Abstract
Isothiocyanates are versatile cancer-preventive compounds. Evidence from animal studies indicates that the anticarcinogenic activities of ITCs involve all the major stages of tumor growth: initiation, promotion and progression. Epidemiological studies have also shown that dietary intake of ITCs is associated with reduced risk of certain human cancers. A number of mechanisms have been proposed for the chemopreventive activities of ITCs. To identify the molecular targets of ITCs is a first step to understand the molecular mechanisms of ITCs. Studies in recent years have shown that the covalent binding to certain protein targets by ITCs seems to play an important role in ITC-induced apoptosis and cell growth inhibition and other cellular effects. The knowledge gained from these studies may be used to guide future design and screen of better and more efficacious compounds. In this review, we intend to cover all potential protein targets of ITCs so far studied and summarize what are known about their binding sites and the potential biological consequences. In the end, we also offer discussions to shed light onto the relationship between protein binding and reactive oxygen species generation by ITCs.
Introduction
Isothiocyanates (ITCs), including benzyl ITC (BITC), phenethyl ITC (PEITC) and sulforaphane (SFN) are versatile compounds that are effective in preventing various cancers including breast, lung, colon and prostate in laboratory animals (1). Epidemiological studies have also shown that dietary intake of ITCs is associated with reduced risk of certain human cancers, and the effects are influenced by genetic polymorphisms in glutathione transferase (2). Evidence from animal studies indicates that the anticarcinogenic activity of ITCs involves all three major stages of tumor growth: initiation, promotion and progression. Several mechanisms have been proposed for the chemopreventive activity of ITCs: (i) inhibition of cytochrome P450s for carcinogen activation; (ii) induction of phase II enzymes for detoxification of carcinogens; (iii) induction of cell cycle arrest and apoptosis and (iv) induction of anti-inflammatory activity. At cellular and molecular level, the cancer preventive activities of ITCs may arise from direct and indirect interactions between ITCs and cellular components. The indirect interactions refer to activation of gene transcription and expression, such as activation of phase II and antioxidant enzymes, whereas direct interactions, which are the focus of this review, denote the covalent binding/modification of certain nucleophile-containing proteins by ITCs postsynthetically. The ‘target’ here is defined as molecules that are subjected to direct binding of ITCs.
ITCs as electrophiles could directly bind to DNA, RNA, proteins and peptides, which may constitute early events-triggering apoptosis. Using 14C-PEITC and 14C-SFN, we found no detectable binding with DNA or RNA in A549 human non-small cell lung cancer cells (3), suggesting that DNA or RNA is unlikely to be a major target of ITCs in cells. These results support previous findings that the genotoxicity induced by ITCs may be caused by reactive oxygen species (ROS), not ITC-DNA adductions, and the genotoxic effects may be attenuated by proteins in vivo, presumably via binding with ITCs (4). The modification of amino acid residues in proteins depends on pH, steric factors, protein microenvironments as well as the chemical properties of modifying reagents (5). Covalent binding to cysteine residues or other nucleophilic amino acids by ITCs may influence enzymatic activity, redox status and signaling transduction. In the examination of ITC binding with cellular proteins, we used both PEITC and SFN; the former contains a hydrophobic phenethyl group and the latter has a hydrophilic sulfinyl group. These studies demonstrated that although PEITC is a stronger inducer of apoptosis than SFN, it causes less ROS generation and oxidative damage than SFN, raising questions about the role of ROS generation in apoptosis induction by ITCs (3,6). Our studies further showed that although the initial cellular uptake of PEITC and SFN is comparable, PEITC binds to proteins at a level 2.7 times greater than that of SFN (3). More importantly, PEITC–protein binding increases with time, whereas protein binding by SFN remains low throughout incubation. Therefore, after 4 h, the level of PEITC–protein conjugate in cells was found to be >6-fold that of the SFN conjugate, indicating that protein binding is the predominant reaction with PEITC, accounting for 87% of its total uptake, as compared with only 12% for SFN. Based on these results, it was concluded that the protein-binding affinities, not the ROS generation, correlated with apoptosis induction, suggesting that proteins may be important targets for ITCs.
To identify the specific protein targets of ITCs in cells, the total proteins from cells treated with 14C-PEITC and 14C-SFN, were analyzed by 2-D gel electrophoresis using proteomics (7). Only a small number of protein spots were found to contain radioactivity, suggesting that ITC binding to cellular proteins is rather selective. Among >30 potential target proteins identified, tubulin was shown to be a major target. Tubulin constitutes cytoskeleton microtubules and is a well-known target for certain cancer chemotherapeutic agents. Subsequently, we showed that binding to tubulin is associated with the cell cycle arrest and apoptosis induced by ITCs (7). More recently, macrophage migration inhibitory factor (MIF) was also identified as another ITC-binding target through affinity chromatography by several laboratories (8–10). Because MIF is a proinflammatory and protumorigenic cytokine, inhibition of MIF by ITCs may constitute a novel mechanism by which dietary ITCs inhibit carcinogenesis. Identification of the protein targets of ITCs is an important first step to understand the molecular mechanisms by which ITCs exert their anticarcinogenic activities. It is likely to provide us the knowledge and tools for the future design and screening of better and more efficacious compounds.
To identify and validate a target of ITCs, one must demonstrate binding to the protein by ITCs, including the specific binding sites; the chemical and biological characteristics of binding, including conformational and functional changes of the protein and most importantly, the functional consequences, including binding-induced signaling changes and downstream effects. This review focuses on most, but not all, potential protein targets that are subjected to direct binding by ITCs. Because the study of ITC molecular targets is an emerging field, most of the proteins discussed in this review may not meet the stringent criteria. Nonetheless, these proteins are considered likely targets of ITCs based on circumstantial evidence. More studies are needed to validate their role as targets of ITCs.
Glutathione as a target
The tripeptide glutathione (GSH), a major cysteine-containing small molecular antioxidant, is an ITC target. ITCs react readily with the cysteine residue in GSH through thiocarbamation, a covalent bond formation between the carbon atom in the ITC moiety and the sulfhydryl group in GSH (Figure 1A). In cells, ITC binding to GSH occurs rapidly and is probably assisted by glutathione transferase (2,11,12). The fast reaction results in significant enrichment of intracellular ITCs. The concentrations of PEITC–GSH and SFN–GSH conjugates in cells can reach up to 5 mM, 100–200 times than that in the incubation medium of murine hepatoma Hepa 1c1c7 cells (13). We showed that PEITC–GSH and SFN–GSH conjugates were detected in A549 cells as early as 15 min upon treatment (3). The peak levels were reached at 30 min and ∼60% of PEITC and 84% of SFN were detected as GSH conjugates. Because of the high concentration of intracellular GSH (1–10 mM), binding to GSH is believed to be a major driving force for the ITC enrichment. This was confirmed using L-buthionine-sulfoximine (BSO) as a GSH depleting agent. Treatment of Hepa 1c1c7 cells with BSO caused a sharp reduction (>60%) in the uptake of BITC and SFN (13). We also observed that after treating A549 cells with 100 μM BSO, the uptakes of PEITC and SFN are reduced by 30% and their conjugation with GSH and cellular proteins are equally reduced (Lixin Mi, Brian L. Hood, Nicolas A. Stewart, Zhen Xiao, Sudha Govind, Xiantao Wang, Thomas P. Conrads, Timothy D. Veenstra, and Fung-Lung Chung, unpublished results). In a material balance study using A549 cells treated with 14C-ITCs (3), we verified that intracellular GSH is a major small molecule target of ITCs (3). Consistent with this, the mercapturic acid pathway is shown to be the predominant route of metabolism of ITCs in rodents and humans (3,13–17). As illustrated in Figure 2, ITC–GSH is enzymatically converted sequentially to ITC–Cys–Gly, ITC–Cys and ITC–N-acetyl–Cys. These conjugates can be expelled from cells through molecular pumps on the cell membrane (13–17).
Fig. 1.
Schematic reactions between ITCs and proteins.
Fig. 2.
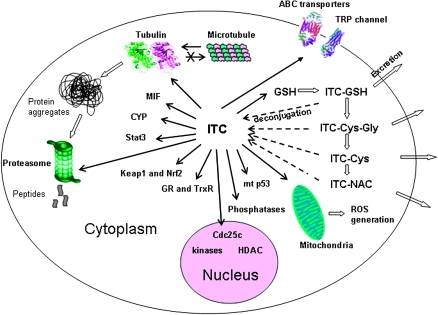
Potential major protein targets of ITCs.
