Abstract
A comprehensive understanding of molecular mechanisms driving cancer onset and progression should provide a basis for improving early diagnosis, biomarker discovery and treatment options. A key value of genetically engineered mice for modeling human cancer is the possibility to analyze the entire process of tumor development. Here, we applied functional genomics approach to study step-by-step development of hepatocellular carcinoma (HCC) in the c-Myc/Tgfα transgenic mouse model of aggressive human liver cancer. We report that coexpression of c-Myc and Tgfα induces progressive and cumulative transcriptional alterations in the course of liver oncogenesis. Functional analysis of deregulated genes at the early stage of HCC disease supports a model of active hepatocyte proliferation on the background of chronic oxidative stress generated by a general metabolic disorder. In addition, early and persistent deregulation of numerous immune-related genes suggested that disruption of immune microenvironment may contribute to oncogenic process in this model of accelerated liver carcinogenesis. In particularly, by flow cytometry analysis, we found loss of the major histocompatibility complex class I expression in dysplastic hepatocytes followed by upregulation of numerous activating ligands for natural killer (NK) cells concomitant with a drastic decrease in hepatic NK cell frequency. In conclusion, our study provides a comprehensive characterization of sequential molecular changes during a stepwise progression of preneoplastic lesions toward HCC and highlights a critical role of metabolic disorders and innate immunity at the early stages of liver cancer.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of visceral cancer worldwide with >550 000 new cases diagnosed each year. HCC also ranks among the deadliest forms of human malignancies with 600 000 deaths annually and a mean survival of 6 months from the time of diagnosis (1,2). Increased incidence and mortality rate along with the lack of effective curative treatment options for advanced HCC has rendered the disease a major health problem worldwide.
Hepatocarcinogenesis in humans is a slow process that may take >30 years after a chronic hepatitis is first diagnosed (3). During this long process, the accumulation of irreversible structural alterations in chromosomes and genes eventually leads to the emergence and the expansion of clonal populations of transformed hepatocytes that evolve toward HCC. Numerous genomic changes, including genomic instability, aberrant methylation and profound alterations in gene expression have been reported in HCC (3,4). However, the precise sequence of molecular events involved in tumor initiation and progression is not well defined due to the limited access to early stages of tumor development in humans (3). In this context, genetically engineered mice represent a valuable experimental system for a sequential analysis of cancer onset and progression. By using a cross-species comparative oncogenomics approach, we identified previously the best-fit mouse models to study human hepatocarcinogenesis (5). Notably, we have demonstrated that transgenic mice which concomitantly overexpress c-Myc and transforming growth factor alpha (Tgfα) in the liver develop HCC with molecular phenotype that mimics a subset of human tumors with poor prognosis (5), suggesting that hepatocarcinogenesis in c-Myc/Tgfα mice and aggressive subsets of human HCC may progress through similar molecular mechanisms.
In the present study, we used the c-Myc/Tgfα transgenic mouse model to provide a comprehensive dynamic characterization of the cellular and molecular alterations involved in HCC onset and progression. We demonstrate that coexpression of the potent oncogene c-Myc with Tgfα generates a broad spectrum metabolic disorder that promotes progressive accumulation of genetic alterations due to chronic stimulation of hepatocyte proliferation in an oxidative stress microenvironment. By using a functional genomics approach focused on the early stages of HCC disease, we further find that disruption of innate immune surveillance mediated by natural killer (NK) and natural killer T (NKT) cells may contribute to oncogenic process in this model of accelerated liver cancer.
Materials and methods
Transgenic and wild-type mice
Male c-Myc/Tgfα double transgenic were produced by crossing homozygous B6CBA Alb/c-Myc (6) and B6CBAxCD1 Mt/Tgfα single transgenic mice (7). Male wild-type (WT) mice were generated in a B6CBAxCD1 background as described previously (8). Liver samples were collected at various time-points ranging from 3 weeks (moderate dysplasia), 3 months (severe dysplasia) and 9 months (HCC). Tissue samples were divided into two parts: one was fixed in 10% formalin for histopathological evaluation and the other was used for RNA analysis. Total RNAs were isolated from dysplastic livers, HCC and surrounding non-tumor livers (5–15 mice per each group, including WT controls). RNAs isolated from the liver of the age-matched WT mice were used as reference for microarray experiments as described (9). All experimental procedures were approved by the NCI-Bethesda Animal Care and Use Committee (ACUC). The NCI animal program meets or exceeds requirements of the Public Health Service Policy on Humane Care and Use of Laboratory Animals and is fully accredited by AAALAC International.
