D risk variant rs7903146 at the TCF7L2 locus (10q25.2). This variant has not only been reported in Europeans and East Asians, but also in African Americans, Latinos and Hawaiians (8). The number of risk SNPs shared between populations is, however, not representative of the actual overlap in genetic background between the populations: report of an association to the same SNP in a region depends largely on the platform used in a study and whether imputation has been performed. Furthermore, slight differences in allele frequency between populations can influence the P-value at a certain SNP, leading authors to report their association within a single shared locus to a different SNP.
Highly linked SNPs at the same locus
To identify shared risk signals while trying to avoid the platform-, power- and publication biases, we considered a risk locus shared if the associated SNPs were in high LD (r2> 0.8) in any population. If multiple index SNPs at the same locus were reported, we chose the closest pair of the European- and East Asian-associated SNPs in terms of r2 and genomic distance. At many of the risk loci, the association signals in Europeans are in a near-perfect LD with the original East Asian association signal, suggesting that these signals originate from the same risk variant. For example, multiple variants in the intron of the CDKAL1 have consistently been reported as risk factors for T2D in individuals of European (rs7754840) and East Asian descent (rs4712523). Even though different SNPs show a genome-wide signal in each population, these SNPs are in LD with each other (r2> 0.98 in both populations) and seem to point to a single T2D causal variant.
Independent SNPs at the same locus
Although the risk loci can be shared between populations, it is still possible that risk variants within these loci are population specific. If the r2 between the association signals was <0.2 in both populations, or if the distance between the signals was >500 kb, we considered the signal to be independent (Table 1 and Fig. 1). We observed several such loci with multiple risk variants, including the T2D risk locus KCNQ1. The association of KCNQ1 was primarily reported in East Asians, with three independent SNPs (rs2237892, rs2237895 and rs2237897) at the intron of the KCNQ1 gene. The strongest association was detected at rs223792 (P = 1.7 × 10−42) (18). Later, in Europeans, the association was reported at another intron SNP rs231362 (17). This SNP is, however, independent of any of the SNPs detected in East Asians (all r2< 0.02 in any populations). This example suggests that multiple and population-specific risk variants can exist at one gene. These independent risk variants within the shared loci could point to population-specific independent causal variants within a region that is functionally important for disease pathogenesis. A recent haplotype analysis on the KCNQ1 region in Indian and European populations suggested that T2D risk associated with KCNQ1 SNPs may be derived from the ‘G’ allele of rs231362 and the ‘C’ allele of rs2237895 and is likely to be mediated by β cell function (19). However, seemingly independent risk variants could also arise as a result of allele frequency differences. The top East Asian-associated KCNQ1 SNP rs2237892 is common (MAF = 0.38) in the HapMap CHB + JPT reference panel (Han Chinese from Beijing and Japanese from Tokyo) but has a relatively low frequency (MAF = 0.08) in the HapMap CEU reference panel (Utah residents with European ancestry). In contrast, the European-associated SNP rs231362 is less common in East Asians than Europeans (MAF = 0.167 in HapMap CHB + JPT and MAF = 0.49 in HapMap CEU) (Fig. 2). The same was observed in the height-associated locus IGF1: the European-associated SNP rs7971536 is frequent in HapMap CEU (MAF = 0.475) but rare in HapMap CHB + JPT (MAF = 0.011). In contrast, the East Asian-associated SNP rs5742692 (425.8 kb distance from rs7971536) has a frequency of 0.222 in the HapMap CHB + JPT, but a frequency of 0.017 in the CEU panel (Fig. 2). This observed allelic heterogeneity likely results from natural selection or genetic drift (5,20) and could result in seemingly population-specific associations (Supplementary Material, Table S2). Most other independent SNPs in shared risk loci are common in both populations but the association is apparently population specific.
CONCLUDING REMARKS
We have explained how the genetic basis of complex traits can be heterogeneous between populations based on GWA findings. At present we can only compare a handful of associated loci for a few traits reported in Europeans and East Asians, which limits how representative our analysis can be considered. First, the studies in East Asians are underpowered and hence the comparison with European studies may be affected by power issues. Secondly, Europeans and East Asians are in fact genetically similar, whereas more major genetic differences are expected to be found between African and non-African populations (21). Unfortunately, the number of GWA studies performed in individuals of African descent is still too limited to enable comparative studies.