A previous study indicated that ITCs are accumulated in cells predominantly as ITC conjugates and the concentrations of free ITCs are relatively low in cells (18). It should also be noted that the all ITC conjugates/metabolites, including ITC–GSH, deconjugate to reach equilibrium with the free ITC (11,18–21). The proportion of the conjugate and free ITC in the reaction medium depends on factors such as pH and concentrations of ITC and thiols. For example, the conjugates are stabilized in acidic medium, whereas basic conditions favor dissociation (19). More importantly, an earlier study showed that ITC conjugates can undergo exchange reactions in the presence of other thiols, such as GSH, cysteine or N-acetylcysteine at pH 7.4 (20), suggesting that high concentration of intracellular ITC conjugates may serve as a major source of free ITC for binding to target proteins. This notion is supported by the results from several studies that ITCs conjugates exhibit biological activities similar to those of free ITCs by acting as prodrugs (21,22) and ITCs rapidly accumulate in cells that are exposed to ITC–GSH conjugates (18).
Binding of GSH with ITCs causes depletion of GSH, ROS generation and oxidative damage on DNA and proteins (3,6). However, the notion that binding to GSH is a major factor that contributes to ITC-induced apoptosis is still questionable because there seems a lack of correlation between ITC-induced apoptosis and ITC binding to GSH. For example, although BITC and PEITC are more potent in inducing apoptosis than SFN at the same concentration, results from our and another lab showed that SFN depletes similar level or a little more GSH than PEITC and BITC in A549 and Hepa 1c1c7 cells (3,18). It was probably due to a higher pseudo-first reaction rate constant between GSH and SFN than PEITC (3). Our recent studies indicated that SFN depletes slightly more GSH than BITC and PEITC in HeLa and U266 cells lines (17), although the potency of apoptosis induction by ITCs in both cell lines follows: BITC > PEITC > SFN. Together, the results suggest that GSH as an ITC target molecule may only have a limited role in apoptosis induction by ITCs.
Proteins as targets
In addition to form thiocarbamates with cysteines (5,19,23), ITCs may react with α-amino groups in N-terminal residues and ϵ-amino containing lysines through alkylation, forming thiourea (Figure 1B). The alkylation is irreversible but the rate of the reaction is at least 1000 times less than thiocarbamation (19). Several recent studies have shown that ITCs react with the N-terminal proline of MIF (8–10), indicating for the first time that ITCs bind to the secondary amine group in cells (Figure 1C). Binding to hydroxyl group-containing residues (Figure 1D), such as tyrosine, may also occur but not at physiological pH (23,24). Like thiol conjugates, the hydroxyl conjugates are unstable and can be deconjugated by dilution or pH changes (23).
The reactivity of nucleophilic residues in a protein toward ITCs depends largely on their pKa, which is known to be affected by surrounding amino acid residues (25). The highly conserved N-terminal proline of MIF has a pKa of 5.9, a significant reduction from 9.4 of an isolated proline, due to its microenvironment of protein structure (26). The reduced pKa not only enables the proline to be a catalytic center for tautomerase activity but also may increase its reactivity to ITCs. The proline has been found to be the only binding site on MIF, despite the fact that it has three conserved cysteines, Cys57, Cys60 and Cys81 (8–10).
Cysteine as the binding site
It is well known that cysteine, which is often located in the catalytic centers or the metal-binding domains of enzymes, plays a pivotal role in a variety of protein functions. Because of its critical roles, modifications of cysteine residues, such as oxidation, glutathiolation, nitrosylation, disulfide formation and formation of thioether and thioester, have been shown to have numerous biological functional consequences, especially in signaling transduction (5,27). Not all cysteine residues are reactive toward electrophiles. The reactivity of a cysteine depends on its protonation state, which is dictated by its pKa value (27). A free cysteine has a pKa of 8.3, whereas reactive cysteines that are often in thiolated form exhibit neutral or even acidic pKa values. The pKa value of a cysteine residue is known to be affected by electrostatic interactions with nearby residues. For example, a neighboring cationic arginine residue can reduce the pKa value and increase the reactivity of the cysteine residue to 4-hydroxynonenal, an electrophile by lipid peroxidation (28). In addition to electrostatic interaction, other aspects of microenvironment, such as hydrophobicity and steric hindrance, may also affect the reactivity of a cysteine residue (27).
The following proteins have been conceived as potential targets of ITCs with binding sites on cysteine residues. Their binding characteristics with ITCs and biological effects are discussed.
Cytochrome P450s.
Phase I enzymes (cytochrome P450s) are a group of heme proteins that participate in oxidation, reduction or hydrolysis of xenobiotics, including carcinogens. They also produce reactive intermediates and electrophiles that may react with cellular DNA, RNA and proteins. Inhibition of P450s is believed to be a valid mechanism for suppressing carcinogenesis. A number of natural and synthetic ITCs, including BITC, PEITC and SFN, have been shown to inhibit the activities of several P450s such as 1A1, 1A2, 2A6, 2A13, 2B1, 2B6, 2C9, 2C19, 2E1, 2D6 and 3A4 in either expressed and purified proteins or in lysate extracted from biological samples (29). Direct binding of one or more nucleophilic residues of P450s by ITCs is believed to be a potential mechanism of P450s inactivation (30). The notion is also supported by observations that there is no detectable loss of native heme and ITC-heme adducts suggests that P450s inhibition is due to ITC modification of a residue in the active site. In a previous study, a mass increase of 115 Da on P450 2E1 was detected after treatment with tert-butyl ITC (molecular weight 118 Da) suggests that tert-butyl ITC covalently binds to one of the four Cys residues of P450 2E1 (31). In another study, BITC at 1 and 2.5 μM inhibited 2A13-mediated nicotine-derived nitrosamine ketone metabolism and 2A6-mediated nicotine metabolism by 40 and 84%, respectively (32). PEITC was also effective in inhibition of these enzymes. Mass spectrometry (MS) analysis showed incremental masses of 159 and 187 Da in P450 2A13 after ITC treatment, corresponding, respectively, to the mass of BITC (149 Da) and PEITC (163 Da) with 1–2 oxygen atoms. Therefore, it was proposed that inhibition mediated by NADPH is also involved in P450s covalent modification by a yet to be identified metabolically activated ITC intermediate (29). Bioactive isocyanate has been identified in vitro as a potential metabolite of ITC in the microsomal system. However, it is unlikely a major metabolite of ITCs in vivo because the mercapturic acid pathway is by far the major metabolic pathway for ITCs and the metabolic rate of conversion from ITC to isocyanate is slow (33,34).
Keap1 and Nrf2.
The Keap1–Nrf2 system is believed to be the major regulatory pathway of cytoprotective gene expression against oxidative stress and electrophiles (35). Several ITCs, including SFN and PEITC, have been shown to induce Keap1–Nrf2-mediated antioxidant responses, involving upregulation of GSH levels and induction of phase II enzymes and antioxidant enzyme expression (12). Keap1, a cysteine-rich protein, is believed to be not only an active redox sensor but also a hypothetical target of ITCs (36). Studies indicated that under unstressed conditions, Nrf2 binds to Keap1–Cullin3 complex and the binding between Keap1 and the DLG motif of Nrf2 is associated with ubiquitin-proteasome degradation (37). Thiol modifications at Cys151 of the BTB region (named for Drosophila broad-complex C, Tramtrack and Bric-a-brac) and Cys273 and Cys288 in the central linker region in Keap1 are essential to Nrf2 translocation to the nucleus where it activates gene expression (38,39). However, others showed that modifications at these sites are not sufficient to release Nrf2 from Keap1 due to the stronger binding at the ETGE motif (40). Therefore, other potential binding sites on Keap1 by electrophiles have been proposed: Cys77 in the BTB domain, Cys226, Cys249 and Cys257 in the intervening domain, Cys489, Cys513, Cys518 and Cys583 in the Kelch domain and Cys624 in the C-terminus (reviewed in ref. (41)). Despite the success in detecting a number of cysteines, including Cys257, Cys273, Cys288 and Cys297, as putative binding sites of electrophiles in vitro, none have been confirmed in vivo.
In addition to the trans-regulation role of Keap1 in Nrf2 activation, a redox-sensitive nuclear exporting signal motif has been reported in the Neh5 domain of Nrf2 (42). Cys183, which is embedded in the nuclear exporting signal motif, appears to mediate the redox sensitivity. Results from site-directed mutagenesis showed that the C183A mutation remarkably reduces Nrf2 translocation kinetics and inhibits Nrf2-mediated transcriptional activity, suggesting that direct modification of the Cys183 by electrophiles such as SFN may serve as a Keap1-independent Nrf2 activation signal.
