Abstract
The tumor suppressor programmed cell death 4 (Pdcd4) is lost in various tumor tissues. Loss of Pdcd4 has been associated with increased tumorigenic potential and tumor progression. While various mechanisms of Pdcd4 regulation have been described, the effect of an inflammatory tumor microenvironment on Pdcd4 protein expression has not been characterized so far. In the present study, we aimed to elucidate the molecular mechanisms of Pdcd4 protein regulation in tumor cells under inflammatory conditions. 12-O-tetradecanoylphorbol 13-acetate-induced differentiation of human U937 monocytes increased the expression and secretion of inflammatory cytokines such as tumor necrosis factor α, interleukin (IL)-6 and IL-8. Exposure to conditioned medium (CM) of these activated macrophages markedly decreased Pdcd4 protein expression in various tumor cells. Similarly, indirect coculture with such activated U937 monocyte-derived macrophages resulted in the loss of Pdcd4 protein in tumor cells. Decreased Pdcd4 protein levels were attributable to enhanced proteasomal degradation, diminishing Pdcd4 protein half-life. Proteasomal degradation required activation of phosphatidylinositol-3-kinase (PI3K)-mammalian target of rapamycin (mTOR) signaling. Since macrophage-CM sufficed to induce Pdcd4 degradation, Pdcd4 downregulation was determined to be an indirect unidirectional effect of the macrophages on the tumor cells. Pdcd4 protein expression was also attenuated in vivo in mouse colon tissue in response to dextran sodium sulfate-induced colitis. In summary, we characterized PI3K–mTOR-dependent proteasome-mediated Pdcd4 degradation in tumor cells in the inflammatory tumor microenvironment. Consequently, stabilization of Pdcd4 protein could provide a promising novel avenue for therapeutics targeting inflammation-associated tumors.
Introduction
Programmed cell death 4 (Pdcd4) was the first tumor suppressor characterized to inhibit translation to exert its tumor suppressive function (1). Specifically, Pdcd4 was shown to attenuate translation initiation by binding to the RNA-helicase eukaryotic initiation factor 4A (eIF4A) via two MA3-domains that are highly homologous to those in eIF4G (2–4). Pdcd4 binding to eIF4A inhibits the activity of the latter and, thus, inhibits translation of messenger RNAs (mRNAs) containing highly structured 5′ untranslated regions, which require unwinding for efficient translation (3,5). Many proteins involved in the growth or proliferation control, i.e. many proto-oncogenes, are encoded by such ‘weak’ mRNAs (6). While the identification of translational Pdcd4 targets is an ongoing endeavor, functional consequences of modulating Pdcd4 levels have been studied extensively in vitro and in vivo. Pdcd4 was shown to inhibit transformation, migration, anchorage-independent growth, intravasation and invasion in vitro (7–11). Furthermore, overexpression of Pdcd4 in keratinocytes was shown to reduce tumor multiplicity in the two-stage skin carcinogenesis model in vivo (12). Interestingly, in this inflammatory tumor model, Pdcd4 protein was lost in all papillomas that developed irrespectively of the genetic background. Importantly, Pdcd4 knockdown sensitized mice to tumor development in the aforementioned inflammatory tumor model and also facilitated spontaneous formation of lymphomas (13,14). The loss of Pdcd4 in the two-stage skin carcinogenesis model was attributed to 12-O-tetradecanoylphorbol 13-acetate (TPA)-induced reduction of Pdcd4 protein half-life (14). p70S6K1- and/or Akt-dependent phosphorylation of Pdcd4 in response to mitogenic stimuli was shown to facilitate binding of the E3-ubiquitin ligase β-TrCP1 and subsequent proteasomal degradation of Pdcd4 (14,15). Recent work further established Pdcd4 as a target of microRNA-21 (miR-21). In line, miR-21 was shown to repress translation of Pdcd4 in various tumor models and Pdcd4 repression appeared to be functionally important for the pro-oncogenic effects of miR-21 (8,16–20).
Analysis of human tumor tissues further revealed that Pdcd4 protein expression is low in most tumor types (19–30). In colorectal cancer, an inverse correlation between Pdcd4 and active Akt was shown and loss of Pdcd4 expression was proposed as an independent prognostic marker for tumor-associated survival in colon carcinogenesis (31). Similarly, Akar et al. (32) recently demonstrated that loss of Pdcd4 protein in breast cancer cells can be overcome by inactivation of p70S6K, which results in the suppression of lung metastasis development. Based on these observations, current approaches aim at identifying compounds stabilizing Pdcd4 to inhibit tumorigenic mechanisms (33).
Here, we present data characterizing the mechanism of Pdcd4 regulation under inflammatory tumorigenic conditions. We provide evidence that activated U937 monocyte-derived macrophages reduce Pdcd4 protein expression in cocultured MCF7 cells. This was exclusively attributable to phosphorylation-dependent reduction of Pdcd4 protein half-life. Consequently, we propose that Pdcd4 degradation is rapidly induced to limit Pdcd4 expression under inflammatory, tumor-associated conditions, therefore constituting a promising target for therapeutic interventions.
