Abstract
von Willebrand factor (VWF) is a large adhesive glycoprotein with established functions in hemostasis. It serves as a carrier for factor VIII and acts as a vascular damage sensor by attracting platelets to sites of vessel injury. VWF size is important for this latter function, with larger multimers being more hemostatically active. Functional imbalance in multimer size can variously cause microvascular thrombosis or bleeding. The regulation of VWF multimeric size and platelet-tethering function is carried out by ADAMTS13, a plasma metalloprotease that is constitutively active. Unusually, protease activity of ADAMTS13 is controlled not by natural inhibitors but by conformational changes in its substrate, which are induced when VWF is subject to elevated rheologic shear forces. This transforms VWF from a globular to an elongated protein. This conformational transformation unfolds the VWF A2 domain and reveals cryptic exosites as well as the scissile bond. To enable VWF proteolysis, ADAMTS13 makes multiple interactions that bring the protease to the substrate and position it to engage with the cleavage site as this becomes exposed by shear. This article reviews recent literature on the interaction between these 2 multidomain proteins and provides a summary model to explain proteolytic regulation of VWF by ADAMTS13.
Many of the coordinated processes involved in hemostasis are driven by proteolytic reactions in which proteases cleave specific substrate bonds. Bond cleavage can lead to activation, propagation, or inactivation of a biochemical process. Control of proteolytic reactions is complex and, in its broadest sense, embraces localization of the protease and substrate, recruitment of cofactors for the purpose of acceleration of cleavage and then, direct or indirect inhibition of the protease to terminate its action. The plethora of proteolytic reactions in hemostasis provides examples of both general and process-specific mechanisms of control, which are all needed to ensure effective coordination. The proteolytic regulation of von Willebrand factor (VWF) function by its cleaving protease, ADAMTS13, falls into the latter category (ie, process-specific control), as it has certain unique features that set it apart from other hemostatic reactions. VWF is a large adhesive glycoprotein, necessary for initial platelet tethering and subsequent platelet adhesion.1 It is a multiadhesive protein that can interact with cell surface, extracellular matrix, and plasma protein ligands through specific domain binding sites.2–6 VWF is synthesized as a multimeric protein, which is central to its physiologic role. VWF function as a vessel wall damage sensor and initiator of primary hemostasis is highly dependent on its multimeric size.7 The larger VWF multimers in plasma are the most hemostatically reactive not only because they contain more ligand binding sites, but also because they are more conformationally responsive to vascular shear forces.1
In circulation, a first level of functional control of VWF is provided by its adoption of a globular conformation.8 The consequence of a globular fold is that certain interaction sites are buried and inaccessible to their ligands, which enables VWF to patrol the intact vasculature without binding unnecessarily to platelets or to plasma proteins. The functional quiescence of circulating VWF is perturbed by vessel damage, which results in the exposure of the collagen-rich matrix that normally underlies the protective endothelial cell monolayer. Globular VWF recognizes this newly exposed collagen leading to its specific recruitment to the damaged vessel wall. VWF then undergoes a unique structural transition whereby it is unfolded by local shear forces exerted on the tethered molecule by the flowing blood.8 In this way, VWF adopts an elongated, “active” conformation that exposes previously hidden platelet binding sites that mediate the capture of circulating platelets to the site of vascular injury. These first steps in the genesis of the primary platelet plug illustrate the importance of VWF conformation for its hemostatic function. Intriguingly, this same process of shear-dependent unfolding that activates VWF into a functional hemostatic protein also represents a primary determinant of the proteolytic regulation that controls VWF function.9
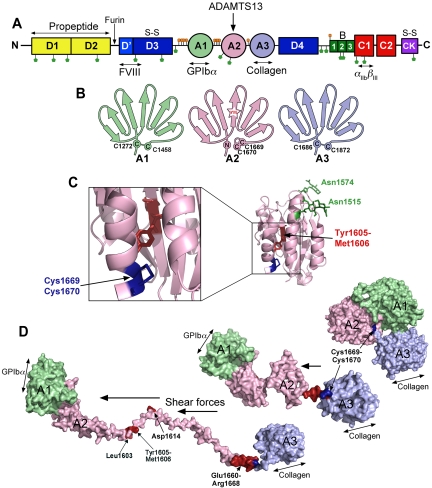
VWF is synthesized in a multimeric form that is essentially too large and thus too reactive for its routine functions. Although the largest plasma VWF multimers have generally been considered to range from 20- to 40-mers, more recent measurements have estimated that 100- or 200-mers may also exist, suggesting that stored endothelial and platelet VWF multimers may even exceed this.10 Control of VWF size, therefore, requires a specific regulatory mechanism. The plasma metalloprotease, ADAMTS13, provides this function by cleavage of a single peptide bond (Tyr1605-Met1606) located within the central VWF A2 domain (Figure 1A-B).11,12 However, while in its globular conformation VWF is essentially resistant to proteolysis, and only when VWF unfolds in response to shear does the scissile bond become accessible for ADAMTS13 to cleave. Proteolysis reduces VWF multimer size and, consequently, also its hemostatic function. The clinical importance of the regulation of VWF multimeric size by ADAMTS13 is exemplified by the clinical manifestation of dysfunction of the VWF-ADAMTS13 axis. ADAMTS13 deficiency (with consequent loss of VWF proteolysis) is associated with thrombotic thrombocytopenia purpura (TTP),13 a disease caused by clumping of platelets by ultra large (UL)–VWF and defined clinically by microangiopathic hemolytic anemia and thrombocytopenia.14 In contrast, excessive ADAMTS13-mediated VWF proteolysis precipitates type 2A von Willebrand disease15 in persons carrying mutations in VWF that increase the cleavage susceptibility of the scissile bond. There have also been suggestions that the hemostatic imbalance caused by reduced ADAMTS13 cleavage of VWF may contribute to thrombosis associated with cardiovascular diseases, but the evidence is currently conflicting.16,17
Figure 1.
The substrate, VWF. (A) Domain organization of VWF. N-linked and O-linked glycosylation sites are represented by green and orange lollipops, respectively. The propeptide (yellow represents D1 and D2) is marked. S-S indicates sites of intermolecular disulphide bond pairing. Vertical arrows indicate the location of cleavage sites (furin and ADAMTS13). Ligand binding sites (FVIII, GPIbα, collagen, and αIIbβIII) are labeled below. (B) Schematic representation of the VWF A1, A2, and A3 domains. The paired cysteines in A1 and A3 are shown. The vicinal disulphide bond that forms the molecular plug in the A2 domain and the Tyr1605-Met1606 (YM) ADAMTS13 cleavage site are also represented. (C) Structure of the VWF A2 domain highlighting the N-linked glycosylation sites (green), vicinal disulphide bond (blue), and the ADAMTS13 cleavage site (red) hidden in the center of the folded domain. (D) Molecular models of the unfolding of the VWF A1-A2-A3 domains. In globular VWF, the A3 domain collagen binding site is exposed. Elevated shear forces on VWF cause uncoupling of the A domains, extraction of the Cys1669-Cys1670 vicinal disulphide plug, and unraveling of the A2 domain. This exposes the GPIbα binding site in the A1 domain, cryptic ADAMTS13 binding sites, and the cleavage site in the A2 domain (red).
Control of VWF function by proteolysis depends on themes that are well rehearsed in hemostasis, including both the interaction of remote exosites and recognition of the scissile bond by the substrate-binding pocket of the protease. However, unlike the vast majority of hemostatic proteases, ADAMTS13 activity is not subject to direct regulation by a specific inhibitor. Although free hemoglobin18 and IL-619 can both inhibit ADAMTS13 function, these do not represent normal physiologic regulators (but may inhibit ADAMTS13 under pathophysiologic circumstances). Certain coagulation proteases, such as thrombin and plasmin,20 might play a role in controlling activity because ADAMTS13 is sensitive to proteolysis by these serine proteases at defined cleavage inactivation sites.21 However, as evidence of ADAMTS13 proteolysis in vivo has only been observed in patients with severe sepsis22 or in a very rare case of α2-antiplasmin deficiency,23 it is probable that this remains a pathologic phenomenon rather than a normal control mechanism. The absence of a specific inhibitor, therefore, implies that in the normal physiologic context, ADAMTS13 is essentially unregulated and that its activity is primarily controlled by both the availability of the scissile bond of VWF and the specificity of the VWF-ADAMTS13 interaction. Substrate control of protease function represents an intriguing and less familiar mechanism in the setting of hemostasis, and yet this is central to control and coordination of platelet tethering. Over the past decade, much attention has been paid to the molecular mechanisms underlying the control of VWF function by ADAMTS13, the focus of this review.