Adenosine triphosphatase.
The adenosine diphosphate (ATP)-binding cassette superfamily of membrane transporters, also called the phase III enzymes, include P-glycoprotein (P-gp) and the multidrug resistance-associated protein family. ATP-binding cassette transporters are responsible for exporting a large variety of compounds, including xenobiotics, anticancer drugs and metabolic intermediates, from cells in an ATP-dependent manner. They are often overexpressed in tumor tissues and have been associated with drug resistance and chemotherapy failure. Numerous ITCs, including BITC and PEITC at 100 μM, have been demonstrated to inhibit P-gp and breast cancer resistance protein (BCRP)-mediated efflux of several anticancer drugs in a variety of cell types (43,44). Further study indicated that BITC, PEITC, the synthetic 1-naphthyl ITC, 4-phenylbutyl ITC and 6-phenylhexyl ITC can inhibit adenosine triphosphatase (ATPase) activity of BCRP (45). Interestingly, the inhibition is selective toward cells with overexpression of BCRP. In fact, fluorescein ITC (FITC) and p-bromophenyl ITC have been demonstrated to almost completely inhibit purified Na+- and K+-ATPase through covalent binding (46,47). The binding of FITC occurs at a stoichiometry of about one FITC per ATP-binding site, and the inhibition can be blocked by ATP, suggesting that the binding site of FITC is probably at the ATP-binding site (48). A later study showed that FITC inhibits Na+/K+-ATPase through specific binding to Lys480 and/or Lys501 at the ATP-binding site to block ATP access (49). However, studies with site-directed mutagenesis excluded the significance of the lysines, which prompted some to consider that binding to the Cys549 residue near the ATP-binding site may be responsible for the ATPase inhibition (50,51).
Tubulin.
Microtubules, one of the major components of the cytoskeleton, are involved in many cellular processes including mitosis, cytokinesis, vesicular transport and structural support (52). They are assembled from dimers of α- and β-tubulin. The dynamic equilibrium between tubulin (dimer) and microtubules (polymer) is not only essential for a variety of cellular functions but also serves as valid therapeutic target. Tubulin-binding agents, like taxanes that stabilize microtubules and colchicine analogs and vincas that destabilize them, are an important class of current chemotherapeutic drugs (52). Recently, both α- and β-tubulin were discovered as potential binding targets of PEITC and SFN in our lab using radiolabeled ITCs and a 2-D gel electrophoresis-based proteomic technique (7). Using the Ellman assay, we showed that ITCs have differential binding affinities to tubulin with an order of BITC > PEITC > SFN. N-methyl phenethylamine, a structural analog of PEITC without the ITC functional group, has no binding affinity for tubulin, indicating that the ITC functional group is essential for the binding and the side groups may dictate the binding activity.
Structural studies using circular dichroism indicated that BITC binding to tubulin modifies the helical content of tubulin, a sign of conformational change (7). Also, monitoring intrinsic tryptophan fluorescence intensity by ITC titration showed that the potency of inducing tubulin tertiary structural changes is BITC > PEITC > SFN. This potency order was also observed in the functional consequences of ITC binding to tubulin, which include (i) inhibition of tubulin polymerization in vitro; (ii) disruption of microtubule networks and (iii) induction of tubulin-containing protein aggregates (53,54). The potencies correlated well with the downstream induction of G2/M phase arrest and apoptosis and ultimately inhibition of cell growth, suggesting that ITC binding to tubulin is a biologically important upstream event (7). The concentration range in cell culture studies to observe microtubule disruption, tubulin aggregation and degradation, and cell growth inhibition by ITCs was from 5 to 15 μM. To confirm that tubulin is the ITC-binding target, we treated purified porcine tubulin with BITC, PEITC and SFN in vitro. MALDI-Tandem MS indicated that all three ITCs bind to Cys303 in β-tubulin. Furthermore, using nanoRPLC-MS/MS, we analyzed tubulin-containing protein aggregates extracted from cell lysates from A549 cells treated with 20 μM BITC and a BITC adduct with Cys347 was detected in α-tubulin (7). This provided the first in vivo evidence of binding between ITC and a cellular target protein.
More recently, we found that ITC binding to tubulin triggers rapid degradation of both α- and β-tubulin leading to apoptosis in a variety of cell types (53). Our results showed that tubulin degradation consists of two sequential steps: firstly, ITC-bound tubulin forms aggresome-like protein aggregates that are insoluble to non-ionic detergents (54) and secondly, these aggregates are degraded through the ubiquitin-proteasome system (53). Despite the fact that a number of proteasome inhibitors and an ATP synthesis inhibitor can block the second step, the precipitation of tubulin can only be blocked by tubulin-binding agents (i.e. colchicine, vinblastine and taxol), supporting the notion that ITC binding to tubulin triggers tubulin misfolding and aggregation. Consistent with this, tubulin aggregation is proportional to concentrations of ITCs. However, tubulin degradation in cells is significantly inhibited at >20 μM of BITC and PEITC (49). These results not only support the two stage tubulin degradation model but also agree with our recent finding that proteasome is another potential target of ITCs (see below, ref. 17).
Transient receptor potential channel.
Transient receptor potential cation channel(TRPA1, subfamily A, member 1), a nonselective cation channel expressed by sensory neurons of the pain pathway, is another potential target of ITCs (55). Both allyl ITC and BITC at 20 μM were shown to activate TRPA1-expressing Xenopus oocytes and HEK293 cells transfected with TRPA1 (55). Further study substituted three closely spaced N-terminal cysteines (Cys619, Cys639 and Cys663) of TRPA1 with either serine or alanine, and the resulting triple mutant showed little response to ITC treatment (55). Although the study did not demonstrate direct ITC binding to specific cysteine residues, the evidence is compelling that electrophiles, including ITCs, may activate TRPA1 mainly through direct covalent modification of N-terminal cysteine residues within the channel protein. The study also identified that lysine 708 can be irreversibly modified by ITCs. However, it is believed that the lysine modification only plays a limited role in TRPA1 activation.
Phosphatase M3/6 and cdc25c.
c-Jun N-terminal kinase (JNK), a member of the mitogen-activated protein kinase family, plays an important role in a variety of cellular stress responses, including mitogenic stimuli, fluid shearing, proinflammatory cytokines and chemical treatments. A previous study showed that PEITC at 10 and 20 μM induces apoptosis through a JNK-mediated pathway (56). The fact that PEITC shows little activity toward MKK4 or MKK7 and JNK suggests that JNK and its upstream kinases are not primary targets of PEITC. Instead, PEITC inhibited JNK dephosphorylation and its deactivation rate, suggesting that JNK-specific phosphatase M3/6 may be a PEITC target. In fact, M3/6 as well as other dual-specificity phosphatases contains an essential cysteine residue (Cys246 for M3/6) in their catalytic cores. Modification of the cysteine residue by an electrophile could lead to inhibition of its activity. Another potential target in the phosphatase family is Cdc25c, an enzyme that dephosphorylates cyclin B-bound Cdc2 and triggers cell entry into mitosis. Its activity was found to be substantially inhibited by PEITC at 10 μM and SFN at 20 μM (57,58). Furthermore, the observation that expression levels of M3/6 and Cdc25c were downregulated by PEITC via a proteasome-dependent mechanism may suggest a mechanism similar to that of tubulin in which the protein is misfolded via ITC binding and further degraded by the proteasome.
Kinases MEKK1, epidermal growth factor receptor and PKC.
In addition to inhibiting phosphatases, ITCs are also effective at inhibiting several kinases (59–62). MEKK1 is an important upstream-signaling molecule that regulates the stress response mechanism of cells, such as the activation of stress-activated protein kinase and JNK. A recent study showed that PEITC at 50, 100 and 200 μM inhibits MEKK1 through direct modification of Cys1238 (59). This claim is consistent with the observation that PEITC treatment has no inhibitory activity against MEKK1 with a C1238V mutation. Since Cys1238 is in the ATP-binding domain of the MEKK1, it is conceivable that other kinases may also be the targets of ITC binding. This notion is supported by reports that both PEITC and alkyl ITCs both at 10 μM can suppress epidermal growth factor receptor kinase activity (60,61). The downregulation of protein level and activity of protein kinase C (PKC) in HeLa cells, including PKC alpha and epsilon, by PEITC and SFN at up to 5 μM has also been reported (62). These results suggest that the inhibition of certain kinases via direct binding may constitute a specific mechanism for the cancer preventive activity of ITCs.