In our analysis using East Asian reported loci as a starting point, we observed considerable heterogeneity of association signals between East Asians and Europeans. Most risk loci seem to be shared, whereas the risk signals often appeared to be population specific.
Our study demonstrates the challenge in assessing the transferability of risk variants between different ethnic populations solely based on the GWA findings. There is no doubt about the existence of the population-specific risk variants, as illustrated by examples like the population-specific association of MYH9 with end-stage renal disease in African Americans (22,23) and the population-specific association of NOD2 with Crohn's disease in Europeans (24,25). However, the assessment of genetic heterogeneity solely based on GWA findings is affected by the limitations of GWA studies. First, the genetic variants identified through GWA studies have small or moderate risk effects and explain only a small part of the heritability of most complex traits. Thus, when comparing the results of GWA studies between different populations, we compare only a small fraction of the total genetic risk present in each population, leading to many seemingly population-specific signals. This would lead us to overestimate the genetic heterogeneity across populations. Secondly, GWA studies are inherently ill suited for detecting population-specific risk variants because the risk variants targeted by such studies are often common variants that are believed to be of ancient origin and shared among different populations. Risk can however also be conveyed by rare variants, which usually have a recent origin. Rare risk variants are more likely to be population-specific and could possibly carry a greater risk effect (Fig. 3). The fact that the current GWA study design does not identify rare risk variants explains part of the missing heritability of complex traits and probably leads us to underestimate the genetic heterogeneity across populations. The recent advances in high-throughput sequencing technology have greatly accelerated the discovery of rare variants and have led to the development of custom-made arrays that specifically include rare variants (26,27). This advance in the discovery of rare variants can greatly aid in detecting the risk contributed by rare variants. Although these rare variants are individually difficult to detect, they can collectively make a substantial contribution to the genetic risk underlying complex traits. Thirdly, the risk variants reported by GWA studies are just proxies of the actual causal variants; finding a population-specific risk variant does not necessarily mean that the causal variant is also population specific. The inter-population difference in LD between tag-SNPs and causal variants, and inter-population differences in allele frequency, can lead to association signals at different SNPs in different populations. This limitation can result in an overestimation of genetic heterogeneity between populations. As the causal variants remain undetermined, the inter-ethnic heterogeneity of the tag-SNPs from GWA studies can interfere significantly with multi-ethnic genome-wide meta-analyses.
Figure 3.
The comparison of MAFs between the population-shared SNPs and the population-specific SNPs.
Although comparing GWA signals from different populations has limited value for assessing the exact extent of the heterogeneity of complex traits across populations, it clearly shows the immense implications of this heterogeneity for GWA studies on multi-ethnic samples. Such studies have several potential applications. In population genetics, an important application of multi-ethnic studies is the mapping of ancestry in modern populations consisting of an admixture of populations with geographically divergent ancestry. Such mapping can, for example, be performed in African Americans and Latinos, revealing a mosaic genome of distinct ancestry. Admixture mapping can provide unforeseen power and resolution in genetic analysis (28–31). In complex trait genetics, the two most well-known applications of multi-ethnic GWA studies are meta-analyses, combining the association signals in different populations to increase the power for detecting new risk loci, and fine-mapping, using the divergent genomic structure between different populations for finding the causal genetic variant in a shared risk locus. If the goal of a multi-ethnic GWA study is a meta-analysis to increase the power to detect new risk loci, it is important that the populations are genetically close enough to assume that the causal variants and the tag-SNPs are shared between the different populations being studied. If the focus of a multi-ethnic GWA study is to fine-map causal variants within shared risk loci, the populations need to be genetically close enough to share such risk loci, but genetically distant enough to have very heterogeneous LD structures. These two approaches both assume that risk variants are shared between populations, whereas our analysis shows that risk variants could be considerably more heterogeneous between populations.