The substrate, VWF
Synthesis of VWF is restricted to endothelial cells and megakaryocytes.24,25 The nascent polypeptide monomer (∼ 310 kDa) contains a signal peptide, a propeptide and the now familiar mature VWF domain organization that contains a variety of specific ligand binding sites (Figure 1A). Covalent dimerization of VWF monomers takes place in the endoplasmic reticulum, mediated through intermolecular disulphide pairing of Cys residues in the cysteine knot domain.26,27 Multimerization of the dimers in the Golgi, in a process catalyzed by the propeptide (D1 and D2 domains), which acts as a protein disulphide isomerase to form disulphide bonds between the N-termini of VWF dimers.28,29 During its synthesis, VWF undergoes extensive glycosylation with 12 N-linked and 10 O-linked glycan side chains on each mature monomer unit (Figure 1A-C). Glycosylation is essential for secretion of the protein and 4 N-linked sites (Asn99, Asn857, Asn2400, and Asn2790) have been identified that are of particular importance for synthesis/secretion.30,31 A small proportion of the N- and O-linked glycans contain the ABO(H) blood sugars.32 Not only do these influence plasma VWF levels, their presence also influences VWF proteolysis by ADAMTS13, as shown by the increased proteolysis of patients with the rare Bombay blood group.33 The most important N-linked glycan in this respect is that attached to Asn1574 within the A2 domain (Figure 1C), which, if removed, appreciably increases the susceptibility of the VWF A2 domain to unfold and be proteolysed.30 This glycan may help stabilize the folding of the VWF A2 domain and therefore also influence the domain unfolding necessary for proteolysis. The O-linked glycans are clustered either side of the VWF A1 domain and probably play a role in stiffening the hinge between the adjacent domains.34
After glycosylation, furin removes the VWF propeptide, which then assists in the trafficking of both the mature multimeric VWF and cleaved propeptide to Weibel-Palade bodies of endothelial cells or α-granules of platelets.35 VWF is stored in these organelles in UL form. These UL-VWF multimers can be released constitutively from the endothelium into the bloodstream,36 or on demand in response to a variety of different physiologic agonists capable of activating the endothelium or platelet.
On secretion, a proportion of VWF released from endothelial cells remains tethered to the cell surface, from which it is proteolytically cleaved and released by ADAMTS13.37 Once in free circulation, VWF adopts its globular conformation. In this respect, the behavior of the VWF A1-A2-A3 domains is central to VWF function. In its globular form, one can envisage that the collagen-binding site within the A3 domain is constitutively exposed on the surface, as it must provide the initial contact point for the newly exposed collagen. Conversely, the glycoprotein Ibα (GPIbα) binding site in the VWF A1 domain remains largely hidden until required to prevent spontaneous or unnecessary platelet binding. The intervening VWF A2 domain that harbors the ADAMTS13 cleavage site (Tyr1605-Met1606) is folded such that the cleavage site and VWF A2 domain exosites are hidden/buried (Figure 1B-C). While in this globular form, VWF exhibits functional quiescence.
Mechanical shear forces in the bloodstream act on the VWF molecule, thereby stretching it and changing its conformation. This modulates the exposure of both VWF A1 domain platelet binding sites and the VWF A2 domain ADAMTS13 binding/cleavage site(s).38,39 Larger VWF multimers unravel more readily under high shear.38 Furthermore, attachment of VWF multimers to collagen and/or platelets further facilitates unfolding because of increased tensile force acting on the VWF molecule. VWF unfolding is thought to involve both the uncoupling of the VWF A1-A2-A3 tridomain cluster, and conformational changes within individual domains,39 most notably the VWF A2 domain40 (Figure 1D).
The protease, ADAMTS13
ADAMTS13 is synthesized in hepatic stellate cells41 and vascular endothelial cells42 as an ∼ 180-kDa glycoprotein. Its domain structure is depicted in Figure 2A. ADAMTS13 is a member of the ADAMTS family of Zn2+-dependent metalloproteases, which all contain (from the mature N-terminus) a metalloprotease, disintegrin-like, thrombospondin type 1 (TSP) repeats, cysteine-rich and spacer domains.13,43–45 Thereafter, different family members contain variable numbers and types of domains.
Figure 2.
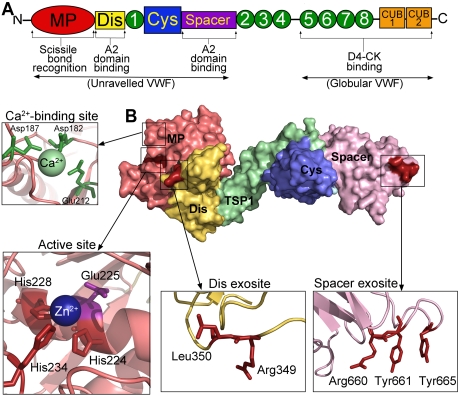
The protease, ADAMTS13. (A) Domain organization of ADAMTS13. From the N-terminus are the metalloprotease domain (MP; red), disintegrin-like domain (Dis; yellow), TSP repeats (1-8; green), cysteine-rich domain (Cys; blue), spacer domain (purple), and CUB domains (orange). Binding sites and function of specific domains are labeled below. (B) Structure of ADAMTS13 N-terminal domains (MDTCS) based on the crystal structure of DTCS and homology modeling of the MP domain. Surface representation is shown. Domains are colored according to panel A. (Insets) Cartoon representation of the location of the high-affinity calcium binding site and coordinating residues (green) in the MP domain, the active site containing 3 His residues (red) and catalytic Glu225 (mauve), the disintegrin-like domain exosite, and spacer domain exosite.
The metalloprotease domains of the ADAMTS family are characterized by a reprolysin-type Zn2+ binding signature (HEXXHXXGXXHD) involving 3 perfectly conserved His residues (Figure 2B). This sequence also contains an active-site Glu residue (at position 225 in ADAMTS13), which polarizes a water molecule that is stabilized by the coordinated Zn2+ ion, and is part of the proteolytic machinery. The Zn2+-binding motif is followed in all ADAMTS metalloprotease domains by a tight “Met-turn,” which is a further structural characteristic of these metalloproteases.
Whereas these common structural features in ADAMTS family members are critical for substrate recognition and hydrolysis of the scissile bond, differences in the substrate binding exosites and in the architecture of the catalytic site allow each ADAMTS family member to recognize and proteolyse its physiologic substrate at a particular site.
ADAMTS13 requires divalent cations for its activity.12 In addition to the Zn2+ ion in the active center, Ca2+ ions are also required for enzyme function. On the basis of ADAMTS family sequence conservation and structural analyses of the metalloprotease domains of ADAMTS1, ADAMTS4, and ADAMTS5,46–48 a double Ca2+ binding site within the ADAMTS13 metalloprotease domain coordinated by Glu83, Asp173, Cys281, and Asp284 was predicted.49,50 However, functional evaluation of this Ca2+ binding site revealed that mutating either Glu83 or Asp173 to Ala did not appreciably diminish proteolysis of a short VWF substrate.50 This suggests that the binding of Ca2+ to this site is not of major importance for ADAMTS13 function. Conversely, mutation of another candidate site composed of Asp182, Asp187, and Glu212 had a major influence on ADAMTS13 activity.50 Single point substitutions of these residues increased the KD(app) for Ca2+ from ∼ 72μM (for wild-type ADAMTS13) up to 720μM, indicating a large reduction in functional binding affinity for Ca2+. That these residues are present in a loop adjacent to the ADAMTS13 active site (Figure 2B) suggests that Ca2+ binding provides structural integrity to this loop that is necessary for efficient proteolysis of the VWF substrate.