GR and TrxR.
Glutathione reductase (GR) and thioredoxin reductase (TrxR) supply the electrons required for the function of a variety of enzymes and maintain redox balance. They play important roles in cell proliferation, cell cycle regulation and apoptosis. A previous study showed that AITC, BITC, PEITC and SFN can significantly inhibit the activities of purified GR and TrxR in a time-dependent manner with IC50s in the lower micromolar range (63). The potency of inhibition follows the order: BITC > AITC > PEITC > SFN. Because the inhibition requires reductases to be in the reduced form and dialysis of ITC-treated reductases cannot recover their activity, it is thought that covalent binding by ITCs to the active sites of GR and TrxR contributes to the inhibition. It has been reported that the disulfide bond between Cys58 and Cys63 in GR has to be reduced before binding to the substrate glutathione disulfide, GSSG (64). Therefore, these cysteines could be potential ITC-binding sites for inhibiting GR activity. There are two redox centers in TrxR, one containing Cys59 and Cys64 and the other Cys497 and SelenoCys498. These residues are all essential for its enzymatic activity and therefore are potential binding sites for electrophiles (65). TrxR was identified as one of the potential ITC targets in our radiolabel-2-D gel electrophoresis mentioned above (Lixin Mi, Brian L. Hood, Nicolas A. Stewart, Zhen Xiao, Sudha Govind, Xiantao Wang, Thomas P. Conrads, Timothy D. Veenstra, and Fung-Lung Chung, unpublished results).
Activator protein 1.
AP-1, a heterodimeric protein composed of components from the c-Fos, c-Jun, ATF and JDP families, is a transcriptional factor that regulates gene expression in response to a variety of stimuli, including cytokines, growth factors, stress and bacterial and viral infections. It is also believed that AP-1 is a potential target for the prevention of certain cancer types (66). A recent study found that SFN either 1 or 2.5 μmol/mouse inhibits ultraviolet B-induced skin carcinogenesis in SKH-1 mice (67). Detailed study indicated that SFN effectively inhibits ultraviolet B-induced AP-1 binding to TRE (12-O-tetradecanoylphorbol-13-acetate response element) in the gene promoter region and suppressed the AP-1-mediated gene transcription. To study whether SFN blocking binding of AP-1 to the TRE is due to modification of some key cysteines at its DNA-binding domain, site-directed mutagenesis was employed to mutate Cys154 in c-Fos and Cys272 in c-Jun to serines. The results showed that mutants are less sensitive to treatments with SFN or diamide, another cysteines modification agent, indicating that direct modification of these cysteines by SFN may underlie its inhibitory activity.
Proteasome.
The ubiquitin-proteasome system plays a critical role in regulating cell growth and survival. The recent Food Drug Administration approval of Bortezomib, the first therapeutic proteasome inhibitor, indicates that inhibition of proteasome function has emerged as a promising strategy for anticancer therapy. The proteasome was also identified as a potential ITC-binding target in our radiolabeled-2-D gel electrophoresis assay. Our recent studies (17) showed that BITC and PEITC at 10–30 μM significantly inhibit proteasome activities, including chymotrypsin-like, trypsin-like and caspase-like, in a variety of cell types. The inhibition against both the 26S and 20S proteasome is probably through direct binding because both ITCs inhibit activity in purified proteasome and N-methyl phenethylamine, an analog without the ITC functional group, did not inhibit proteasome activity under the same conditions. The results also showed that ROS generation and ITC-induced protein aggregation are less probably the triggering events for proteasome inhibition by ITCs and the potency of proteasome inhibition correlates with the rapid accumulation of p53 and IκB. More importantly, BITC and PEITC, the two strong proteasome inhibitors, significantly suppressed cell growth of multiple myeloma, a blood cancer type that is sensitive to proteasome inhibition, through the induction of G2/M cell cycle arrest and apoptosis. The half maximal growth inhibitory concentration values for BITC at 24 and 48 h were 8.3 and 4.7 μM, respectively. Taken together, these data support the idea that the proteasome, like tubulin, is a potential target of ITCs.
Histone deacetylase.
Histone deacetylases (HDAC) are another possible target of ITCs. SFN has been conceived as an HDAC inhibitor (68). Specifically, HDAC activity in SFN-treated cells is inhibited with a concomitant increase in global acetylation of histones, including histones H3 and H4. The inhibition coincides with transactivation of p21 and Bax and induction of G2/M cell cycle arrest and apoptosis. Biochemical assays showed that the metabolites of SFN-GSH conjugate, including SFN–NAC and SFN–Cys, rather than the parent compound, inhibited HDAC activity in vitro. Based on computer modeling, the carboxylate groups of SFN-Cys and SFN-NAC interact with a zinc atom and the surrounding amino acids at the HDAC active site, whereas the rest of their structures physically fit into the channel of the active site. Although a unique model other than the one involving covalent modification by ITCs was proposed in this study, this model needs additional verification. Firstly, the Cys and NAC conjugates of ITCs are known to be unstable and dissociate to parent ITCs; secondly, little or no inhibition was observed when purified recombinant HDAC1 and HDAC2 were incubated with up to 30 μM of BITC, PEITC or SFN for up to 4 h in our lab (7), suggesting that HDAC may not be a direct-binding target of ITCs. Whether ITC-induced gene expression (indirect effects) underlies histone hyperacetylation deserves further investigation.
Signal transducer and activator of transcription factor 3.
A recent study reported that signal transducer and activator of transcription factor 3 (Stat3) may be a target of ITCs (69). The study found that both the activated form and the total level of Stat3 protein are significantly and rapidly decreased after BITC treatment in several pancreatic cancer cell lines, such as BxPC-3, AsPC-1, Capan-2 and MiaPaCa-2. The effective concentration range was 10–30 μM. The decrease in protein level seems to be caused by both transcriptional suppression and proteasome-dependent protein degradation. The results also indicated that the BITC-induced Stat3 depletion correlates well with the induction of apoptosis and the overexpression of Stat3 restores Stat3 activity and inhibits BITC-induced apoptosis. More importantly, Stat3 depletion was also observed in vivo, suggesting that Stat3 is targeted by BITC both in vitro and in vivo. Interestingly, the reduction in protein level has also been observed with tubulin (53) PKC isoforms (62) and phosphatases (56–58), suggesting that it may be a common fate of ITC-targeted proteins. More recently, two independent studies also discovered that PEITC and SFN inhibit Stat3 activation through inhibition of the interleukin-6-mediated Jak2/Stat3 pathway (70,71) in prostate cancer cells.
Mutant p53.
Our recent observations that BITC and PEITC at 10 and 15 μM can selectively deplete mutant p53 protein, but not the wild-type, in a transcription-independent manner, points to the possibility that mutant p53 may serve as another target of ITCs (72). Although the detailed mechanisms remain unclear, we proposed based on the tubulin binding/depletion model that the effects on mutant p53 may also be due to direct binding by ITCs, followed by its conformational changes. We demonstrated using a monochlorobimane fluorometric assay that cysteines in mutant (G245C) p53 DNA-binding domain are modified by ITCs. We showed that ITC treatment decreases the intrinsic tryptophan fluorescence intensity as an indicator of conformational changes. Interestingly, cells with p53 mutations appear to be more sensitive to ITC-induced cytotoxicity than those with the wild-type. Furthermore, the structure–activity relationships studies using naturally occurring and synthetic ITCs indicated that ITC side chain moieties dictate the potency of the depletion. Specifically, 2,2-diphenylethyl ITC, the most potent depletor of mutant p53 studied, has one of the highest affinities for mutant p53 as well as one of the strongest inducers of apoptosis in mutant breast cancer cells. More studies are needed to verify these interactions; nonetheless, the results suggest that mutant p53 may be a potential novel target for ITCs.