Material and methods
Materials
All chemicals were purchased from Sigma–Aldrich (Schnelldorf, Germany), if not indicated otherwise. Peptide-purified anti-Pdcd4 antibody was described earlier (34). Anti-actin came from Sigma–Aldrich, anti-Pdcd4 (phospho S67) from Abcam (Cambridge, UK) and IRDyes 680LT and 800CW secondary antibodies from Li-COR Biosciences GmbH (Bad Homburg, Germany). LY294002 and rapamycin were purchased from Cell Signaling Technology (Frankfurt, Germany), PD98059 and SB203580 from Calbiochem (Darmstadt, Germany).
Cell culture
All cell lines came from LGC Standards GmbH (Wesel, Germany). RKO and MCF7 cells were maintained in Dulbecco's modified Eagle's medium, T47D and U937 cells in RPMI medium and DU145 cells in modified Eagle's medium. All media were supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. Additionally, DU145 media contained 1 mM sodium pyruvat and 1× modified Eagle's medium non-essential amino acids. Cells were kept at 37°C in a humidified atmosphere with 5% CO2. Medium and supplements were purchased from PAA (Linz, Austria). Fetal bovine serum came from Biochrom (Berlin, Germany).
Macrophage differentiation and conditioned medium
U937 monocytes (1 × 107/25 ml) were exposed to TPA [10 nM] for 48 h. The resulting adherent activated U937 monocyte-derived macrophages (differentiated U937) were trypsinized, pelleted and washed with phosphate-buffered saline. For control purposes, undifferentiated U937 monocytes (3 × 106/25 ml) were incubated with dimethyl sulfoxide (0.1%) for 48 h, pelleted by centrifugation and washed with phosphate-buffered saline. Subsequently, control and differentiated U937 were treated equally. For the generation of conditioned medium (CM), U937 cells were reseeded at a concentration of 2 × 106/5 ml. Cells were allowed to condition medium for 24 h unless specified otherwise. For harvesting the CM, supernatant was centrifuged, passed through sterile 0.45 μm filters and stored at −80°C until further use. Conditioned medium from differentiated U937 (CM) was always prepared in parallel to CM from undifferentiated U937 (Ctr). For coculture experiments, control or differentiated U937 cells were added indirectly (in a Boyden chamber) to MCF7 cells at concentrations equivalent to those used for CM generation (undiluted = 2 × 106/5 ml). All experiments were carried out in RPMI medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine.
Western blotting
For western blot analysis, cells were sonicated and then lysed on ice for 30 min in lysis buffer [50 mM Tris–HCl, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor mix (Roche, Mannheim, Germany)]. Fifty micrograms protein were separated on sodium dodecyl sulfate gels and transferred onto nitrocellulose membranes. Proteins were detected using specific antibodies and appropriate secondary antibodies. Then, they were visualized using the Odyssey infrared imaging system (Li-COR Biosciences GmbH, Bad Homburg, Germany).
Protein stability determination
For the evaluation of protein stability, MCF7 cells were pre-incubated with the respective medium (CM versus Ctr) for 4 or 20 h. Then, cycloheximide (CHX) [10 μM] was added to stop translation and protein levels were followed for 1–4 h. To determine the kinetics and half-life, Pdcd4 protein levels were quantified densitometrically, normalized to the respective actin and analyzed relative to levels at the beginning of the CHX treatment. For alternative determination of protein stability, pulse-chase experiments were performed. Briefly, MCF7 cells were incubated in methionine/cysteine-free medium for 30 min. Then, 0.45 mCi EasyTag Express 35S-Protein Labeling mix (PerkinElmer, Groningen, The Netherlands) was added for 2 h to label newly synthesized proteins (pulse). After washing off the labeling medium, cells were either harvested directly after labeling or incubated in Ctr or CM for 2 h (chase) before harvesting. Cells were lysed in immunoprecipitation-buffer [50 mM Tris–HCl, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 0.5% NP-40, 20% glycerin, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor mix (Roche)]. Following lysis, Pdcd4 was immunoprecipitated and separated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis. For enhancement of autoradiography detection, gels were fixed in fixing solution (7% acetic acid, 25% methanol), incubated in EN3HANCE solution (PerkinElmer), soaked in soaking solution (1% glycerol, 5% PEG8000) and dried on a filter. Autoradiographic detection was performed using an intensifier screen (Fujifilm, Düsseldorf, Germany) and analyzed using the Bio-imaging analyzer system 1500 (Fujifilm). For the determination of the impact of proteasomal degradation, cells were treated with MG132 [10 μM] for the last 8 h of the respective treatment.