Adjacent to the metalloprotease domain in ADAMTS family members are disintegrin-like domains, which share similarity with snake venom disintegrins but lack the canonical cysteine arrangement. Despite its name, the disintegrin-like domain of ADAMTS family members does not appear to function like a disintegrin and bears greater structural similarity to the cysteine-rich domains51: this is evident from the recently published crystal structure of the ADAMTS13 noncatalytic domains spanning the disintegrin to spacer domains.52 In addition, and unlike the ADAM family metalloproteases, the disintegrin-like domain of the ADAMTS family contains a linker region that extends around the back of the metalloprotease domain and positions the disintegrin-like domain at one end of the active-site cleft.
The TSP repeats of ADAMTS13 are homologous to the type 1 repeat of thrombospondin-1 and -2. The first TSP repeat (48-54 residues) is very similar in all members and generally contains 6 cysteines. The downstream TSP repeats (after the spacer domain) are much more variable in sequence. The 4th TSP repeat of ADAMTS13 contains just 4 Cys residues, 2 of which are predicted to remain unpaired. Four TSP domains in ADAMTS13 contain a CSVSCG motif in which the second serine is O-linked glycosylated and forms a consensus binding sequence for the CD36 cell surface receptor.53
The cysteine-rich domain of ADAMTS13 demonstrates high sequence homology with the other ADAMTS family members, which contain 10 conserved cysteines (except ADAMTS10). This precedes the spacer domain, which contains no cysteines and is of highly variable length among family members. It is the least homologous of all the domains and is globular with 10 β-strands in a jelly roll topology, forming 2 antiparallel β-sheets.
ADAMTS13 is the only member of the ADAMTS family to contain CUB domains, of which there are 2 located at the C-terminus. CUB domains are present on proteins known to be important for developmental regulation, such as bone morphogenetic protein-1.
ADAMTS13 recognition of globular VWF
Recognition of VWF by ADAMTS13 is complex, yet highly specific and involves multiple interactions between distinct domains of both ADAMTS13 and VWF, with some of the interaction sites on VWF only becoming exposed during shear. Much of the structure-function work on ADAMTS13 has used domain deletion mutants that retained the metalloprotease domain but were progressively truncated from the C-terminus. Despite the limitations of potential conformational changes induced by domain deletion, these mutants have provided valuable initial tools for exploring ADAMTS13 domain function. Early results with deletion mutants of ADAMTS13, however, provided inconsistent and condition-dependent evidence for a functional role for the C-terminal TSP repeats and CUB domains. For example, it was widely held that truncation mutants of ADAMTS13 with deletions after the spacer domain (MDTCS) retained most of their activity in cleaving VWF under static denaturing conditions, implying that these domains might be dispensable.54–56 Another study reported activity of deletion mutants in flow based, but not in static assays.57 However, the C-terminal TSP and CUB domains have been claimed to be important for optimal cleavage activity of ADAMTS13 under flow conditions,58 against full-length murine VWF59 and for cleaving platelet decorated strings in vivo.60 Finally, a peptide sequence from first CUB domain was found to inhibit VWF proteolysis under flow conditions.61 To resolve such conflicting results, Zanardelli et al prepared deletion mutants of both VWF and of ADAMTS13 and performed binding and activity studies under both static and flow conditions.62 These experiments showed that full-length ADAMTS13 could bind with a KD ∼ 86nM to VWF in its quiescent/globular (as opposed to its unraveled) conformation. This binding is mediated in part by the VWF D4 domain and the TSP5–8 and/or the CUB domains of ADAMTS13. Similar conclusions were drawn by Feys et al using immunoprecipitation of VWF-ADAMTS13 complexes in solution to determine a KD ∼ 79nM.63 This interaction was dependent on the ADAMTS13 TSP2–8 repeats. Both studies demonstrated the specific binding of ADAMTS13 to globular VWF, and that in the absence of shear-induced unfolding of VWF this interaction is nonproductive in terms of VWF proteolysis.
Feys et al demonstrated that a proportion of globular VWF and ADAMTS13 normally circulate in plasma in complex with each other.63 The stoichiometry of ADAMTS13 to globular VWF is low (∼ 1 ADAMTS13 molecule per 250 VWF molecules).63 This suggests that the larger, more hemostatically active VWF multimers are the most probably globular VWF forms to circulate in complex with ADAMTS13.64 It is also these larger species that contribute most to platelet plug formation. Consequently, although only a small percentage of plasma ADAMTS13 (∼ 3%) actually circulates bound to VWF, this pool may be particularly effective at colocalizing VWF and ADAMTS13 to the site of vessel damage. Evidence that this may be of functional significance was provided by Banno et al,65 who examined mice with an Adamts13 gene that expresses a truncated form of the enzyme (ADAMTS13S) that lacks the TSP(7/8) and CUB domains.66 Neither the Adamts13+/+ nor Adamts13S/S mice had discernible differences in their plasma VWF multimers, suggesting that the TSP(7/8) and the CUB domains may not be necessary for modulating normal plasma VWF size.65 However, mice expressing the truncated ADAMTS13 were more thrombogenic in experimental thrombus formation under high shear conditions, suggesting that the loss of the TSP(7/8) and CUB domains impaired the ability of ADAMTS13 to regulate VWF-dependent development of the platelet plug.
VWF A2 domain unfolding is required for proteolysis by ADAMTS13
Although ADAMTS13 can bind globular VWF, it cannot cleave it12 because further binding sites and the scissile bond are hidden inside the folded VWF A2 domain.67,68 The folded VWF A2 domain adopts an atypical Rossman fold in which amphipathic α-helices surround a central β-sheet in which the ADAMTS13 cleavage site is buried67 (Figure 1B-C). Although the VWF A2 domain is highly homologous to the VWF A1 and A3 domains, it is more prone to unfolding because it lacks an intradomain disulphide bond that connects the N- and C-termini and further destabilized because one of the amphipathic α-helices has been replaced by a less-ordered loop (α4-less loop)67 (Figure 1B-C). The VWF A2 domain crystal structure has also revealed a very rare vicinal disulphide bond between adjacent Cys1669 and Cys1670 at the C-terminus of the last α-helix in the A2 domain. This vicinal disulphide bond forms an 8-membered ring that bends the peptide backbone in an unusually strained conformation and forms a “molecular plug” that directly interacts with hydrophobic residues in the core of the domain.67 This disulphide bond was therefore hypothesized to stabilize the VWF A2 domain fold. Luken et al indeed showed that when this disulphide bond is removed by mutagenesis, the VWF A2 domain more readily unfolds and is more susceptible to proteolysis by ADAMTS13.69 When VWF encounters elevated rheologic shear forces, this “molecular plug” is pulled out, allowing water molecules to enter and destabilize the hydrophobic core resulting in VWF A2 domain unfolding (Figure 1D).
Of all the VWF A2 domains that are present in a long VWF multimer, it is those in the middle that are most likely to unfold and therefore be cleaved. Under shear the tensile forces that are applied on VWF increase with the distance from the nearest end of the multimer.38 This means that the force is highest in the middle, but also that an VWF A2 domain in the middle of a large multimer is more prone to unfolding than one in the middle of a short multimer. This provides a mechanism that distinguishes VWF multimers that need to be cleaved (larger ones) from those that do not (shorter ones).
Zhang et al investigated the forces necessary to unfold the VWF A2 domain using laser tweezers.38 They applied force to a single VWF A2 domain and measured the increase in length at different forces. They found that the VWF A2 domain typically unfolds at approximately 11pN. Their analysis predicted that this tensile force might correspond to that encountered in the middle of a free flowing 200-mer VWF multimer in arterioles and capillaries. Apart from during transit through arterioles and capillaries, such high shear rates are also encountered when VWF is bound to platelets, collagen or during secretion from endothelial cells. VWF bound to platelets has indeed be shown to be more susceptible to cleavage by ADAMTS13,70 and VWF multimers are cleaved by ADAMTS13 on secretion from endothelial cells.37
Zhang et al also found that when force was relaxed after domain unfolding, the VWF A2 domains could refold.38 Refolding after a high shear encounter in the circulation is probably important in preventing excessive cleavage. Interestingly, the VWF A2 domain contains a cis-proline (Pro1645) that when exposed to high forces (eg, when VWF is bound to platelets) could potentially become a trans-proline, which could last long enough (100-1000 seconds) to delay refolding and, in turn, enhance the opportunity for proteolysis by ADAMTS13. The biophysical balance between unfolding and refolding represents a key determinant of VWF function.