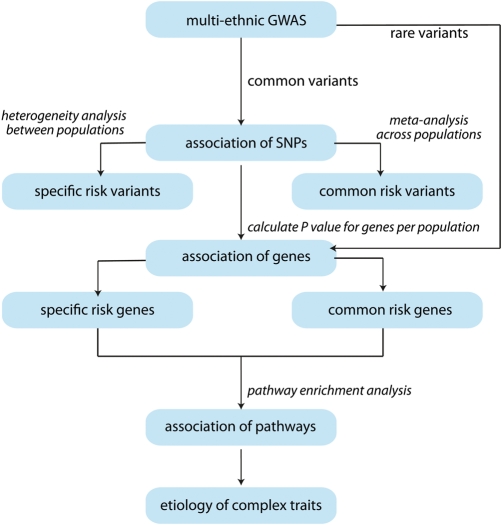
To optimize the power of multi-ethnic GWA studies, we propose an analysis framework combining SNP-, gene- and pathway-based analyses, which will deal with the heterogeneity between populations by assuming that most population-specific risk variants affect risk genes that converge to the same disease pathways (Box 1 and Fig. 4). We expect this framework to contribute greatly to the effectiveness of multi-ethnic GWA studies. The knowledge gained from such studies will eventually aid advances in clinical intervention and disease prevention worldwide.
Box 1. Three-stage analysis framework for multi-ethnic GWA studies.
Through the evolution of the human genome, complex traits are characterized by marked heterogeneity across populations, as can be seen in the comparison of association signals across populations. The overall genetic basis and pathological mechanisms underlying complex traits may be identical between different ethnic populations, but the variants representing this risk can be population specific.
To cope with the genetic heterogeneity of complex traits and to gain maximal power in multi-ethnic GWA studies, we propose to incorporate gene- and pathway-based association analyses into the analysis framework for multi-ethnic GWA studies (Fig. 4). Unlike the traditional SNP-based approach, the gene- and pathway-based approaches can take into account both the consistency and inconsistency of association between populations at different levels.
SNP-based approach
The meta-analysis on a single SNP level is currently the standard approach for combining association signals from multiple GWA studies. The procedure includes genotype imputation based on the appropriate reference panel, testing the association per imputed and directly genotyped SNP in each GWA study, performing a meta-analysis to combine the association signals from different studies, and testing for between-study heterogeneity and correcting for this (32). Applying this meta-analysis method to multi-ethnic association studies has the disadvantage that it is targeted at the shared and more common risk variants among populations, whereas signals of population-specific risk variants will be diluted. This is due to the fact that imputation of the GWA data for the different ethnic samples needs to be performed with a reference panel from the same ethnic origin. Most SNPs are shared, but some will be population specific and relatively rare. The meta-analysis on a single SNP level can naturally only test the overlapping set of common SNPs.
We therefore propose that the SNP-based analysis should be followed by a gene-based analysis.
Gene-based approach
With the gene-based approach, the association between a trait and all markers in the intragenic and regulatory regions of a gene are considered (33). The definition of gene regulatory regions is still arbitrary, but we propose to define this as 100 kb upstream and 40 kb downstream of a gene, as suggested by expression quantitative trait locus analysis (34). There are several advantages of the gene-based approach over the SNP-based approach. Genes are the functional units of the human genome and causal variants should somehow influence gene function, either by affecting the expression or by affecting the resulting protein. The identification of the causal genes can therefore provide a direct entry to functional information. Secondly, risk variants and their haplotype structure can vary across populations, so that meta-analysis and replication on a single variant level can thus be not only underpowered, but also misleading. Gene function and pathomechanisms are, however, highly consistent across human populations. Thirdly, the gene-based approach can be more powerful than the SNP-based approach because genes (or their functional units) are less numerous than SNPs, which diminishes the need for multiple testing. Fourthly, the gene-based approach can cope with population-specific risk variants, allele frequency differences between populations and haplotype structure differences. It can also successfully tackle the problem of rare variants present within a single population. Although these rare variants are individually difficult to detect, they can collectively contribute substantially to the genetic risk underlying complex traits (35). The gene-based analysis for the detection of rare variants should be different from that for detecting common variants. To detect gene association at the level of common variants, the gene-based P-value should be computed based on the P-value of the individual SNPs within the gene (36). To detect gene association at the level of rare variants, the gene-based P-value should be computed by the comparing the collective frequency of rare variants within a gene between cases and controls.