mRNA expression analysis
To determine the expression of inflammatory cytokine mRNAs in U937 cells, mRNA was isolated from differentiated U937 or undifferentiated U937 monocytes at the time of CM or Ctr harvest, respectively. Specifically, U937 cells were treated with TPA [10 nM] or dimethyl sulfoxide for 48 h, washed and reseeded at the appropriate concentration for conditioning medium (see above). Twenty-four hours after reseeding, U937 cells were harvested for mRNA isolation. mRNA was isolated using PeqGold RNAPure (peqLab, Erlangen, Germany) according to the manufacturer’s manual. RNA was transcribed using the Maxima First Strand cDNA synthesis kit from Fermentas (St. Leon-Rot, Germany) and expression of individual mRNAs was analyzed using real-time PCR with Absolute qPCR SYBR Green Fluorescent mix (Abgene, Surrey, UK). Specific primers were individually designed (ribosomal protein 18S RNA: forward 5′-GTAACCCGTTGAACCCC-ATT-3′ and reverse 5′-CCATCCAATCGGTAGTA GCG-3′), alternatively QuantiTect primer assays were used [tumor necrosis factor (TNF) α, interleukin (IL)-6, IL-8] (Qiagen, Hamburg, Germany). Expression was normalized to expression of ribosomal protein 18S.
Quantification of cytokine secretion
To determine the secretion of inflammatory cytokines from undifferentiated or differentiated U937 cells, Ctr and CM were analyzed for TNFα, IL-6 and IL-8 using the BD Cytometric Bead Array (CBA) Human Inflammation Kit according to the manufacturer’s manual. Samples were measured using the BD LSRFortessa flow cytometer and analyzed with BD Biosciences FCAP software (BD Biosciences, Heidelberg, Germany).
Animal studies
Mice were housed and cared for and all animal studies were conducted in accordance with National Cancer Institute-Frederick Animal Care and Use Committee guidelines. Seven- to eight-week-old mice (129/C57Bl/6/CD1-mixed background) received 0, 1 or 2% dextran sodium sulfate (DSS) in the drinking water for 7 days followed by 7 days of untreated drinking water. Mice received three cycles of DSS and were killed at the end of the last cycle of DSS treatment. Colons were cut, rinsed and snap frozen in liquid nitrogen. For lysis, colon tissue was thawed in protein lysis buffer, disrupted and homogenized with 1.0 mm glass beads on a Mini-Beadbeater instrument from Biospec. After centrifugation, the clear supernatant was used for western blot analysis (see above).
Statistical analysis
Each experiment was performed at least three times. Representative blots are shown. Data are presented as mean values ± SEMs. Statistical analysis was performed using Student’s t-test.
Results
Inflammatory conditions attenuate Pdcd4 protein expression
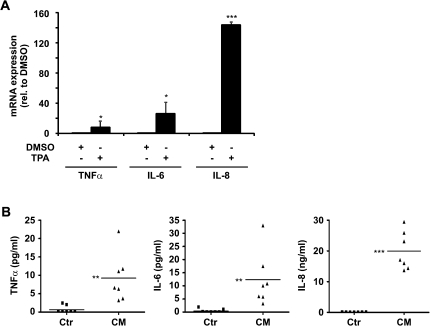
To determine how an inflammatory environment affects Pdcd4 protein expression in tumor cells, we first aimed at establishing an in vitro model for inflammatory pro-tumorigenic conditions. To this end, human monocytic U937 cells were differentiated to macrophages with TPA for 48 h. To verify that the resulting macrophages generate an inflammatory microenvironment, we analyzed the expression of inflammatory cytokine mRNAs in TPA-differentiated U937 cells. TPA-induced U937 monocyte-derived macrophages showed a significantly elevated mRNA expression of the pro-inflammatory mediators TNFα, IL-6 and IL-8 as compared with dimethyl sulfoxide-treated control cells (Figure 1A). Since expression and secretion of many cytokines is regulated post-transcriptionally, we further analyzed protein levels of pro-inflammatory mediators secreted from undifferentiated U937 and from TPA-differentiated U937 cells. Levels of TNFα, IL-6 and IL-8 were significantly higher in CM from differentiated U937 cells as compared with CM from undifferentiated U937 cells (CM and Ctr, respectively) (Figure 1B). Furthermore, the environment provided by differentiated U937 elicited pro-tumorigenic effects. Specifically, differentiated U937 and CM induced activator protein-1 activity in MCF7 cells and CM increased invasion of MCF7 cells (supplementary Figure S1 is available at Carcinogenesis Online). Thus, we deemed the conditions generated by TPA-differentiated U937 cells as inflammatory and pro-tumorigenic.
Fig. 1.