The ADAMTS13 spacer domain binds to a cryptic VWF A2 domain exosite that is revealed on unfolding
Once the VWF A2 domain is unfolded, additional binding sites for ADAMTS13 on VWF are revealed. To investigate where within the VWF A2 domain these binding sites reside, short VWF A2 domain fragments, such as VWF115 and VWF73, have been used as substrates.68,71 C-terminal deletion mutants of these fragments demonstrated that VWF A2 domain residues Glu1660-Arg1668 appreciably contribute to the cleavage of the Tyr1605-Met1606 scissile bond.71 Gao et al found that the ADAMTS13 spacer domain binds to this sequence, referred to as a VWF A2 domain exosite, and that deletion of either the spacer domain from ADAMTS13, or deletion of the VWF A2 domain exosite reduced cleavage of VWF73 by approximately 20-fold.72 Wu et al isolated short peptides containing the VWF A2 domain exosite that were able to inhibit cleavage of short substrates and of full-length multimeric VWF.72,73 This VWF A2 domain exosite is cryptic inasmuch as it is not exposed in the isolated folded VWF A2 domain or in globular VWF.62,69 Exposure of this cryptic exosite requires the extraction of the adjacent molecular plug formed by the vicinal disulphide bond, Cys1669-Cys1670 (Figure 1D) and uncoupling of the C-terminal α-helix in which residues Glu1660-Arg1668 reside. As this region is predicted to unfold first, even partial unfolding of the VWF A2 domain could be sufficient for the spacer to bind.
Further insight into the nature of the ADAMTS13 spacer domain exosite that binds this VWF A2 domain site came from the study of autoantibodies that arise in acquired TTP. The ADAMTS13 Cys and spacer domains were shown to be the primary autoimmune target in TTP.74 Using both monoclonal and polyclonal antibodies isolated from patients with acquired TTP, Pos et al demonstrated that ADAMTS13 spacer domain residues Arg660, Tyr661, and Tyr665 represent a core binding site for autoantibodies.75 The ADAMTS13 spacer domain residues Arg660, Tyr661, and Tyr665 reassuringly align as a surface cluster on the structure of the noncatalytic domains,52 available for both antibody and VWF A2 exosite interactions75 (Figure 2B). Substitution of these amino acids resulted in a 12-fold reduction in catalytic efficiency (kcat/Km) of VWF115 proteolysis and a 25-fold reduction in VWF115 binding.75 This suggested that the core binding site for TTP autoantibodies overlapped with a functional spacer domain exosite. Jin et al used similar mutagenesis approaches and suggested that Arg659 of ADAMTS13 might also contribute to ADAMTS13 spacer-VWF A2 interactions.76 Although ADAMTS13 can bind globular VWF with a KD of ∼ 80nM via interactions of the C-terminal TSP and CUB domains, once VWF is unfolded this binding increases appreciably (KD ∼ 5-10nM) because of the cooperative effects of the spacer domain and the C-terminal TSP and CUB domains.
An interesting point is that, although the ADAMTS13 spacer domain exosite undoubtedly engages with the cryptic VWF A2 exosite and is a major determinant of short substrate binding and proteolysis, mutation of this exosite in full-length ADAMTS13 (rather than in the ADAMTS13 truncation, MDTCS) does not greatly influence proteolysis of full-length multimeric VWF.75 Therefore, the interaction between the ADAMTS13 spacer domain and the VWF A2 domain exosites makes a modest, rather than pivotal, contribution to the proteolysis of full-length VWF. This may suggest some functional redundancy between the spacer domain and C-terminal TSP and CUB domain binding sites and further highlights the importance of additional functional exosites in both molecules.
The ADAMTS13 disintegrin-like domain interacts with a VWF exosite to orient the scissile bond toward the active-site
The shear induced extraction of the vicinal Cys1669-Cys1670 plug from the hydrophobic core of the VWF A2 domain not only exposes the VWF Glu1660-Arg1668 A2 domain exosite, but also destabilizes the domain fold to enable exposure of additional sequences that act as additional exosites. Gao et al used deletion mutants of VWF73 to propose a number of discreet interactions between ADAMTS13 domains and the VWF polypeptide.77 Similar conclusions were drawn by Akiyama et al based on their crystal structure of the ADAMTS13 DTCS domains and functional analyses of ADAMTS13 mutants.52 Both studies suggested that the cysteine-rich domain contains a functional exosite that may interact with residues adjacent to the VWF A2 domain exosite that binds the spacer domain.52,77 However, at the time of writing, it is not yet clear what the nature of this interaction is, or what its relative contribution to the proteolysis of full-length VWF might be.
The role of the ADAMTS13 disintegrin-like domain was investigated in detail by de Groot et al using both deletion and substitution mutagenesis.78 A role for this domain had been suggested by earlier reported domain deletion studies54,55 and confirmed by de Groot et al.78 The use of molecular modeling and sequence homology alignments with ADAMTS family members helped identify potential exosite residues in this domain that might make interactions with VWF. Kinetic analysis of point substitutions revealed that mutation of Arg349 or Leu350 in the ADAMTS13 disintegrin-like domain reduced cleavage of VWF115 by approximately 10- to 20-fold because of both an increase in Km and decrease in kcat, suggesting changes in both functional substrate binding and substrate turnover.78 The effect of mutating this exosite is very similar to the magnitude of the disruption of the spacer domain exosite in proteolysis of short A2 domain fragments, but appreciably greater when examining the proteolysis of full-length multimeric VWF.75
The proximity of these amino acids in the disintegrin-like domain to the active site in the adjacent ADAMTS13 metalloprotease domain (Figures 2B, 3A) suggested that they would likely interact with VWF residues approximately 26Å C-terminal to the scissile bond.78 Fortuitously, previous work by Zanardelli et al had identified Asp1614 (located 9 residues C-terminal to the cleavage site) in the VWF A2 domain as functionally important to the cleavage of VWF115.68 Based on this, de Groot et al explored whether Asp1614 in VWF forms a charged interaction with Arg349. They showed that substituting either residue produced an equivalent reduction in VWF115 cleavage, suggesting a mutual and dependent interaction between these residues. This defined a functional exosite on the ADAMTS13 disintegrin-like domain that interacts with a complementary exosite on VWF involving Asp1614 and that this helps orientate the scissile bond toward the active center of ADAMTS1378 (Figure 3).
Figure 3.
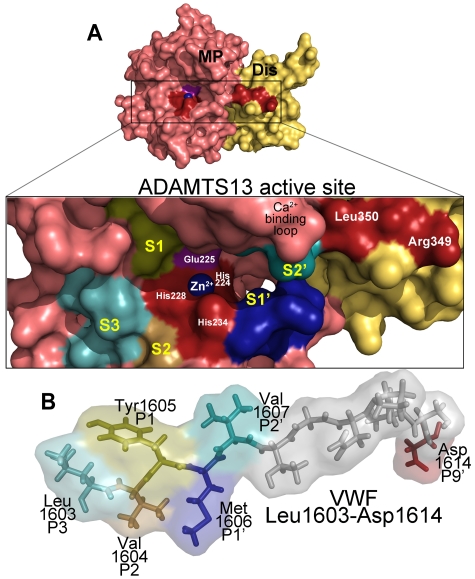
ADAMTS13 recognition of the VWF cleavage site. (A) Molecular model of the ADAMTS13 metalloprotease (MP; light red) and disintegrin-like (Dis; yellow) domains. (Inset) The active site cleft. Active site residues (Zn2+ ion, its 3 coordinating His residues His224, His228, and His234, as well as the catalytic Glu225 residue) and disintegrin-like domain exosite residues are labeled. The high-affinity functional Ca2+-binding loop is also marked. Regions of the metalloprotease domain that are predicted to harbor the S3, S2, S1, S1′, and S2′ subsites are labeled marked in different colors. (B) Below the active site cleft, VWF A2 domain residues Leu1603-Asp1614 are depicted as sticks and transparent spacefill. The P3, P2, P1, P1′, P2′, and P9′ residues in VWF that are important for proteolysis are labeled and colored according to their predicted complementary subsites (S) in the metalloprotease domain.