Some possible disadvantages of the gene-based approach are that many associations in GWA studies are to areas without any genes (gene deserts) or to areas with a large number of genes. Associations to gene deserts are notoriously hard to interpret and will, to some extent, be detected in the traditional SNP-based part of the proposed analysis framework. Associations to gene-rich areas will become easier to interpret with a gene-based approach, as for each gene in the area the association load, and hence the likelihood of each gene being the causal gene, can be calculated.
Pathway-based approach
It has been widely observed that genes associated with complex diseases can converge to the same pathway (37,38). Hence it follows that the risk genes for a trait, whether they are shared between populations or are specific to a certain population, are also expected to converge to common pathways shared between all populations. This convergence to global disease pathways means that a pathway-based approach, which simultaneously considers multiple risk genes from different populations, can aid the interpretation of the associated loci (39). Furthermore, the pathway-based approach has more power to detect risk variants with a small effect that do not reach the stringent genome-wide significance level.
We suggest using a gene-set enrichment analysis which tests the association of modules of functional related genes within pathways and thus increases the power to detect genes or variants with small effect size (40).
Obviously, the results of each step in this analysis framework will have to be validated in replication studies to prevent the publication of false-positive results. In addition, the results from each step within the analysis frame-work should be considered and replicated separately to prevent the magnification of errors in the initial steps. Since the results of this analysis framework are likely to give more insight into the mechanisms underlying the traits studied, replication should not be limited to the genetic level but should be expanded towards functional studies.
Figure 4.
The three-stage framework of a multi-ethnic GWA study.
SUPPLEMENTARY MATERIAL
FUNDING
The authors are supported by the Netherlands Organization for Scientific Research (NWO-VENI grant 863.09.007 to J.F., NWO-AGIKO grant 92.003.533 to E.F. and NWO-VICI grant 918.66.620 to C.W.). Funding to pay the Open Access publication charges for this article was provided by Netherlands Organization for Science Research (NWO-VENI grant 863.09.007 to J.F.).
ACKNOWLEDGEMENTS
We thank Jackie Senior for editing the text.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeyemo A., Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13:72–79. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad D.F., Jakobsson M., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 4.Evans D.M., Cardon L.R. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am. J. Hum. Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg N.A., Huang L., Jewett E.M., Szpiech Z.A., Jankovic I., Boehnke M. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClellan J., King M.C. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 8.Waters K.M., Stram D.O., Hassanein M.T., Le Marchand L., Wilkens L.R., Maskarinec G., Monroe K.R., Kolonel L.N., Altshuler D., Henderson B.E., et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sim X., Ong R.T., Suo C., Tay W.T., Liu J., Ng D.P., Boehnke M., Chia K.S., Wong T.Y., Seielstad M., et al. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from Southeast Asia. PLoS Genet. 2011;7:e1001363. doi: 10.1371/journal.pgen.1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamauchi T., Hara K., Maeda S., Yasuda K., Takahashi A., Horikoshi M., Nakamura M., Fujita H., Grarup N., Cauchi S., et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat. Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 11.Nordenstedt H., White D.L., El-Serag H.B. The changing pattern of epidemiology in hepatocellular carcinoma. Dig. Liver Dis. 2010;42:S206–S214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herszenyi L., Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur. Rev. Med. Pharmacol. Sci. 2010;14:249–258. [PubMed] [Google Scholar]