TPA-induced differentiation of U937 monocytes increases the expression of inflammatory cytokines. U937 monocytes were incubated with dimethyl sulfoxide or TPA [10 nM] for 48 h. After washing, cells were cultured for 24 h in fresh medium. (A) mRNA expression of inflammatory cytokines (TNFα, IL-6 and IL-8) in the U937 cells was analyzed using reverse transcription (RT)-quantitative polymerase chain reaction (qPCR). Expression was normalized to 18S ribosomal RNA. (B) Secretion of inflammatory mediators (TNFα, IL-6 and IL-8) in Ctr and CM was analyzed using CBA assays (n > 3, *P < 0.05, **P < 0.01 and ***P < 0.001).
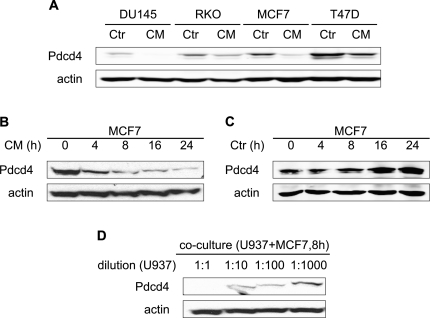
We next assessed the effects of CM on Pdcd4 protein expression in a panel of tumor cell lines representing a variety of tumor entitities (prostate: DU145; colon: RKO; breast: T47D and MCF7). Interestingly, Pdcd4 protein expression was diminished in all tumor cell lines tested after 24 h of CM incubation (Figure 2A). Downregulation of Pdcd4 occurred irrespective of the initial Pdcd4 status. For further mechanistical analysis of CM-induced loss of Pdcd4 protein expression, MCF7 mammary carcinoma cells were chosen as a model system. To determine the temporal pattern of Pdcd4 protein disappearance, MCF7 cells were incubated with CM for 4–24 h. Pdcd4 protein expression was attenuated already at 4 h of CM treatment and further decreased toward 24 h (Figure 2B). In contrast, Ctr did not reduce Pdcd4 protein expression at any time point tested (Figure 2C). In an attempt to identify a critical concentration of macrophages needed to secrete Pdcd4-attenuating stimuli, differentiated U937 were indirectly cocultured with MCF7 cells for 8 h at various dilutions. At a concentration of differentiated U937 per milliliter that was equivalent to the concentration of differentiated U937 cells used for the conditioning protocol (1:1), coculture caused the strongest decrease of Pdcd4 protein in MCF7 cells. Downregulation was lower at 1:10 to 1:100 and lost at a dilution of 1:1000 (Figure 2D). Similary, dilution of CM or differentiated U937 in a coculture approach led to a markedly lower activator protein-1 activation as compared with the undiluted form (supplementary Figure S1A is available at Carcinogenesis Online). To exclude a potential TPA carryover from the differentiation procedure into the CM, CM of MCF7 cells was prepared using the same protocol as for U937-CM, i.e. MCF7 cells were incubated for 48 h with TPA, washed, reseeded at the same concentration as U937 cells for conditioning purposes and allowed to condition medium for 24 h. Since the resulting MCF7-CM did not elicit changes on Pdcd4 protein level in MCF7 target cells (data not shown), TPA carryover was ruled out as responsible factor.
Fig. 2.
Differentiated U937 cells secrete factors that regulate Pdcd4 protein levels in tumor cells. (A) DU145, RKO, T47D and MCF7 tumor cells were incubated with CM from differentiated U937 for 24 h. (B + C) MCF7 cells were incubated with CM (B) or Ctr (C) for the indicated times. (D) Differentiated U937 were seeded in the upper well of a boyden chamber assay with MCF7 cells in the lower chamber at the indicated dilutions for 8 h. Whole-cell extracts were subjected to western blot analysis and probed with the indicated antibodies. Blots are representative of at least three independent experiments.
Thus, we conclude that macrophage-associated inflammatory conditions reduce Pdcd4 protein expression in tumor cells.