Role of the VWF P1-P1′ scissile bond residues and the ADAMTS13 S1-S1′ pockets
Notwithstanding the importance of the aforementioned remote exosite interactions for binding and for guiding the substrate to the active site of ADAMTS13, the metalloprotease domain itself must also necessarily interact with the substrate in the vicinity of the cleavage site and therefore harbor sites that contribute to scissile bond specificity. Indeed, interactions between the metalloprotease domain and VWF scissile bond have been demonstrated to be critical for VWF proteolysis, as mutagenesis of the Tyr1605 (P1) and Met1606 (P1′) residues to alanine predictably greatly reduce proteolytic efficiency.79,80 The comparatively normal proteolysis of VWF115 Tyr1605Phe and Tyr1605Trp variants79 demonstrated the importance of the aromatic side chain of the P1 residue. Undoubtedly, the P1 and P1′ residues must be accommodated in specific complementary binding pockets on ADAMTS13. The ADAMTS13 S1 pocket that accommodates the P1 residue probably involves hydrophobic residues Leu151 and Val195.80 Furthermore, molecular modeling has suggested that this may also be made up by elements of the loop containing the high-affinity Ca2+ binding site (Figure 3).
Similar analysis using VWF115 Met1606Leu highlighted the requirement for a large and hydrophobic amino acid in the P1′ position.79 The nature of the S1′ subsite that accommodates the P1′ residue of the substrate has been recently investigated by de Groot et al.81 This report showed that a VWF115 P1′ variant, Met1606Ala, was proteolysed 15- to 18-fold less efficiently than wild-type VWF115. However, when ADAMTS13 residues Asp252-Pro256 were substituted with the corresponding residues of ADAMTS1, the variant exhibited ADAMTS1-like P1′ specificity by efficiently cleaving a VWF115 mutant with Ala at the P1′ site.81 This strongly implied that amino acids Asp252-Pro256 of ADAMTS13 are important in shaping the docking site of the P1′ (Met) residue of the VWF scissile bond and contribute to the S1′ pocket (Figure 3).
Positioning of the VWF scissile bond: role of the P3 residue
It is interesting to note that, in the literature to date, most attention has been paid to regions and residues of VWF that are C-terminal to the cleavage site. This approach has successfully identified the importance of the VWF cleavage site residues, the ADAMTS13 disintegrin-like domain binding site and the VWF A2 domain exosite that binds the spacer domain. Xiang et al hypothesized that another necessary feature of enzymatic cleavage of VWF must be a docking point for ADAMTS13 that is N-terminal to the scissile bond.80 They considered the cleavage susceptibilities of previously reported short VWF A2 domain fragments that span the cleavage site. One, VWF64 (1605-1668), cannot be cleaved by ADAMTS13,77 whereas others, such as VWF73 (1596-1668), VWF76 (1593-1668), and VWF115 (1554-1668) are all proteolysed efficiently.68,71,82 The difference between the substrates that are, and those that are not cleaved, resides in the sequence N-terminal to the scissile bond. Therefore, Xiang et al conjectured that the VWF1596-1604 sequence immediately N-terminal to the cleavage site must contain a structural determinant that is essential for proteolysis.80 Accordingly, by systematic substitution of all of the amino acids in this region, they showed that VWF P3 residue, Leu1603, has an essential role in proteolysis of the scissile bond, and the adjacent P2 residue (Val1604) plays an additional minor role. Remarkably, substitution of Leu1603 alone to Ser, Asn, or Lys all reduced the cleavage efficiency, up to ADAMTS13 > 400-fold.80 Molecular modeling identified candidate interacting residues for VWF Leu1603 on the surface of ADAMTS13 adjacent to the active site. Mutagenesis studies identified ADAMTS13 residues Leu198, Leu232, and Leu274 as elements that together may make up the S3 subsite (Figure 3).
Because the VWF P3 residue is functionally so important, it suggested a role in the cleavage mechanism of the VWF scissile bond. Xiang et al have proposed a model of cleavage in which the VWF P3 (Leu1603), P2 (Val1604), and P9′ (Asp1614) residues act as docking sites on either side of the scissile bond.80 This brings the P1 and P1′ residues into position over the active site-coordinated Zn2+ ion and the catalytic Glu225 of ADAMTS13 for cleavage. A model of the ADAMTS13 metalloprotease-disintegrin domains aligned with the VWF sequence, including and surrounding the scissile bond, is shown in Figure 3.
A multidomain, conformation-driven model of VWF bond proteolysis by ADAMTS13
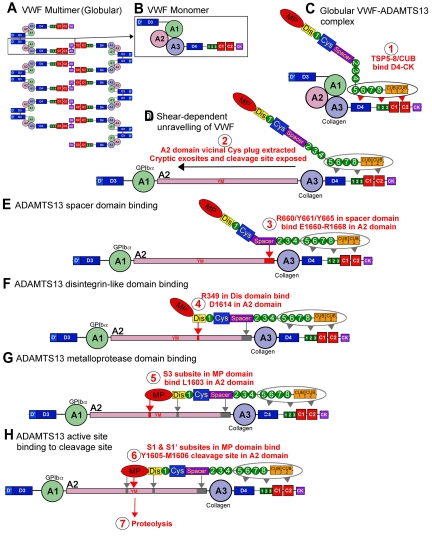
A summary model of VWF and ADAMTS13 interactions, encompassing binding at multiple sites, ultimately leading to scissile bond cleavage can now be proposed that involves 7 potentially distinct steps. Under normal physiologic conditions, VWF circulates in plasma in a globular conformation in which the VWF A2 domain is hidden within the core of the molecule. A binding site for ADAMTS13 within the D4-cysteine knot domains of VWF is nevertheless constitutively exposed, and a small proportion of ADAMTS13 can reversibly associate with the globular protein (step 1), with binding mediated by its TSP5-CUB domains (KD ∼ 80nM; Figure 4A-C). When shear forces begin to induce the transition of VWF from a globular to a fibrillar conformation (ie, when secreted, when tethered to the site of vessel injury by its A3 domain or during passage through the microvasculature under high shear), additional exosite binding sites on VWF are revealed that present themselves (step 2) to complementary sites on ADAMTS13. The ADAMTS13 spacer domain recognizes residues VWF residues Glu1660-Arg1668 (step 3), revealed when the vicinal Cys disulphide bond plug is extracted from the hydrophobic core of the A2 domain. This increases the affinity of ADAMTS13 for VWF (KD ∼ 10nM). An ADAMTS13 disintegrin-like domain exosite involving Arg349 recognizes a complementary exosite on VWF composing Asp1614 in a critical, but low affinity, interaction (step 4). Thereafter, an essential contact is made between VWF Leu1603 and a complementary S3 subsite in ADAMTS13 involving Leu198, Leu232, and Leu274 (step 5). Together, these interactions bring the Tyr1605-Met1606 scissile bond over the ADAMTS13 active site. This allows the P1 and P1′ residues to engage with their respective S1 and S1′ subsite pockets, involving ADAMTS13 Leu151/Val195 (S1) and Asp252-Pro256 (S1′), respectively (step 6). Steps 3 to 6 can be envisaged to represent a “molecular zipper” that ends in the precise delivery of the scissile bond to the active site. Once cleavage has taken place (step 7), there is a consequent reduction in affinity of protease and substrate, enabling the protease to recycle.
Figure 4.