Genome-wide messenger RNA analysis
Total RNAs were purified using a CsCl gradient-density centrifugation method (9). A genome-wide set of mouse oligonucleotides obtained from Illumina (San Diego, CA) and printed at the Advanced Technology Center (NCI/NIH) was used in this study. Gene expression profiling was performed as described (9). MIAME compliant microarray data have been deposited to the GEO database (GSE25587). Differentially expressed genes were clustered using K-mean algorithm and visualized using TreeView. Data mining was performed using Gene Ontology annotations and relevant gene networks were identified by Ingenuity Pathway Analysis (Ingenuity Systems, Mountain View, CA).
Hepatocyte isolation and flow cytometry analysis
Hepatocytes were isolated by a 2-step collagenase perfusion of mouse liver followed by isodensity purification in Percoll gradient (10). Cell viability was >90% as determined by trypan blue exclusion. For major histocompatibility complex (MHC)-I and Raet1 staining, a single-cell suspension of freshly isolated hepatocytes was incubated for 20 min at 4°C with phycoerythrin-conjugated anti-mouse H-2Kb/H-2Db (Catalog #114607; BioLegend, San Diego, CA) and allophycocyanin-conjugated anti-mouse-Raet-1 pan-specific (Catalog #FAB17582A; R&D Systems, Minneapolis, MN) monoclonal antibodies. Hepatocytes were washed twice with phosphate-buffered saline and analyzed on a FACSCalibur G4 flow cytometer using Flowjo software (BD Bioscience, San Jose, CA).
Analysis of liver NK and NKT cells
Mouse liver mononuclear cells were isolated as described previously (11). Hepatic NK and NKT cells were analyzed by flow cytometry using phycoerythrin-conjugated anti-mouse CD49b/Pan-NK (Pharmingen, San Diego, CA) and allophycocyanin-conjugated anti-mouse-CD3e monoclonal antibodies. After exclusion of dead cells using propidium iodide labeling, mononuclear cells were gated to evaluate the proportion of Cd49b+CD3e− (NK) and Cd49b+Cd3e+ (NKT) cells at 3 and 12 weeks. Data were acquired from three to six experiments per group.
Statistical analysis
Microarray data were analyzed using BRB-ArrayTools software developed by Richard Simon and BRB-ArrayTools Development Team (12). Differentially expressed genes were identified by a univariate two-sample t-test with a random variance model (10). Permutation P-values for significant genes (P < 0.001) were computed based on 10 000 random permutations with a false discovery rate <1% and a 95% confidence interval. Expression of Raet1 and MHC-1 proteins was evaluated by the non-parametric two-tailed Mann–Whitney test using Prism software, version 5.01 (GraphPad Software, Inc., La Jolla, CA).
Results
Progressive and cumulative transcriptional alterations during HCC development in c-Myc/Tgfα transgenic mice
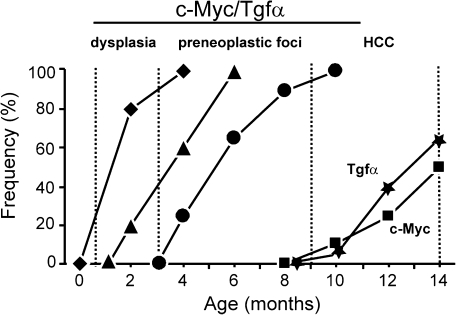
Our earlier work has established that hepatic co-expression of c-Myc and Tgfα accelerates HCC development as compared with each parental strain (Figure 1) (8,13). We also showed that neoplastic lesions in c-Myc/Tgfα mice develop through a distinct sequence of events including early (3 weeks) and widespread (3 months) dysplasia followed by the appearance of preneoplastic foci that advance to HCC stage with 100% penetrance by 10 months (Figure 1).
Fig. 1.