Inflammatory conditions increase proteasomal degradation to limit the half-life of Pdcd4 protein
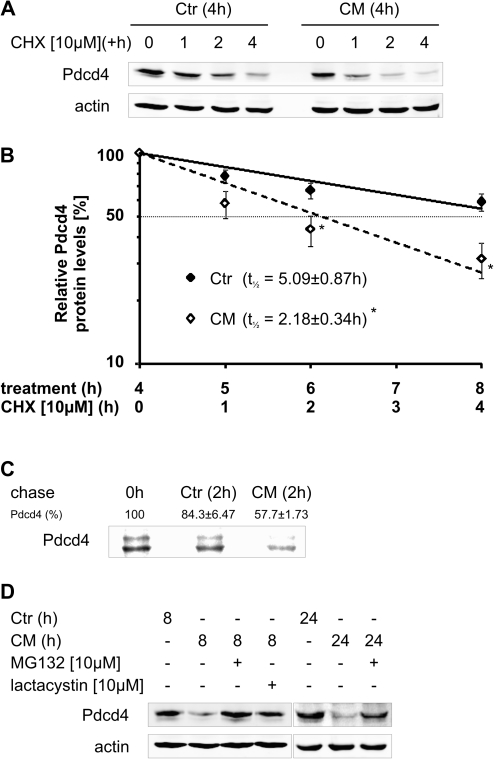
The next set of experiments was designed to elucidate the molecular mechanisms underlying the loss of Pdcd4 protein under inflammatory conditions. To determine changes in Pdcd4 protein stability, translation was inhibited using CHX [10 μM] in MCF7 cells pre-exposed to CM for 4 h. While in Ctr-treated cells Pdcd4 protein remained stable, CM markedly reduced Pdcd4 protein stability, i.e. Pdcd4 protein amount decreased markedly already 1 h after the addition of CHX in CM-treated cells, whereas Pdcd4 protein levels only slightly decreased starting at 2 h of translational inhibition in Ctr-treated cells (Figure 3A). Taken together, the kinetics of the Pdcd4 decrease after addition of CHX translated into half-lives of 5.09 ± 0.87 h for Ctr and 2.18 ± 0.34 h for CM (Figure 3B). To exclude potential indirect effects of CHX, we next determined Pdcd4 protein stability using pulse-chase experiments. As shown in Figure 3C, pulse-labeled Pdcd4 protein levels were markedly reduced after 2 h of CM as compared with 2 h Ctr (57.7 versus 84.3% of the starting level, respectively). As Pdcd4 destabilization was previously associated with increased proteasomal degradation (14), we next assessed the involvement of the proteasome. CM-induced loss of Pdcd4 protein at 8 h was completely prevented by the proteasome inhibitor MG132 (Figure 3D). Similarly, the specific proteasome inhibitor lactacystin rescued Pdcd4 from degradation. To determine, if proteasomal degradation was still effective at extended CM incubations, MG132 was added to cells pre-incubated with CM for 16 h and incubations continued for 8 h. Similarly, blocking proteasomal degradation for 8 h in the presence of CM after 16 h CM pretreatment allowed for an almost complete recovery of Pdcd4 protein levels (Figure 3D).
Fig. 3.
CM attenuates Pdcd4 protein expression by increasing proteasomal degradation. (A) MCF7 cells were incubated with Ctr or CM for 4 h. Then, CHX [10 μM] was added to block translation and incubations continued for 1, 2 or 4 h. (B) Protein levels determined in (A) were quantified densitometrically and expression was normalized to actin. Data are presented relative to 4 h pretreated samples (n > 3, *P < 0.05). (C) MCF7 cells were subjected to a pulse-chase experiment, in the presence of Ctr or CM during the 2 h chase period. Pdcd4 was immunoprecipitated and detected by autoradiography. The autoradiographic picture is representative for at least three independent experiments and the remaining labeled Pdcd4 levels are presented relative to 0 h (n = 3). (D) MCF7 cells were incubated with Ctr or CM for 8 or 24 h. MG132 [10 μM] or lactacystin [10 μM] were added for the last 8 h of the incubations to block proteasomal degradation. Whole-cell extracts were subjected to western blot analysis and probed with the indicated antibodies. Blots are representative of at least three independent experiments.
Based on these observations, we propose that enhanced proteasomal degradation predominantly accounts for Pdcd4 regulation under macrophage-associated inflammatory conditions.
Phosphatidylinositol-3-kinase-mammalian target of rapamycin signaling is required for inflammation-induced Pdcd4 protein loss
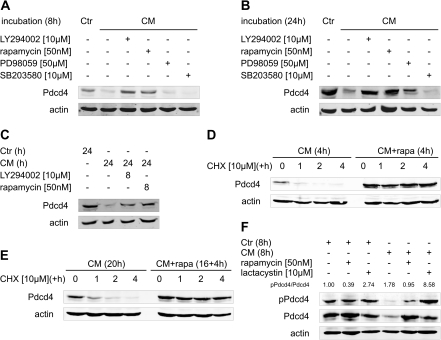
p70S6K1-dependent phosphorylation was shown previously to be critical for proteasomal degradation of Pdcd4 (14,15). Therefore, we addressed the signaling pathways involved in Pdcd4 degradation under inflammatory conditions. Figure 4A and B show that inhibition of phosphatidylinositol-3-kinase (PI3K) (by LY294002) or mammalian target of rapamycin (mTOR) (by rapamycin) completely prevented the loss of Pdcd4 at 8 and 24 h. Inhibitors of mitogen-activated protein kinase/extracellular signal regulated kinase- (PD98059) or p38-mitogen-activated protein kinase signaling (SB203580) appeared largely ineffective in rescuing Pdcd4 protein from CM-induced downregulation. Interestingly, inhibition of PI3K or mTOR for 8 h after 16 h pre-incubation with CM, almost completely restored the Pdcd4 protein level to control (Figure 4C). To verify that the increase in Pdcd4 protein in response to inhibitors of PI3K–mTOR signaling was indeed a result of increased protein stability, we next determined the impact of mTOR inhibition on Pdcd4 protein stability. As expected, rapamycin co-treatment with CM for 4 h (Figure 4D, right side) markedly increased Pdcd4 protein levels as compared with CM only (Figure 4D, left side). Pdcd4 protein rapidly decreased when translation was blocked in CM-only treated cells, whereas Pdcd4 protein levels remained high in CM/rapamycin co-treated cells (Figure 4D). Cells pre-incubated with CM for 16 h before rapamycin addition for another 4 h followed by a subsequent block of the translation by CHX, displayed a very similar development of Pdcd4 protein levels (Figure 4E). That is, Pdcd4 protein was stabilized in the presence of mTOR inhibitor irrespective of the duration of the CM exposure. To further connect phosphorylation and proteasomal degradation of Pdcd4, we next determined phosphorylation of Pdcd4 at the p70S6K-phosphorylation site. MCF7 cells treated for 8 h with CM displayed increased phospho-Pdcd4 to total Pdcd4 ratios as compared with Ctr-treated cells (Figure 4F). Phosphorylation of Pdcd4 was attenuated by rapamycin co-treatment, yet, further increased when proteasomal degradation was blocked by lactacystin. Further proof for the efficacy of PI3K and mTOR inhibitors to inhibit p70S6K activity came from the observation that phosphorylation of ribosomal protein S6 was completely inhibited after co-treatment of CM with either LY294002 or rapamycin for 8 or 24 h (supplementary Figure S2 is available at Carcinogenesis Online).