Proteolysis of VWF by ADAMTS13. VWF circulates in plasma as a multimeric molecule (A) that adopts a quiescent globular conformation. Each multimer is composed of disulphide linked VWF monomers (B). In its globular conformation, the A3 domain collagen binding site is exposed. ADAMTS13 can bind to this globular VWF via its TSP (5-8) and CUB domains (C), step 1. This enables VWF and ADAMTS13 complexes to form and circulate in plasma. Under elevated shear forces (which can occur on secretion, collagen binding, or passage through the microvasculature), VWF can unravel to expose A1 domain binding site for GPIbα. These shear forces also remove molecular plug formed by the vicinal disulphide bond in the A2 domain, which causes A2 domain unfolding (D), step 2 (see Figure 1). This unfolding reveals cryptic exosites that enable residues in the ADAMTS13 spacer domain to bind to the unfolded A2 domain (E), step 3 (see Figures 1D, 2B). Thereafter, a critical low-affinity interaction between D1614 and the Dis domain helps approximate and position the cleavage site (F), step 4 (see Figure 3). This enables further interactions between the MP domain to occur, including an essential interaction via an S3 subsite with L1603 in VWF (G), step 5 (see Figure 3). Together, these interactions allow the MP to engage via S1 and S1′ subsites with the cleavage site (YM; H), step 6, after which proteolysis can occur, step 7.
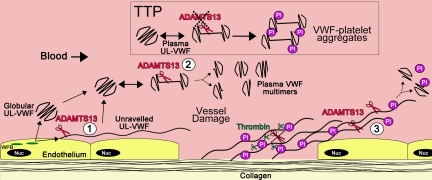
Location specific cleavage of VWF by ADAMTS13
Three distinct locations of VWF proteolysis by ADAMST13 can be considered: (1) newly secreted VWF strings from the endothelial cell surface, (2) UL-VWF in free circulation, and (3) unraveled VWF at sites of platelet plug formation (Figure 5). Although proteolysis in each of these instances could fulfill a somewhat different purpose, the cleavage reaction is driven by local shear forces that reveal the VWF A2 exosites that enable productive ADAMTS13 binding. That ADAMTS13 proteolysis is dependent on shear-induced unfolding may seem paradoxical inasmuch as the very unfolding process that is required for platelet tethering is also the primary determinant of proteolysis that counteracts this function. However, the recent advances in our understanding of the biochemistry of the interactions of ADAMTS13 and VWF have provided a potential explanation for this.
Figure 5.
Location of VWF cleavage by ADAMTS13. Cartoon depicting the sites of VWF proteolysis by ADAMTS13. (UL)VWF is synthesized by the endothelium and stored within Weibel-Palade bodies (WPB; green). VWF multimers of various sizes, including UL-VWF, can be secreted directly into the circulation. (1) Alternatively, a proportion of UL-VWF may attach to the endothelial surface during exocytosis and unravel in response to shear forces. Under such circumstances, the VWF A2 domain unfolds to enable ADAMTS13 (red scissors) to cleave VWF and release the VWF string. Whether directly secreted or proteolytically released, VWF can adopt a globular fold in the plasma circulation. However, during passage through the microvasculature, globular UL-VWF in free circulation will probably unravel (at least partially/transiently). (2) Such unraveling permits the processing of the largest, most hemostatically active forms of VWF, resulting in their conversion to smaller plasma VWF multimers. Mutations in VWF that precipitate type 2A von Willebrand disease are particularly influenced by such proteolysis. This group of mutations enhances the propensity of VWF to unfold in free circulation, leading to excessive proteolysis and loss of hemostatically functional VWF. Conversely, ADAMTS13 deficiency results in the loss of such plasma processing. Under these circumstances, platelets (Pl) can become bound to transiently unraveled VWF, leading to the accumulation of VWF-platelet aggregates that occlude the microvasculature, as seen in patients presenting with TTP. At sites of vessel damage, endothelial damage results in exposure of subendothelial collagen. Plasma VWF binds to this, unravels, and, in turn, recruits platelets. The presence of collagen and thrombin induces rapid platelet activation, which consolidates the platelet plug. Thrombin further stabilizes this through the deposition of fibrin and the proteolytic inactivation of ADAMTS13. (3) Downstream of the site of injury (ie, in the absence of collagen and thrombin), VWF-platelet strings may still be proteolysed by ADAMTS13, which in turn limits/regulates platelet plug formation. Low or reduced ADAMTS13 levels may impair this process and consequently influence the pathophysiology of arterial thrombosis.
Vessel damage exposes collagen, to which VWF binds and subsequently unravels in response to shear forces. This, in turn, allows circulating platelets to bind. The binding of GPIbα to VWF occurs rapidly, involving both fast on- and off-rates that exceed the rate of VWF proteolysis by ADAMTS13.39,83 It should be noted that, despite the high-affinity binding of ADAMTS13 to VWF (KD ∼ 10nM), the Km for proteolysis is appreciably higher (Km = 0.16-1.6μM).38,68,78,81 Moreover, at sites of vessel damage, the presence of collagen and thrombin generated by activation of the coagulation cascade can act as potent local activators of recruited platelets that prompt further VWF-independent adhesion and aggregation. These recruited platelets can thereby become resistant to the consequences of ADAMTS13-mediated VWF proteolysis. As the platelet plug develops, however, it grows beyond the site of injury (and also the primary source of collagen and thrombin). In these locations, although unraveled VWF may tether platelets through GPIbα, binding is not consolidated as effectively by the effects of collagen and thrombin. This probably provides more time for ADAMTS13 to cleave VWF, and so regulate platelet plug growth by confining it to the site of vessel damage. This is further supported by the visualization of ADAMTS13 function on the surface of developing thrombi using whole blood flow models.84
The scenario is different for VWF that is either newly secreted from the endothelium, or larger VWF multimers that are present in free circulation, which may naturally unravel to expose their GPIbα binding site. In these cases, such unraveling takes place in the absence collagen or thrombin that could otherwise activate bound platelets. As platelet tethering in these instances is not consolidated by the effects of such agonists, and, furthermore, as this process also promotes VWF A2 domain unfolding, the proteolysis of VWF by ADAMTS13 can occur, albeit relatively slowly. The consequent decrease in VWF multimer size reduces the shear forces exerted on the proteolysed molecules, enabling them to adopt a quiescent globular conformation. This plasma proteolysis thereby counteracts the formation of unwanted platelet-rich thrombi in the microvasculature, thrombi such as occurs in persons with TTP/ADAMTS13 deficiency.
Acknowledgments
This work was supported by the British Heart Foundation (grants RG/06/007 and PG/09/038).
Authorship
Contribution: All authors contributed to writing and revising the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Lane, Centre for Haematology, Imperial College London, 5th Floor Commonwealth Building, Hammersmith Hospital Campus, Du Cane Road, London, W12 0NN, United Kingdom; e-mail: d.lane@imperial.ac.uk.