Lysine as a binding site
Perhaps, the Edman degradation, a method of sequencing amino acids in a peptide, is the most well-known reaction between ITC and proteins (73). However, this reaction occurs only at nonphysiological conditions. Phenyl ITC reacts with N-terminal amino groups under basic conditions to form a cyclic phenylthiocarbamoyl derivative, which is further hydrolyzed and cleaved as a thiazolinone derivative under acidic and high-temperature conditions. In addition to N-terminal amino groups, several studies have indicated that under physiological conditions, ITCs may bind to the primary amino side chain of the lysine residue (74,75). The conjugated adducts between AITC and lysine residues in bovine serum albumin (BSA) were identified using liquid chromatography-tandem MS (74). The results indicated that the conjugates between AITC and thiols are unstable and that the deconjugated AITC may react with the amino groups of lysine forming stable conjugates. The in vivo adducts formed between ITCs, including PEITC and SFN, and lysine residues in albumin have recently been reported (75). Albumin was first purified from blood from either a human subject regularly eating watercress (rich in PEITC) and broccoli (rich in SFN) or mice fed with a PEITC–NAC conjugate. Isolated albumin was then digested by proteinase E to amino acids before analysis by MS. While the biological consequences of ITC-lysine binding are unclear, the long half-life of albumin suggests that ITC-lysine adducts may provide a more reliable biomarker than the currently used urinary ITC metabolites to assess dietary ITC uptake and its relationship with cancer prevention in prospective studies. The binding of ITCs to lysine residues in albumin was also demonstrated in our lab. BSA was treated with ITCs and trypsin digested to peptides before analyzed by nanoflow reversed-phase liquid chromatography-MS/MS (Lixin Mi, Brian L. Hood, Nicolas A. Stewart, Zhen Xiao, Sudha Govind, Xiantao Wang, Thomas P. Conrads, Timothy D. Veenstra, and Fung-Lung Chung, unpublished results). The results indicated that BITC, PEITC and SFN covalently modify Lys 235, 437 and 548 in BSA. However, the modification of Cys58, the only supposedly free cysteine residue in BSA, was not observed. The absence of the ITC-Cys58 adduct suggests several explanations including (i) ITCs possess selectivity in binding to certain thiols; (ii) the thiocarbamates are less stable than thiourea and (iii) Cys58 exists in the oxidized form.
Proline as a binding site
MIF is a proinflammatory cytokine that is implicated in the pathogenesis of a number of inflammatory and autoimmune diseases. It is overexpressed in cancer cells and its expression level correlates with tumor aggressiveness (76). Recently, three independent studies reported that ITCs, including BITC and PEITC, at the micromolar concentration range irreversible inhibit tautomerase activity of MIF through covalent binding to the N-terminal catalytic proline residue (8–10). The ITC modification of N-terminal proline was probably due to its low pKa, presumably a result of a positively charged potential by the surrounding residues Lys-32 and Lys-66 (77). MIF was discovered as a potential target of ITCs in cells by two separate studies using affinity chromatography, one with biotin-labeled pentyl ITC (8) and the other with a meta-aminoethyl group-labeled PEITC (10). Proof of binding comes from: (i) the ITC-proline adduct detected by MS in ITC-treated recombinant MIF (8–10); (ii) mutation of the N-terminal proline to glycine inhibited ITC binding to MIF (8) and (iii) a number of ITC analogs, including AITC, ethyl ITC, methallyl ITC, piperidinoethyl ITC and cyclopropyl ITC, also formed adducts with the proline (9). The ITC induced conformational change in MIF was indicated by a gradual decrease of the immunoaffinity of MIF by a monoclonal antibody specific to the intact form (10). A nuclear magnetic resonance study found that BITC bound to the proline of MIF dramatically changed the local conformation surrounding the active site. However, the binding has no effects on MIF quaternary structure distribution—trimerization status. Functional changes induced by ITCs, including inhibition of MIF tautomerase activity (8–10) and interference with binding of MIF to its receptor CD74 (9), indicate that ITCs are inflammation inhibitors. Also, BITC was found to inhibit MIF-mediated glucocorticoid overriding activity and inhibit MIF-induced Akt phosphorylation (9). Interestingly, human volunteers who consumed 50 g of PEITC-rich watercress (10) showed a significant decrease (45%) in plasma immunoreactive MIF levels, coupled to a steady increase of ITCs and their corresponding dithiocarbamates in plasma, indicating that MIF is an in vivo ITC target at physiologically achievable concentrations. Although together these studies confirm that MIF is a target of ITCs, one study noted that BITC also has MIF-independent anti-inflammatory activity (9). This activity is consistent with results from previous studies that ITCs inhibit lipopolysaccharide-induced inflammation through inhibiting nuclear factor-kappaB activity, a pathway independent of MIF (78,79). However, the upstream targets through which ITCs inhibit nuclear factor-kappaB activity are still unknown.
Mitochondria and ROS
ROS and endogenous oxidative stress have long been implicated in cancer development. ROS generation may also be involved in the induction of apoptosis and growth inhibition in cancer cells (80,81). ITCs are known to induce ROS generation in cells (3,6,11,13); but, the mechanisms by which ITCs induce ROS generation are not entirely clear. Three major mechanisms for ITC-induced ROS generation have been conceived: (i) rapid conjugation and depletion of GSH by ITCs with and without glutathione transferases results in redox imbalance (3,11,12,16,18); (ii) the inhibition of GSH reductase and TrxR (63) since both are heavily involved in redox homeostasis and (iii) binding to mitochondrial protein targets. Several studies indicate that BITC and PEITC can induce mitochondrial transmembrane potential change and caspase-9-dependent apoptosis (82–85). An earlier study, using purified rat mitochondria, showed that BITC can directly induce rapid mitochondria swelling and release of cytochrome c through inhibition of mitochondrial respiration (82), suggesting that binding to mitochondrial proteins may be an upstream event. Consistent with this notion, several mitochondrial proteins such as NADH dehydrogenase (Complex I), cytochrome c oxidase (Complex IV), ATP synthase (Complex V), mitochondrial import inner membrane translocase subunit TIM13, cytochrome b5, Hsp60, Hsp10 and thioredoxin-dependent peroxide reductase have been identified as potential ITC targets in our 2-D gel electrophoresis assay (Lixin Mi, Brian L. Hood, Nicolas A. Stewart, Zhen Xiao, Sudha Govind, Xiantao Wang, Thomas P. Conrads, Timothy D. Veenstra, and Fung-Lung Chung, unpublished results). Two recent studies using BITC and PEITC further indicated that complex III activity was significantly inhibited by ITCs, presumably through covalent binding, resulting in ROS generation (84,86). Taken together, mitochondria may be an important source of ITC protein targets that require further studies.
A number of studies have reported that ROS generation underlies ITC-induced apoptosis and cell death (6,82,84,86–88). A key piece of evidence in some studies supporting this mechanism comes from the observations that pretreatment of cells with NAC diminishes ITC-induced downstream effects, including GSH depletion, ROS accumulation and oxidative damage in mitochondria, various signaling pathways and eventually apoptosis and cell death induced by ITCs. Since NAC is an antioxidant, it is presumed that ITC-induced downstream effects are ROS dependent. However, our studies have shown that the rapid ITC conjugate formation with NAC in a culture medium can significantly decrease the cellular uptake (16). Therefore, it is conceivable that the decreased uptake of ITCs caused by NAC is attributed to the abrogation of the downstream effects by NAC, as observed in some of these studies. Recently, ectopic expression of antioxidative enzymes, Cu, Zn-superoxide dismutase and catalase, was observed to significantly suppress apoptosis- and autophagy-mediated cell death by BITC and PEITC, indicating that ROS play a key role in ITC-induced cytotoxicity (84,86). Considering that ROS generation may be a result of ITC binding to mitochondrial proteins, the results also support that binding to target proteins is the upstream trigger for apoptosis induction by ITCs.
ITC concentration and target selection
An array of studies, including cytochrome P450s (29), BCRP and other ATPases (45,47), tubulin (7,53,54), cdc25c (57,58), proteasome (17), PKC (62), mutant p53 (72), mitochondria (83) and MIF (9), have observed a correlation between molecular structure of ITCs and their activity. For instance, BITC and PEITC demonstrated stronger potencies than SFN in some of these studies. It should be noted that these studies focused on a single target or pathway and were carried out in vitro. However, it is conceivable that ITCs, once inside a cell, may react with multiple targets simultaneously and affect a variety of signaling pathways. Therefore, the anticarcinogenesis activity of ITCs is presumably a collective consequence of all affected targets and pathways. This may explain that little potency difference between PEITC and SFN was shown in preventing smoking-related carcinogen benzo(a)pyrene- and nicotine-derived nitrosamine ketone-induced lung tumor incidence in A/J mice (21).