Fig. 4.
CM-induced Pdcd4 protein degradation requires intact PI3K–mTOR signaling. MCF7 cells were incubated with CM in combination with LY294002 [10 μM], rapamycin [50 nM], PD98059 [50 μM] or SB203580 [10 μM] for (A) 8 h or (B) 24 h. (C) MCF7 cells were incubated with CM or Ctr for 24 h. LY294002 [10 μM] or rapamycin [50 nM] were added for the last 8 h of the incubations. (D) MCF7 cells were incubated with CM (±rapamycin [50 nM]) for 4 h and (E) pre-incubated with CM for 16 h before incubations continued in the presence or absence of rapamycin [50 nM] for 4 h. Then, CHX [10 μM] was added to block translation and incubations continued for 1, 2 or 4 h (D + E). (F) MCF7 cells were incubated with Ctr or CM for 8 h in combination with rapamycin [50 nM] or lactacystin [10 μM]. Whole-cell extracts were subjected to western blot analysis and probed with the indicated antibodies. Blots are representative of at least three independent experiments. Ratios of pPdcd4 to Pdcd4 were determined after densitometric analysis of the expression and are presented as means of four independent experiments (F).
To determine if inflammatory conditions also affect Pdcd4 protein expression in vivo, we employed DSS, which is known to induce inflammation of the colon. Initial experiments indicate that after three cycles of DSS (1 or 2%), Pdcd4 protein levels were attenuated in whole colon homogenates at 2% DSS as compared with untreated controls, whereas 1% DSS-receiving mice showed unaltered Pdcd4 expression (Figure 5). Interestingly, only at 2% DSS signs of inflammation, i.e. mild rectal bleeding, was observed. Even though these data are preliminary, they can be taken as an indicator that the concept of Pdcd4 degradation under inflammatory conditions might hold true in vivo as well.
Fig. 5.
Pdcd4 protein is lost in response to DSS-induced colitis. Colitis was induced by exposing mice to DSS via the drinking water for 7 days followed by 7 days of DSS-free water. After the third cycle, mice were killed, whole colon homogenates were subjected to western blot analysis and probed with the indicated antibodies.
Taken together, our data indicate that PI3K–mTOR-dependent destabilization of Pdcd4 protein is the key event limiting Pdcd4 protein levels in response to short term as well as extended exposure to inflammatory microenvironmental conditions.
Discussion
Inflammatory conditions are increasingly acknowledged to contribute to tumor formation. Inflammation was even introduced as a seventh hallmark of cancer development (35,36). In this study, we demonstrate that Pdcd4 protein expression is strongly attenuated in tumor cells when they are exposed to macrophage-associated inflammatory conditions. The inflammation-induced loss of Pdcd4 protein was due to increased proteasomal degradation, which critically required activation of PI3K–mTOR signaling. Initial results from a colitis model indicate that loss of Pdcd4 might occur under inflammatory conditions in vivo as well.