- 13.Clifford R.J., Zhang J., Meerzaman D.M., Lyu M.S., Hu Y., Cultraro C.M., Finney R.P., Kelley J.M., Efroni S., Greenblum S.I., et al. Genetic variations at loci involved in the immune response are risk factors for hepatocellular carcinoma. Hepatology. 2010;52:2034–2043. doi: 10.1002/hep.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Zhai Y., Hu Z., Wu C., Qian J., Jia W., Ma F., Huang W., Yu L., Yue W., et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat. Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 15.Kumar V., Kato N., Urabe Y., Takahashi A., Muroyama R., Hosono N., Otsuka M., Tateishi R., Omata M., Nakagawa H., et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat. Genet. 2011;43:455–458. doi: 10.1038/ng.809. [DOI] [PubMed] [Google Scholar]
- 16.Shu X.O., Long J., Cai Q., Qi L., Xiang Y.B., Cho Y.S., Tai E.S., Li X., Lin X., Chow W.H., et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda K., Miyake K., Horikawa Y., Hara K., Osawa H., Furuta H., Hirota Y., Mori H., Jonsson A., Sato Y., et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 19.Been L.F., Ralhan S., Wander G.S., Mehra N.K., Singh J., Mulvihill J.J., Aston C.E., Sanghera D.K. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Med. Genet. 2011;12:18. doi: 10.1186/1471-2350-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman S.R., Shylakhter I., Karlsson E.K., Byrne E.H., Morales S., Frieden G., Hostetter E., Angelino E., Garber M., Zuk O., et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oleksyk T.K., Nelson G.W., An P., Kopp J.B., Winkler C.A. Worldwide distribution of the MYH9 kidney disease susceptibility alleles and haplotypes: evidence of historical selection in Africa. PLoS One. 2010;5:e11474. doi: 10.1371/journal.pone.0011474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao W.H., Klag M.J., Meoni L.A., Reich D., Berthier-Schaad Y., Li M., Coresh J., Patterson N., Tandon A., Powe N.R., et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue N., Tamura K., Kinouchi Y., Fukuda Y., Takahashi S., Ogura Y., Inohara N., Nunez G., Kishi Y., Koike Y., et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q.S., Xia B., Jiang Y., Qu Y., Li J. NOD2 3020insC frameshift mutation is not associated with inflammatory bowel disease in Chinese patients of Han nationality. World J. Gastroenterol. 2004;10:1069–1071. doi: 10.3748/wjg.v10.i7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb E., Broderick P., Lubbe S., Chandler I., Tomlinson I., Houlston R.S. A genome-wide scan of 10 000 gene-centric variants and colorectal cancer risk. Eur. J. Hum. Genet. 2009;17:1507–1514. doi: 10.1038/ejhg.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos A.M., Crooijmans R.P., Affara N.A., Amaral A.J., Archibald A.L., Beever J.E., Bendixen C., Churcher C., Clark R., Dehais P., et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Hildesheim A., Wang S.S., Herrero R., Gonzalez P., Burdette L., Hutchinson A., Thomas G., Chanock S.J., Yu K. Genetic admixture and population substructure in Guanacaste Costa Rica. PLoS One. 2010;5:e13336. doi: 10.1371/journal.pone.0013336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper R.S., Tayo B., Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum. Mol. Genet. 2008;17:R151–R155. doi: 10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shriner D., Adeyemo A., Ramos E., Chen G., Rotimi C.N. Mapping of disease-associated variants in admixed populations. Genome Biol. 2011;12:223. doi: 10.1186/gb-2011-12-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasaniuc B., Zaitlen N., Lettre G., Chen G.K., Tandon A., Kao W.H., Ruczinski I., Fornage M., Siscovick D.S., Zhu X., et al. Enhanced statistical tests for GWAS in admixed populations: assessment using African Americans from CARe and a Breast Cancer Consortium. PLoS Genet. 2011;7:e1001371. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale B.M., Sham P.C. The future of association studies: gene-based analysis and replication. Am. J. Hum. Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veyrieras J.B., Kudaravalli S., Kim S.Y., Dermitzakis E.T., Gilad Y., Stephens M., Pritchard J.K. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bansal V., Libiger O., Torkamani A., Schork N.J. Statistical analysis strategies for association studies involving rare variants. Nat. Rev. Genet. 2010;11:773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., International Inflammatory Bowel Disease Genetics C., Cotsapas C., Daly M.J. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Li M., Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 39.Wang K., Li M., Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina I., Montaner D., Bonifaci N., Pujana M.A., Carbonell J., Tarraga J., Al-Shahrour F., Dopazo J. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–W344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]