The concentration of ITCs may be another critical factor in determining its activity. At low concentration, ITCs by inducing mild cellular stress activate the cytoprotective systems, including Keap1–Nrf2-mediated antioxidant response (36) and Hsf1-mediated heat shock response (89). However, the gene regulation-dependent interaction by ITCs decreases when ITC concentration exceeds a certain limit, as suggested by a previous study (90). In our own study, SFN-induced G1 phase arrest in A549 cells at 10 μM, a concentration for activation of both antioxidant and heat shock responses (7). At 30 μM, however, SFN was found to inhibit tubulin polymerization in vitro and in vivo, inducing G2/M phase arrest and consequently apoptosis. In the same cells, PEITC at 10 μM induced significant G2/M phase arrest and apoptosis after 24 and 48 h, respectively; at 15 and 20 μM, PEITC-induced apoptosis in 24 h and at 30 μM, PEITC can cause necrosis-dominant cell death within 12 h (7,53). The finding that ITCs induce tubulin degradation serves as another example illustrating that the concentration of ITCs plays a key role in binding to multiple targets causing different downstream effects (53,54). As ITC concentration increased, more tubulin was modified by ITCs and misfolded to form aggregates. Therefore, tubulin aggregation is proportional to ITC concentration in a range of 5–30 μM. However, the degradation of aggregated tubulin is mediated by ubiquitin-proteasome system, which can be substantially inhibited when ITC concentrations exceeded 20 μM (17). As a result, tubulin degradation was clearly evident in a concentration window of 5–20 μM in cultured HeLa and A549 cells.
Since concentration of ITCs is closely related to their downstream effects, whether the potential targets mentioned above are biologically significant may very well depend on the ITC concentration that can be reached in vivo. In a recent study, up to 2 μM plasma ITC was detected 2 h after human volunteers each consumed 50 g of watercress (10). A previous phase I trial also indicated that up to 10 μM, PEITC can be attainable in plasma after trial subjects took pills containing 200 mg of PEITC (91). An earlier study, treating rats orally with a dose of 10 μmol PEITC per 200 g bodyweight indicated that a peak of 18 μM of PEITC can be achieved in rat plasma (92). Although the ITC concentrations used in most of the studies reviewed above are physiological attainable, evidence obtained from preclinical and clinical studies will be one of the most important criteria in validation of these potential targets. In most animal studies, ITCs have been demonstrated to be safe and effective cancer-preventive agents (1). However, ITCs were also shown to have tumor promoting and carcinogenic activities in the rat urinary bladder (93). This is probably due to dosage level, tissue sensitivity and/or ITC metabolism. Although no evidence of cellular DNA binding by ITCs was found in our previous study (3), further investigation is requires answering whether binding to target proteins underlies ITC genotoxicity.
Concluding remarks
The chemopreventive potential of ITCs is demonstrated by a plethora of studies in cells and animals and by prospective and case–control studies in humans (1). Numerous studies have uncovered a complex set of signaling pathways that are associated with anticancer activity of ITCs. ITCs are electrophiles capable of covalently modifying proteins and these modifications may cause functional changes in proteins constituting downstream effects at cellular level. The notion of direct binding to proteins by ITCs was proposed based on previous studies of cytochrome P450s and phase II enzymes (29–32,36–41). However, the identification of binding targets for ITC-induced cell cycle arrest and apoptosis has not been fully investigated until recent years. The protein-binding model was reinforced by studies that demonstrate binding to intracellular proteins is by far the predominant intracellular reaction of ITCs, and the protein-binding affinities correlate with their activities to induce apoptosis (3). Subsequent identification of tubulin (7,53), proteasome (17) and MIF (8–10) as functionally important binding targets of ITCs highlighted the importance of the protein binding model in cancer-preventive activity of ITCs.
This review covers a wide range of proteins that may serve as ITC targets. These protein targets are generally identified by at least one of the following methods: (i) proteomic screening for potential targets using either radiolabeled-2-D gel electrophoresis or affinity chromatography; (ii) protein target verification by functional and conformational assays and (iii) binding site identification by either MS or site-directed mutagenesis. The site-directed mutagenesis, coupled with a functional assay, not only confirm a protein target but also determine the specific binding site, if the mutated residue plays an essential role in protein function, and if the modification of the residue nullifies the activity. While the site-directed mutagenesis studies are perhaps one of the most definitive approaches, it has limitations. For example, the cysteine residues, especially those located in the hydrophilic DNA-binding domain of some transcriptional factors, may be directly modified by ITCs but also by ITC-induced ROS, including superoxide radicals and hydrogen peroxide. Therefore, site-directed mutagenesis is unable to provide unequivocal evidence of ITC binding. Likewise, MS also has its weaknesses, as the binding demonstrated by it would not indicate functional changes. Still, the MS study, particularly in vivo, present other major challenges because it relies on the stability of the protein adducts, the abundance of target proteins and stringent protein purity and quantity (94). Needless to say, many of the potential targets so far identified need to be verified and further evaluated for their specific functional roles. To fully understand the molecular events that initiate apoptosis by ITCs, it is critical to identify the key protein targets that possess important functional roles through a combination of these approaches. Only with this knowledge can we begin to design mechanism-based studies for the discovery of more efficacious ITC-related chemopreventive and therapeutic agents.
Funding
National Institutes of Health (CA-100853 to F.-L. C.).
Acknowledgments
The authors also thank Dr Noriyuki Nagahara of Nippon Medical School, Tokyo, Japan for fruitful discussion.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP-1
activator protein
- ATP
adenosine diphosphate
- ATPase
adenosine triphosphatase
- BCRP
breast cancer resistance protein
- BITC
benzyl isothiocyanate
- BSA
bovine serum albumin
- BSO
L-buthionine-sulfoximine
- FITC
fluorescein isothiocyanate
- GSH
glutathione
- GR
glutathione reductase
- HDAC
histone deacetylase
- ITC
isothiocyanate
- JNK
c-Jun N-terminal kinase
- MS
mass spectrometry
- MIF
migration inhibitory factor
- PEITC
phenethyl isothiocyanate
- PKC
protein kinase C
- ROS
reactive oxygen species
- SFN
sulforaphane
- Stat3
signal transducer and activator of transcription factor
- TrxR
thioredoxin reductase
References
- 1.WHO. IARC Handbook on Cancer Prevention 9. Lyon: IARC; 2004. [Google Scholar]
- 2.London SJ, et al. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: a prospective study of men in Shanghai, China. Lancet. 2000;356:724–729. doi: 10.1016/S0140-6736(00)02631-3. [DOI] [PubMed] [Google Scholar]
- 3.Mi L, et al. The role of protein binding in induction of apoptosis by phenethyl isothiocyanate and sulforaphane in human non-small lung cancer cells. Cancer Res. 2007;67:6409–6416. doi: 10.1158/0008-5472.CAN-07-0340. [DOI] [PubMed] [Google Scholar]
- 4.Kassie F, et al. Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC) Chem. Biol. Interact. 2000;127:163–180. doi: 10.1016/s0009-2797(00)00178-2. [DOI] [PubMed] [Google Scholar]
- 5.Palumaa P. Biological redox switches. Antioxid. Redox Signal. 2009;11:981–983. doi: 10.1089/ars.2009.2468. [DOI] [PubMed] [Google Scholar]