Inflammatory conditions have been causally connected to many prevalent tumor types (37,38). Importantly, although inflammation initially has been associated primarily with the emergence of neoplastic lesions, the interaction between tumor cells and the inflammatory microenvironment also appears to be critical for the progression of tumors (39,40). Macrophages constitute a significant proportion of the immune cell infiltrate and contribute to tumor progression (41,42). The importance of macrophages is underlined by clinical studies correlating high macrophage contents in tumors with poor prognosis (43). The tumor suppressor Pdcd4 was shown previously to be lost in various tumor types and loss of Pdcd4 was associated with increased tumorigenic potential and tumor progression (19–31). Lack of Pdcd4 protein was even put forward as a prognostic marker for colon cancer-associated survival (31). To determine the connection between macrophage-dependent inflammatory conditions and Pdcd4 protein amount in tumor cells, we used an in vitro model based on activated U937 monocyte-derived macrophages, which display increased expression and secretion of pro-inflammatory cytokines such as TNFα, IL-6 and IL-8 [Figure 1 and (44)]. The fact that CM attenuated Pdcd4 protein expression in a panel of tumor cell lines, representing a selection of prominent tumor sites (colon, prostate and breast) is indicative for a general mode of action (Figure 2A). The observation that coculture with differentiated U937 and CM elicited comparable effects on Pdcd4 expression and activator protein-1 transactivation further indicates that secreted factor(s) are involved and that the mechanism is unidirectional in nature (Figure 2; supplementary Figure S1 is available at Carcinogenesis Online). Further studies are required to identify those factors in the CM that contribute to the downregulation of Pdcd4. Identification of such factors may allow to specifically target the Pdcd4-regulating components in the tumor micro-environment.
Mechanistically, we identify PI3K–mTOR-dependent proteasomal degradation as key pathway for Pdcd4 downregulation in our macrophage-associated model (Figure 4). Similarly, Sheedy et al. (45) recently demonstrated that Pdcd4 is lost in macrophages in response to inflammatory stimuli such as lipopolysaccharide. They determined proteasomal degradation of Pdcd4 as responsible for the loss of Pdcd4 up to 6 h only. Extended incubations were associated with miR-21-dependent repression of Pdcd4, in contrast. Our data corroborate their short-term effects only. The observation that Pdcd4 protein can be recovered by inhibition of proteasomal degradation or PI3K–mTOR signaling even after extended CM incubations, where Pdcd4 protein was almost undetectable (Figures 3C and 4), is indicative for ongoing degradation as the prime mechanism in our model. Furthermore, our previous findings that Pdcd4 mRNA levels are significantly reduced in response to CM (44) have to be extended in that the loss of Pdcd4 mRNA appears not to be sufficient to repress Pdcd4 protein levels under inflammatory conditions. This is further corroborated by the fact that while Pdcd4 protein regulation was sensitive to PI3K–mTOR inhibition (Figure 4), Pdcd4 mRNA regulation was not (44). While Pdcd4 transcription and translation appear to play a lesser role under these conditions, it cannot be ruled out that under chronic inflammatory conditions Pdcd4 mRNA regulation or translational repression by miR-21 may contribute to the repression of Pdcd4 expression.
Provided that inflammation-induced reduction of Pdcd4 protein in vivo can be substantiated by larger experimental cohorts and other inflammatory conditions, loss of Pdcd4 might serve as an indicator of inflammatory microenvironments. Interestingly, Pdcd4 degradation might even occur at early stages where no tumorigenic lesions are detectable. In line, reduction of Pdcd4 was previously established as an early prognostic marker for colon cancer-associated survival (31). Taking the tumor suppressive function of Pdcd4 into account, it will be interesting to see if Pdcd4 might not only serve as an early biomarker, but whether its stabilization could be exploited for therapeutic intervention. We have shown previously that a loss of Pdcd4 occurred in all papillomas that developed in the inflammation-associated two-stage skin carcinogenesis model, even if Pdcd4 was transgenically overexpressed (12,14). The fact that a complete loss of Pdcd4 also occurred in tumors developing in transgenically Pdcd4 overexpressing mice is a further proof that mechanisms targeting the protein directly are critical for the regulation rather than transcriptional or mechanisms requiring the untranslated regions of the mRNA such as miR-dependent effects. Importantly, loss of Pdcd4 not only serves as a potent marker for tumorigenesis but also appears critical to facilitate tumor development. This notion is corroborated by the fact that, while Pdcd4 levels in the papillomas were strongly attenuated, inherent Pdcd4 protein expression of the skin determined the susceptibility of the mice to this inflammatory tumor model.
In conclusion, we propose that downregulation of Pdcd4 under inflammatory conditions establishes Pdcd4 protein stabilization as a promising target for the development of novel tumor therapeutics. Importantly, while stabilization of Pdcd4 can be predicted to occur in various currently used therapeutic approaches (e.g. mTOR or proteasome inhibitors), it will be important to further elucidate the functional relevance of Pdcd4 stabilization under such settings and, eventually, it might be beneficial to develop specific Pdcd4-targeted therapeutic approaches.
Supplementary material
Supplementary Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported by the DFG (BR999, GRK1172); LOEWE Schwerpunkt OSF [III L 4-518/55.004 (2009)] funded by the Hessian Ministry of Higher Education, Research and Arts. Part of the work was supported by the Intramural Research Program of the National Institutes of Health; National Cancer Institute; Center for Cancer Research.