References
- 1.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Ruggeri ZM, De Marco L, Gatti L, Bader R, Montgomery RR. Platelets have more than one binding site for von Willebrand factor. J Clin Invest. 1983;72(1):1–12. doi: 10.1172/JCI110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockenstedt P, Greenberg JM, Handin RI. Structural basis of von Willebrand factor binding to platelet glycoprotein Ib and collagen: effects of disulfide reduction and limited proteolysis of polymeric von Willebrand factor. J Clin Invest. 1986;77(3):743–749. doi: 10.1172/JCI112369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth GJ, Titani K, Hoyer LW, Hickey MJ. Localization of binding sites within human von Willebrand factor for monomeric type III collagen. Biochemistry. 1986;25(26):8357–8361. doi: 10.1021/bi00374a004. [DOI] [PubMed] [Google Scholar]
- 5.Hamer RJ, Koedam JA, Beeser-Visser NH, Bertina RM, Van Mourik JA, Sixma JJ. Factor VIII binds to von Willebrand factor via its Mr-80,000 light chain. Eur J Biochem. 1987;166(1):37–43. doi: 10.1111/j.1432-1033.1987.tb13480.x. [DOI] [PubMed] [Google Scholar]
- 6.Mohri H, Yoshioka A, Zimmerman TS, Ruggeri ZM. Isolation of the von Willebrand factor domain interacting with platelet glycoprotein Ib, heparin, and collagen and characterization of its three distinct functional sites. J Biol Chem. 1989;264(29):17361–17367. [PubMed] [Google Scholar]
- 7.Moake JL, Turner NA, Stathopoulos NA, Nolasco LH, Hellums JD. Involvement of large plasma von Willebrand factor (vWF) multimers and unusually large vWF forms derived from endothelial cells in shear stress-induced platelet aggregation. J Clin Invest. 1986;78(6):1456–1461. doi: 10.1172/JCI112736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siedlecki CA, Lestini BJ, Kottke-Marchant KK, Eppell SJ, Wilson DL, Marchant RE. Shear-dependent changes in the three-dimensional structure of human von Willebrand factor. Blood. 1996;88(8):2939–2950. [PubMed] [Google Scholar]
- 9.Tsai HM, Sussman II, Nagel RL. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83(8):2171–2179. [PubMed] [Google Scholar]
- 10.Schneider SW, Nuschele S, Wixforth A, et al. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci U S A. 2007;104(19):7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furlan M, Robles R, Lammle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87(10):4223–4234. [PubMed] [Google Scholar]
- 12.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87(10):4235–4244. [PubMed] [Google Scholar]
- 13.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 14.Sadler JE. Von Willebrand factor, ADAMTS13, and thrombotic thrombocytopenic purpura. Blood. 2008;112(1):11–18. doi: 10.1182/blood-2008-02-078170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai HM, Sussman II, Ginsburg D, Lankhof H, Sixma JJ, Nagel RL. Proteolytic cleavage of recombinant type 2A von Willebrand factor mutants R834W and R834Q: inhibition by doxycycline and by monoclonal antibody VP-1. Blood. 1997;89(6):1954–1962. [PubMed] [Google Scholar]
- 16.Crawley JT, Lane DA, Woodward M, Rumley A, Lowe GD. Evidence that high von Willebrand factor and low ADAMTS-13 levels independently increase the risk of a non-fatal heart attack. J Thromb Haemost. 2008;6(4):583–588. doi: 10.1111/j.1538-7836.2008.02902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chion CK, Doggen CJ, Crawley JT, Lane DA, Rosendaal FR. ADAMTS13 and von Willebrand factor and the risk of myocardial infarction in men. Blood. 2007;109(5):1998–2000. doi: 10.1182/blood-2006-07-038166. [DOI] [PubMed] [Google Scholar]
- 18.Studt JD, Hovinga JA, Antoine G, et al. Fatal congenital thrombotic thrombocytopenic purpura with apparent ADAMTS13 inhibitor: in vitro inhibition of ADAMTS13 activity by hemoglobin. Blood. 2005;105(2):542–544. doi: 10.1182/blood-2004-06-2096. [DOI] [PubMed] [Google Scholar]
- 19.Bernardo A, Ball C, Nolasco L, Moake JF, Dong JF. Effects of inflammatory cytokines on the release and cleavage of the endothelial cell-derived ultralarge von Willebrand factor multimers under flow. Blood. 2004;104(1):100–106. doi: 10.1182/blood-2004-01-0107. [DOI] [PubMed] [Google Scholar]
- 20.Crawley JT, Lam JK, Rance JB, Mollica LR, O'Donnell JS, Lane DA. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005;105(3):1085–1093. doi: 10.1182/blood-2004-03-1101. [DOI] [PubMed] [Google Scholar]
- 21.Lam JK, Chion CK, Zanardelli S, Lane DA, Crawley JT. Further characterization of ADAMTS-13 inactivation by thrombin. J Thromb Haemost. 2007;5(5):1010–1018. doi: 10.1111/j.1538-7836.2007.02514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono T, Mimuro J, Madoiwa S, et al. Severe secondary deficiency of von Willebrand factor-cleaving protease (ADAMTS13) in patients with sepsis-induced disseminated intravascular coagulation: its correlation with development of renal failure. Blood. 2006;107(2):528–534. doi: 10.1182/blood-2005-03-1087. [DOI] [PubMed] [Google Scholar]
- 23.Feys HB, Vandeputte N, Palla R, et al. Inactivation of ADAMTS13 by plasmin as a potential cause of thrombotic thrombocytopenic purpura. J Thromb Haemost. 2010;8(9):2053–2062. doi: 10.1111/j.1538-7836.2010.03942.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagner DD, Olmsted JB, Marder VJ. Immunolocalization of von Willebrand protein in Weibel-Palade bodies of human endothelial cells. J Cell Biol. 1982;95(1):355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sporn LA, Chavin SI, Marder VJ, Wagner DD. Biosynthesis of von Willebrand protein by human megakaryocytes. J Clin Invest. 1985;76(3):1102–1106. doi: 10.1172/JCI112064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner DD, Marder VJ. Biosynthesis of von Willebrand protein by human endothelial cells: processing steps and their intracellular localization. J Cell Biol. 1984;99(6):2123–2130. doi: 10.1083/jcb.99.6.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsumi A, Tuley EA, Bodo I, Sadler JE. Localization of disulfide bonds in the cystine knot domain of human von Willebrand factor. J Biol Chem. 2000;275(33):25585–25594. doi: 10.1074/jbc.M002654200. [DOI] [PubMed] [Google Scholar]
- 28.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc Natl Acad Sci U S A. 1992;89(8):3531–3535. doi: 10.1073/pnas.89.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise RJ, Pittman DD, Handin RI, Kaufman RJ, Orkin SH. The propeptide of von Willebrand factor independently mediates the assembly of von Willebrand multimers. Cell. 1988;52(2):229–236. doi: 10.1016/0092-8674(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 30.McKinnon TA, Chion AC, Millington AJ, Lane DA, Laffan MA. N-linked glycosylation of VWF modulates its interaction with ADAMTS13. Blood. 2008;111(6):3042–3049. doi: 10.1182/blood-2007-06-095042. [DOI] [PubMed] [Google Scholar]
- 31.McKinnon TA, Goode EC, Birdsey GM, et al. Specific N-linked glycosylation sites modulate synthesis and secretion of von Willebrand factor. Blood. 2010;116(4):640–648. doi: 10.1182/blood-2010-02-267450. [DOI] [PubMed] [Google Scholar]
- 32.Canis K, McKinnon TA, Nowak A, et al. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J Thromb Haemost. 8(1):137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell JS, McKinnon TA, Crawley JT, Lane DA, Laffan MA. Bombay phenotype is associated with reduced plasma-VWF levels and an increased susceptibility to ADAMTS13 proteolysis. Blood. 2005;106(6):1988–1991. doi: 10.1182/blood-2005-02-0792. [DOI] [PubMed] [Google Scholar]
- 34.Schulte am Esch J, 2nd, Robson SC, Knoefel WT, Eisenberger CF, Peiper M, Rogiers X. Impact of O-linked glycosylation of the VWF-A1-domain flanking regions on platelet interaction. Br J Haematol. 2005;128(1):82–90. doi: 10.1111/j.1365-2141.2004.05253.x. [DOI] [PubMed] [Google Scholar]
- 35.Wagner DD, Saffaripour S, Bonfanti R, et al. Induction of specific storage organelles by von Willebrand factor propolypeptide. Cell. 1991;64(2):403–413. doi: 10.1016/0092-8674(91)90648-i. [DOI] [PubMed] [Google Scholar]
- 36.Giblin JP, Hewlett LJ, Hannah MJ. Basal secretion of von Willebrand factor from human endothelial cells. Blood. 2008;112(4):957–964. doi: 10.1182/blood-2007-12-130740. [DOI] [PubMed] [Google Scholar]
- 37.Dong JF, Moake JL, Nolasco L, et al. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100(12):4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324(5932):1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Zhang CZ, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466(7309):992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu T, Lin J, Cruz MA, Dong JF, Zhu C. Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood. 2010;115(2):370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uemura M, Tatsumi K, Matsumoto M, et al. Localization of ADAMTS13 to the stellate cells of human liver. Blood. 2005;106(3):922–924. doi: 10.1182/blood-2005-01-0152. [DOI] [PubMed] [Google Scholar]