- 6.Singh SV, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J. Biol. Chem. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 7.Mi L, et al. Covalent binding to tubulin by isothiocyanates. A mechanism of cell growth arrest and apoptosis. J. Biol. Chem. 2008;283:22136–22146. doi: 10.1074/jbc.M802330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross JV, et al. Nutrient isothiocyanates covalently modify and inhibit the inflammatory cytokine macrophage migration inhibitory factor (MIF) Biochem. J. 2009;423:315–321. doi: 10.1042/BJ20091170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouertatani-Sakouhi H, et al. A new class of isothiocyanate-based irreversible inhibitors of macrophage migration inhibitory factor. Biochemistry. 2009;48:9858–9870. doi: 10.1021/bi900957e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown KK, et al. Direct modification of the proinflammatory cytokine macrophage migration inhibitory factor by dietary isothiocyanates. J. Biol. Chem. 2009;284:32425–32433. doi: 10.1074/jbc.M109.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Reversible conjugation of isothiocyanates with glutathione catalyzed by human glutathione transferases. Biochem. Biophys. Res. Commun. 1995;206:748–755. doi: 10.1006/bbrc.1995.1106. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa T. Glutathione S-transferases in the metabolism of foreign compounds. Ecotoxicol. Environ. Saf. 1977;1:305–309. doi: 10.1016/0147-6513(77)90022-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YS, et al. Mechanism of differential potencies of isothiocyanates as inducers of anticarcinogenic Phase 2 enzymes. Cancer Res. 1998;58:4632–4639. [PubMed] [Google Scholar]
- 14.Eklind KI, et al. Distribution and metabolism of the natural anticarcinogen phenethyl isothiocyanate in A/J mice. Carcinogenesis. 1990;11:2033–2036. doi: 10.1093/carcin/11.11.2033. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro TA, et al. Human metabolism and excretion of cancer chemoprotective glucosinolates and isothiocyanates of cruciferous vegetables. Cancer Epidemiol. Biomarkers Prev. 1998;7:1091–1100. [PubMed] [Google Scholar]
- 16.Mi L, et al. A cautionary note on using N-acetylcysteine as an antagonist to assess isothiocyanate-induced ROS-mediated apoptosis. Anal. Biochem. 2010;405:269–271. doi: 10.1016/j.ab.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi L, et al. Isothiocyanates inhibit proteasome activity and proliferation of multiple myeloma cells. Carcinogenesis. 2010;32:216–223. doi: 10.1093/carcin/bgq242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y. Role of glutathione in the accumulation of anticarcinogenic isothiocyanates and their glutathione conjugates by murine hepatoma cells. Carcinogenesis. 2000;21:1175–1183. [PubMed] [Google Scholar]
- 19.Podhradský D, et al. Reaction of Cysteine, its derivatives, glutathione, coenzyme A, and dihydrolipoic acid with isothiocyanates. Experientia. 1979;35:154–155. doi: 10.1007/BF01920581. [DOI] [PubMed] [Google Scholar]
- 20.Jiao D, et al. Inhibition of N-nitrosodimethylamine demethylase in rat and human liver microsomes by isothiocyanates and their glutathione, L-cysteine, and N-acetyl-L-cysteine conjugates. Chem. Res. Toxicol. 1996;9:932–938. doi: 10.1021/tx9502094. [DOI] [PubMed] [Google Scholar]
- 21.Conaway CC, et al. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Res. 2005;65:8548–8557. doi: 10.1158/0008-5472.CAN-05-0237. [DOI] [PubMed] [Google Scholar]
- 22.Tang L, et al. The principal urinary metabolites of dietary isothiocyanates, N-acetylcysteine conjugates, elicit the same anti-proliferative response as their parent compounds in human bladder cancer cells. Anticancer Drugs. 2006;17:297–305. doi: 10.1097/00001813-200603000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Drobnica L, et al. Use of isothiocyanates as “reporter” groups in modification of enzymes. In: Fox JL, et al., editors. (eds) Protein Structure and Evolution, New York, NY: Dekker; 1976. pp. 105–115. [Google Scholar]
- 24.Murthy NVKK, et al. Interaction of allyl isothiocyanate with mustard 12S protein. J. Agric. Food Chem. 1986;34:448–452. [Google Scholar]
- 25.Snyder GH, et al. Electrostatic influence of local cysteine environments on disulfide exchange kinetics. Biochemistry. 1981;20:6509–6519. doi: 10.1021/bi00526a001. [DOI] [PubMed] [Google Scholar]
- 26.Soares TA, et al. Internal dynamics and ionization states of the macrophage migration inhibitory factor: comparison between wild-type and mutant forms. Biopolymers. 2002;65:313–323. doi: 10.1002/bip.10252. [DOI] [PubMed] [Google Scholar]
- 27.Nagahara N, et al. Protein cysteine modifications: (2) reactivity specificity and topics of medicinal chemistry and protein engineering. Curr. Med. Chem. 2009;16:4490–4501. doi: 10.2174/092986709789760643. [DOI] [PubMed] [Google Scholar]
- 28.Doorn JA, et al. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 29.Hollenberg PF, et al. Mechanism-based inactivation of human cytochromes p450s: experimental characterization, reactive intermediates, and clinical implications. Chem. Res. Toxicol. 2008;21:189–205. doi: 10.1021/tx7002504. [DOI] [PubMed] [Google Scholar]
- 30.Rando RR. Mechanism-based enzymes inactivators. Pharmacol. Rev. 1984;36:111–142. [PubMed] [Google Scholar]
- 31.Kent UM, et al. Spectral studies of tert-butyl isothiocyanate-inactivated P450 2E1. Biochemistry. 2001;40:7253–7261. doi: 10.1021/bi0102712. [DOI] [PubMed] [Google Scholar]
- 32.von Weymarn LB, et al. Effects of benzyl and phenethyl isothiocyanate on P450s 2A6 and 2A13: potential for chemoprevention in smokers. Carcinogenesis. 2006;27:782–790. doi: 10.1093/carcin/bgi301. [DOI] [PubMed] [Google Scholar]
- 33.Lee MS. Enzyme induction and comparative oxidative desulfuration of isothiocyanates to isocyanates. Chem. Res. Toxicol. 1996;9:1072–1078. doi: 10.1021/tx950213f. [DOI] [PubMed] [Google Scholar]
- 34.Egner PA, et al. Quantification of sulforaphane mercapturic acid pathway conjugates in human urine by high-performance liquid chromatography and isotope-dilution tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:1991–1996. doi: 10.1021/tx800210k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi A, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong KI, et al. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto T, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi M, et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, et al. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong F, et al. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem. Res. Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 42.Li W, et al. Nrf2 possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J. Biol. Chem. 2005;280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 43.Tseng E, et al. Effect of organic isothiocyanates on the P-glycoprotein- and MRP1-mediated transport of daunomycin and vinblastine. Pharm. Res. 2002;19:1509–1515. doi: 10.1023/a:1020460700877. [DOI] [PubMed] [Google Scholar]
- 44.Ji Y, et al. Effect of organic isothiocyanates on breast cancer resistance protein (ABCG2)-mediated transport. Pharm. Res. 2004;21:2261–2269. doi: 10.1007/s11095-004-7679-1. [DOI] [PubMed] [Google Scholar]
- 45.Ji Y, et al. Membrane transport of dietary phenethyl isothiocyanate by ABCG2 (breast cancer resistance protein) Mol. Pharm. 2005;2:414–419. doi: 10.1021/mp050029f. [DOI] [PubMed] [Google Scholar]
- 46.Karlish SJ. Na+, K+-ATPase Structure and Kinetics. 1979. In Skou, J. C. and Norby J. G. (eds) Academic Press, London, UK, pp. 115–128. [Google Scholar]
- 47.Breier A, et al. Inhibition of (Na/K)-ATPase by electrophilic substances: functional implications. Mol. Cell. Biochem. 1995;147:187–192. doi: 10.1007/BF00944800. [DOI] [PubMed] [Google Scholar]
- 48.Carilli CT, et al. The active site structure of Na+- and K+-stimulated ATPase. Location of a specific fluorescein isothiocyanate reactive site. J. Biol. Chem. 1982;257:5601–5606. [PubMed] [Google Scholar]
- 49.Taniguchi K, et al. Microenvironment of two different extrinsic fluorescence probes in Na+ K1-ATPase changes out of phase during sequential appearance of intermediates. J. Biol. Chem. 1988;263:12943–12947. [PubMed] [Google Scholar]
- 50.Breier A, et al. “Lysine is the Lord”, thought some scientists in regard to the group interacting with fluorescein isothiocyanate in ATP-binding sites of P-type ATPases but, is it not cysteine? Gen. Physiol. Biophys. 2000;19:253–263. [PubMed] [Google Scholar]
- 51.Kubala M, et al. Eight amino acids form the ATP recognition site of Na(+)/K(+)-ATPase. Biochemistry. 2003;42:6446–6452. doi: 10.1021/bi034162u. [DOI] [PubMed] [Google Scholar]
- 52.Wilson L, et al. Modulation of microtubule dynamics by drugs: a paradigm for the actions of cellular regulators. Cell Struct. Funct. 1999;24:329–335. doi: 10.1247/csf.24.329. [DOI] [PubMed] [Google Scholar]