Supplementary Material
Acknowledgments
We thank Marina Talamini and Glen Hegamyer for excellent technical assistance and Chris Perella for animal support.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CHX
cycloheximide
- CM
conditioned medium
- DSS
dextran sodium sulfate
- IL
interleukin
- mTOR
mammalian target of rapamycin
- miR-21
microRNA-21
- mRNA
messenger RNA
- Pdcd4
programmed cell death 4
- PI3K
Phosphatidylinositol-3-kinase
- TNF
tumor necrosis factor
- TPA
12-O-tetradecanoylphorbol 13-acetate
References
- 1.Yang HS, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 2003;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang JH, et al. Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl Acad. Sci. USA. 2009;106:3148–3153. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaRonde-LeBlanc N, et al. Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol. Cell. Biol. 2007;27:147–156. doi: 10.1128/MCB.00867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh PG, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 2009;28:274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svitkin YV, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 7.Yang HS, et al. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol. Cell. Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 9.Leupold JH, et al. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 10.Santhanam AN, et al. Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion. Oncogene. 2010;29:3921–3932. doi: 10.1038/onc.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, et al. Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene. 2008;27:1527–1535. doi: 10.1038/sj.onc.1210793. [DOI] [PubMed] [Google Scholar]
- 12.Jansen AP, et al. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 13.Hilliard A, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol. 2006;177:8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 14.Schmid T, et al. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 15.Dorrello NV, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 16.Frankel LB, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 17.Lu Z, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 18.Folini M, et al. miR-21: an oncomir on strike in prostate cancer. Mol. Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motoyama K, et al. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int. J. Oncol. 2010;36:1089–95. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]
- 20.Reis PP, et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol. Cancer. 2010;9:238. doi: 10.1186/1476-4598-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit. Rev. Oncol. Hematol. 2010;73:185–91. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Bredel M, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carinci F, et al. Potential markers of tongue tumor progression selected by cDNA microarray. Int. J. Immunopathol. Pharmacol. 2005;18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- 24.Fang W, et al. Transcriptional patterns, biomarkers and pathways characterizing nasopharyngeal carcinoma of Southern China. J. Transl. Med. 2008;6:32. doi: 10.1186/1479-5876-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fassan M, et al. Programmed cell death 4 protein in esophageal cancer. Oncol. Rep. 2010;24:135–9. doi: 10.3892/or_00000838. [DOI] [PubMed] [Google Scholar]
- 26.Gao F, et al. Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol. Rep. 2007;17:123–128. [PubMed] [Google Scholar]
- 27.Hayashi A, et al. Pdcd4 expression in intraductal papillary mucinous neoplasm of the pancreas: its association with tumor progression and proliferation. Hum. Pathol. 2010;41:1507–1515. doi: 10.1016/j.humpath.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Matsuhashi S, et al. Expression patterns of programmed cell death 4 protein in normal human skin and some representative skin lesions. Exp. Dermatol. 2007;16:179–184. doi: 10.1111/j.1600-0625.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 29.Wen YH, et al. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol. Rep. 2007;18:1387–1393. [PubMed] [Google Scholar]
- 30.Zhang Z, et al. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20:4450–4456. doi: 10.1038/sj.onc.1204588. [DOI] [PubMed] [Google Scholar]
- 31.Mudduluru G, et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–1707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 32.Akar U, et al. Targeting p70S6K prevented lung metastasis in a breast cancer xenograft model. Mol. Cancer Ther. 2010;9:1180–1187. doi: 10.1158/1535-7163.MCT-09-1025. [DOI] [PubMed] [Google Scholar]
- 33.Blees JS, et al. Development of a high-throughput cell-based reporter assay to identify stabilizers of tumor suppressor Pdcd4. J. Biomol. Screen. 2010;15:21–29. doi: 10.1177/1087057109351028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen AP, et al. Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol. Cancer Ther. 2004;3:103–110. [PubMed] [Google Scholar]
- 35.Colotta F, et al. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 36.Hanahan D, et al. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Balkwill F, et al. Cancer: an inflammatory link. Nature. 2004;431:405–406. doi: 10.1038/431405a. [DOI] [PubMed] [Google Scholar]
- 38.Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 39.Pages F, et al. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 40.Sica A, et al. Cancer related inflammation: the macrophage connection. Cancer Lett. 2008;267:204–215. doi: 10.1016/j.canlet.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 41.Solinas G, et al. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol. 2009;86:1065–1073. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 42.Porta C, et al. Cellular and molecular pathways linking inflammation and cancer. Immunobiology. 2009;214:761–777. doi: 10.1016/j.imbio.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Mantovani A, et al. Inflammation and cancer: breast cancer as a prototype. Breast. 2007;16(suppl. 2):S27–S33. doi: 10.1016/j.breast.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Yasuda M, et al. Downregulation of programmed cell death 4 by inflammatory conditions contributes to the generation of the tumor promoting microenvironment. Mol. Carcinog. 2010;49:837–848. doi: 10.1002/mc.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheedy FJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.