- 42.Turner N, Nolasco L, Tao Z, Dong JF, Moake J. Human endothelial cells synthesize and release ADAMTS-13. J Thromb Haemost. 2006;4(6):1396–1404. doi: 10.1111/j.1538-7836.2006.01959.x. [DOI] [PubMed] [Google Scholar]
- 43.Fujikawa K, Suzuki H, McMullen B, Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98(6):1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 44.Soejima K, Mimura N, Hirashima M, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem (Tokyo) 2001;130(4):475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 45.Zheng X, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276(44):41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 46.Gerhardt S, Hassall G, Hawtin P, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373(4):891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 47.Mosyak L, Georgiadis K, Shane T, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17(1):16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shieh HS, Mathis KJ, Williams JM, et al. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2). J Biol Chem. 2008;283(3):1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- 49.Anderson PJ, Kokame K, Sadler JE. Zinc and calcium ions cooperatively modulate ADAMTS13 activity. J Biol Chem. 2006;281(2):850–857. doi: 10.1074/jbc.M504540200. [DOI] [PubMed] [Google Scholar]
- 50.Gardner MD, Chion CK, de Groot R, Shah A, Crawley JT, Lane DA. A functional calcium-binding site in the metalloprotease domain of ADAMTS13. Blood. 2009;113(5):1149–1157. doi: 10.1182/blood-2008-03-144683. [DOI] [PubMed] [Google Scholar]
- 51.Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin Cell Dev Biol. 2009;20(2):146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106(46):19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis AK, Makar RS, Stowell CP, Kuter DJ, Dzik WH. ADAMTS13 binds to CD36: a potential mechanism for platelet and endothelial localization of ADAMTS13. Transfusion. 2009;49(2):206–213. doi: 10.1111/j.1537-2995.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- 54.Soejima K, Matsumoto M, Kokame K, et al. ADAMTS-13 cysteine-rich/spacer domains are functionally essential for von Willebrand factor cleavage. Blood. 2003;102(9):3232–3237. doi: 10.1182/blood-2003-03-0908. [DOI] [PubMed] [Google Scholar]
- 55.Zheng X, Nishio K, Majerus EM, Sadler JE. Cleavage of von Willebrand factor requires the spacer domain of the metalloprotease ADAMTS13. J Biol Chem. 2003;278(32):30136–30141. doi: 10.1074/jbc.M305331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ai J, Smith P, Wang S, Zhang P, Zheng XL. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280(33):29428–29434. doi: 10.1074/jbc.M505513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao Z, Wang Y, Choi H, et al. Cleavage of ultralarge multimers of von Willebrand factor by C-terminal-truncated mutants of ADAMTS-13 under flow. Blood. 2005;106(1):141–143. doi: 10.1182/blood-2004-11-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang P, Pan W, Rux AH, Sachais BS, Zheng XL. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von Willebrand factor under flow. Blood. 2007;110(6):1887–1894. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W, Bouhassira EE, Tsai HM. An IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Blood. 2007;110(3):886–893. doi: 10.1182/blood-2007-01-070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Maeyer B, De Meyer SF, Feys HB, et al. The distal carboxyterminal domains of murine ADAMTS13ba influence proteolysis of platelet-decorated VWF strings in vivo. J Thromb Haemost. 2010;8(10):2305–2312. doi: 10.1111/j.1538-7836.2010.04008.x. [DOI] [PubMed] [Google Scholar]
- 61.Tao Z, Peng Y, Nolasco L, et al. Recombinant CUB-1 domain polypeptide inhibits the cleavage of ULVWF strings by ADAMTS13 under flow conditions. Blood. 2005;106(13):4139–4145. doi: 10.1182/blood-2005-05-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zanardelli S, Chion AC, Groot E, et al. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114(13):2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7(12):2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crawley JT, de Groot R, Luken BM. Circulating ADAMTS-13-von Willebrand factor complexes: an enzyme on demand. J Thromb Haemost. 2009;7(12):2085–2087. doi: 10.1111/j.1538-7836.2009.03621.x. [DOI] [PubMed] [Google Scholar]
- 65.Banno F, Chauhan AK, Kokame K, et al. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009;113(21):5323–5329. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Identification of strain-specific variants of mouse Adamts13 gene encoding von Willebrand factor-cleaving protease. J Biol Chem. 2004;279(29):30896–30903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Q, Zhou YF, Zhang CZ, Zhang X, Lu C, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106(23):9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281(3):1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 69.Luken BM, Winn LY, Emsley J, Lane DA, Crawley JT. The importance of vicinal cysteines, C1669 and C1670, for von Willebrand factor A2 domain function. Blood. 2010;115(23):4910–4913. doi: 10.1182/blood-2009-12-257949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shim K, Anderson PJ, Tuley EA, Wiswall E, Sadler JE. Platelet-VWF complexes are preferred substrates of ADAMTS13 under fluid shear stress. Blood. 2008;111(2):651–657. doi: 10.1182/blood-2007-05-093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kokame K, Matsumoto M, Fujimura Y, Miyata T. VWF73, a region from D1596 to R1668 of von Willebrand factor, provides a minimal substrate for ADAMTS-13. Blood. 2004;103(2):607–612. doi: 10.1182/blood-2003-08-2861. [DOI] [PubMed] [Google Scholar]
- 72.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci U S A. 2006;103(50):19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu JJ, Fujikawa K, McMullen BA, Chung DW. Characterization of a core binding site for ADAMTS-13 in the A2 domain of von Willebrand factor. Proc Natl Acad Sci U S A. 2006;103(49):18470–18474. doi: 10.1073/pnas.0609190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klaus C, Plaimauer B, Studt JD, et al. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103(12):4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 75.Pos W, Crawley JT, Fijnheer R, Voorberg J, Lane DA, Luken BM. An autoantibody epitope comprising residues R660, Y661, and Y665 in the ADAMTS13 spacer domain identifies a binding site for the A2 domain of VWF. Blood. 2010;115(8):1640–1649. doi: 10.1182/blood-2009-06-229203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin SY, Skipwith CG, Zheng XL. Amino acid residues Arg(659), Arg(660), and Tyr(661) in the spacer domain of ADAMTS13 are critical for cleavage of von Willebrand factor. Blood. 2010;115(11):2300–2310. doi: 10.1182/blood-2009-07-235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112(5):1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Groot R, Bardhan A, Ramroop N, Lane DA, Crawley JT. Essential role of the disintegrin-like domain in ADAMTS13 function. Blood. 2009;113(22):5609–5616. doi: 10.1182/blood-2008-11-187914. [DOI] [PubMed] [Google Scholar]
- 79.Pruss CM, Notley CR, Hegadorn CA, O'Brien LA, Lillicrap D. ADAMTS13 cleavage efficiency is altered by mutagenic and, to a lesser extent, polymorphic sequence changes in the A1 and A2 domains of von Willebrand factor. Br J Haematol. 2008;143(4):552–558. doi: 10.1111/j.1365-2141.2008.07266.x. [DOI] [PubMed] [Google Scholar]
- 80.Xiang Y, de Groot R, Crawley JT, Lane DA. Mechanism of von Willebrand factor scissile bond cleavage by a disintegrin and metalloproteinase with a thrombospondin type I motif, member 13 (ADAMTS13) [published online ahead of print June 24, 2011]. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1018559108. doi: 10.1073/pnas.1018559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Groot R, Lane DA, Crawley JT. The ADAMTS13 metalloprotease domain: roles of subsites in enzyme activity and specificity. Blood. 2010;116(16):3064–3072. doi: 10.1182/blood-2009-12-258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129(1):93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 83.Miura S, Li CQ, Cao Z, Wang H, Wardell MR, Sadler JE. Interaction of von Willebrand factor domain A1 with platelet glycoprotein Ibalpha-(1-289): slow intrinsic binding kinetics mediate rapid platelet adhesion. J Biol Chem. 2000;275(11):7539–7546. doi: 10.1074/jbc.275.11.7539. [DOI] [PubMed] [Google Scholar]
- 84.Shida Y, Nishio K, Sugimoto M, et al. Functional imaging of shear-dependent activity of ADAMTS13 in regulating mural thrombus growth under whole blood flow conditions. Blood. 2008;111(3):1295–1298. doi: 10.1182/blood-2007-09-110700. [DOI] [PubMed] [Google Scholar]