- 53.Mi L, et al. Cancer preventive isothiocyanates induce selective degradation of cellular alpha- and beta-tubulins by proteasomes. J. Biol. Chem. 2009;284:17039–17051. doi: 10.1074/jbc.M901789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi L, et al. Aggresome-like structure induced by isothiocyanates is novel proteasome-dependent degradation machinery. Biochem. Biophys. Res. Commun. 2009;388:456–462. doi: 10.1016/j.bbrc.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinman A, et al. TRP channel activation by reversible covalent modification. Proc. Natl. Acad. Sci. USA. 2006;103:19564–19568. doi: 10.1073/pnas.0609598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YR, et al. Phenylethyl isothiocyanate induces apoptotic signaling via suppressing phosphatase activity against c-Jun N-terminal kinase. J. Biol. Chem. 2002;277:39334–39342. doi: 10.1074/jbc.M202070200. [DOI] [PubMed] [Google Scholar]
- 57.Xiao D, et al. Proteasome-mediated degradation of cell division cycle 25C and cyclin-dependent kinase 1 in phenethyl isothiocyanate-induced G2-M-phase cell cycle arrest in PC-3 human prostate cancer cells. Mol. Cancer Ther. 2004;3:567–575. [PubMed] [Google Scholar]
- 58.Singh SV, et al. Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J. Biol. Chem. 2004;279:25813–25822. doi: 10.1074/jbc.M313538200. [DOI] [PubMed] [Google Scholar]
- 59.Cross JV, et al. The isothiocyanate class of bioactive nutrients covalently inhibit the MEKK1 protein kinase. BMC Cancer. 2007;7:183. doi: 10.1186/1471-2407-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JH, et al. Inhibition of EGFR signaling in human prostate cancer PC-3 cells by combination treatment with beta-phenylethyl isothiocyanate and curcumin. Carcinogenesis. 2006;27:475–482. doi: 10.1093/carcin/bgi272. [DOI] [PubMed] [Google Scholar]
- 61.Nomura T, et al. Alkyl isothiocyanates suppress epidermal growth factor receptor kinase activity but augment tyrosine kinase activity. Cancer Epidemiol. 2009;33:288–292. doi: 10.1016/j.canep.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 62.Mukherjee S, et al. Isothiocyanates sensitize the effect of chemotherapeutic drugs via modulation of protein kinase C and telomerase in cervical cancer cells. Mol. Cell. Biochem. 2009;330:9–22. doi: 10.1007/s11010-009-0095-4. [DOI] [PubMed] [Google Scholar]
- 63.Hu Y, et al. Glutathione- and thioredoxin-related enzymes are modulated by sulfur-containing chemopreventive agents. Biol. Chem. 2007;388:1069–1081. doi: 10.1515/BC.2007.135. [DOI] [PubMed] [Google Scholar]
- 64.Krauth-Siegel RL, et al. Role of active site tyrosine residues in catalysis by human glutathione reductase. Biochemistry. 1998;37:13968–13977. doi: 10.1021/bi980637j. [DOI] [PubMed] [Google Scholar]
- 65.Lee SR, et al. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc. Natl Acad. Sci. USA. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matthews CP, et al. AP-1 a target for cancer prevention. Curr. Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- 67.Dickinson SE, et al. Inhibition of activator protein-1 by sulforaphane involves interaction with cysteine in the cFos DNA-binding domain: implications for chemoprevention of UVB-induced skin cancer. Cancer Res. 2009;69:7103–7110. doi: 10.1158/0008-5472.CAN-09-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Myzak MC, et al. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 69.Sahu RP, et al. The role of STAT-3 in the induction of apoptosis in pancreatic cancer cells by benzyl isothiocyanate. J. Natl Cancer Inst. 2009;101:176–193. doi: 10.1093/jnci/djn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong A, et al. Phenethyl isothiocyanate inhibits STAT3 activation in prostate cancer cells. Mol. Nutr. Food Res. 2009;53:878–886. doi: 10.1002/mnfr.200800253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hahm ER, et al. Sulforaphane inhibits constitutive and interleukin-6-induced activation of signal transducer and activator of transcription 3 in prostate cancer cells. Cancer Prev. Res. 2010;3:484–494. doi: 10.1158/1940-6207.CAPR-09-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, et al. Selective. depletion of mutant p53 by cancer chemopreventive isothiocyanates and its structure-activity relationships. J. Med. Chem. 2010 doi: 10.1021/jm101199t. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edman P. Method for the determination of the amino acid sequence in peptides. Acta Chem. Scand. 1950;4:283–293. [Google Scholar]
- 74.Nakamura T, et al. Covalent modification of lysine residues by allyl isothiocyanate in physiological conditions: plausible transformation of isothiocyanate from thiol to amine. Chem. Res. Toxicol. 2009;22:536–542. doi: 10.1021/tx8003906. [DOI] [PubMed] [Google Scholar]
- 75.Kumar A, et al. New biomarkers for monitoring the levels of isothiocyanates in humans. Chem. Res. Toxicol. 2010;23:756–765. doi: 10.1021/tx900393t. [DOI] [PubMed] [Google Scholar]
- 76.Campa MJ, et al. Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res. 2003;63:1652–1656. [PubMed] [Google Scholar]
- 77.Stamps SL, et al. Characterization of the role of the amino-terminal proline in the enzymatic activity catalyzed by macrophage migration inhibitory factor. Biochemistry. 1998;37:10195–10202. doi: 10.1021/bi9806955. [DOI] [PubMed] [Google Scholar]
- 78.Rose P, et al. Beta-phenylethyl and 8-methylsulphinyloctyl isothiocyanates, constituents of watercress, suppress LPS induced production of nitric oxide and prostaglandin E2 in RAW 264.7 macrophages. Nitric Oxide. 2005;12:237–243. doi: 10.1016/j.niox.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 79.Prawan A, et al. Anti-NF-kappaB and anti-inflammatory activities of synthetic isothiocyanates: effect of chemical structures and cellular signaling. Chem. Biol. Interact. 2009;179:202–211. doi: 10.1016/j.cbi.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 82.Nakamura Y, et al. Involvement of the mitochondrial death pathway in chemopreventive benzyl isothiocyanate-induced apoptosis. J. Biol. Chem. 2002;277:8492–8499. doi: 10.1074/jbc.M109760200. [DOI] [PubMed] [Google Scholar]
- 83.Tang L, et al. Mitochondria are the primary target in isothiocyanate-induced apoptosis in human bladder cancer cells. Mol. Cancer Ther. 2005;4:1250–1259. doi: 10.1158/1535-7163.MCT-05-0041. [DOI] [PubMed] [Google Scholar]
- 84.Xiao D, et al. Benzyl isothiocyanate targets mitochondrial respiratory chain to trigger reactive oxygen species-dependent apoptosis in human breast cancer cells. J. Biol. Chem. 2008;283:30151–30163. doi: 10.1074/jbc.M802529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown KK, et al. Mitochondrial respiratory chain involvement in peroxiredoxin 3 oxidation by phenethyl isothiocyanate and auranofin. FEBS Lett. 2010;584:1257–1262. doi: 10.1016/j.febslet.2010.02.042. [DOI] [PubMed] [Google Scholar]
- 86.Xiao D, et al. Phenethyl isothiocyanate inhibits oxidative phosphorylation to trigger reactive oxygen species-mediated death of human prostate cancer cells. J. Biol. Chem. 2010;285:26558–26569. doi: 10.1074/jbc.M109.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shankar S, et al. Sulforaphane enhances the therapeutic potential of TRAIL in prostate cancer orthotopic model through regulation of apoptosis, metastasis, and angiogenesis. Clin. Cancer Res. 2008;14:6855–6866. doi: 10.1158/1078-0432.CCR-08-0903. [DOI] [PubMed] [Google Scholar]
- 88.Zhang H, et al. Effective killing of Gleevec-resistant CML cells with T315I mutation by a natural compound PEITC through redox-mediated mechanism. Leukemia. 2008;22:1191–1199. doi: 10.1038/leu.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gan N, et al. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. J. Biol. Chem. 2010;285:35528–35536. doi: 10.1074/jbc.M110.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang L, et al. Dietary isothiocyanates inhibit the growth of human bladder carcinoma cells. J. Nutr. 2004;134:2004–2010. doi: 10.1093/jn/134.8.2004. [DOI] [PubMed] [Google Scholar]
- 91.Liebes L, et al. Phase I safety and pharmacokinetics clinical trials of PEITC in chronic smokers. Proceedings of the American Association for Cancer Research. 2000;42 NCI CN-55120. [Google Scholar]
- 92.Conaway CC, et al. Disposition and pharmacokinetics of phenethyl isothiocyanate and 6-phenylhexyl isothiocyanate in F344 rats. Drug Metab. Dispos. 1999;27:13–20. [PubMed] [Google Scholar]
- 93.Akagi K, et al. Involvement of toxicity as an early event in urinary bladder carcinogenesis induced by phenethyl isothiocyanate, benzyl isothiocyanate, and analogues in F344 rats. Toxicol. Pathol. 2003;31:388–396. doi: 10.1080/01926230390202326. [DOI] [PubMed] [Google Scholar]
- 94.Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ. Health Perspect. 2002;110(1) Suppl:3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]