Incidence and kinetics of hepatic lesions in c-Myc/Tgfα mice. Combined hepatic overexpression of c-Myc with Tgfα resulted in tumor development with a shorten latency and a higher incidence (left 3 curves) when compared with single transgenic strains (right 2 curves, Tgfα HCC and c-Myc HCC). Hepatocarcinogenesis in c-Myc/Tgfα mice progressed from early and severe hepatocyte dysplasia (closed diamonds) to preneoplastic focal lesions (closed triangles) and HCC (closed circles) within 4 months. By 9 months, >90% c-Myc/Tgfα mice developed HCC in a background of severe dysplasia, whereas single transgenic strains did not exhibit any tumors. Mice were killed at various times-points (vertical dotted bars; 5–15 mice per each group) and macrodissected liver samples were submitted to a gene expression profiling study. Only male mice were used in this study.
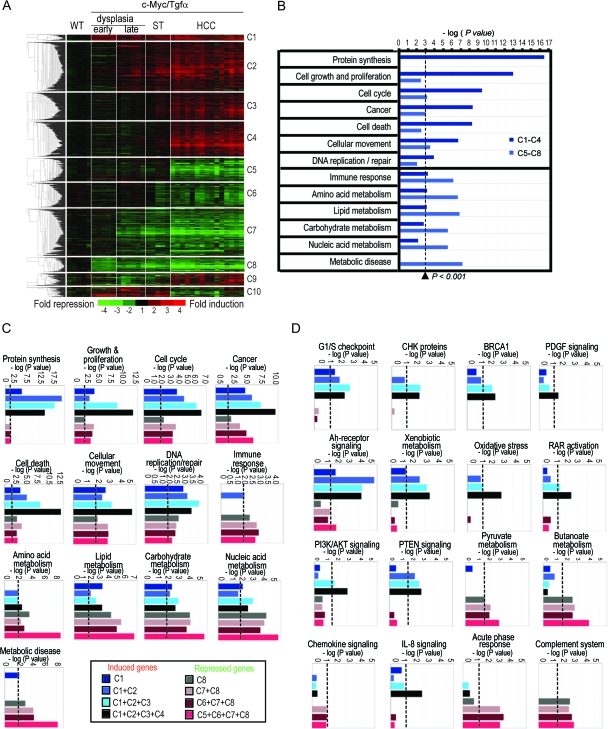
To define the sequential changes in transcriptome during a stepwise progression toward malignancy, we performed a genome-wide expression profiling at various stages of tumor development (3 weeks, 3 months and 9 months, Figure 1). As shown in Table I, the number of genes differentially expressed in c-Myc/Tgfα transgenic mice versus WT mice progressively increased from early dysplasia (3 weeks) to tumor stage (9 months). In total, the expression of 2802 genes was altered (P < 0.001, false discovery rate < 1%). All significant genes were further organized into 10 clusters (C1–C10) using a K-means algorithm (Figure 2A). C1–C4 and C5–C8 clusters contained genes that were, respectively, induced and repressed at specific stages of tumor development. For example, clusters C1 and C8 included genes with alterations initiated at an early stage of dysplasia and maintained up to the final tumor stage, whereas clusters C4 and C5 included genes which expression levels were altered exclusively in the fully developed tumors (Figure 2A). Clusters C9 and C10 included genes with opposite regulation in HCC versus other stages. Thus, the unsupervised clustering analysis based on the expression of all significant genes revealed that the transcriptional changes induced in c-Myc/Tgfα livers were progressive and cumulative during the course of HCC development.
Table I.
Number of genes differentially expressed during the course of HCC development in c-Myc/Tgfα transgenic mice
| c-Myc/Tgfα transgenic mice |
|||||
| 3 wk | 3 mo | 9 mo (ST) | 9 mo (HCC) | ||
| WT mice | 197* | 613 | 1175 | 2896 | |
| c-Myc/Tgfα | 3 Weeks | — | 910 | 841 | 2044 |
| 3 Months | — | — | 267 | 2152 | |
| 9 Months (ST) | — | — | — | 1245 | |
ST, surrounding non-tumor tissue; mo, months; wk, weeks.
*P < 0.001, random-variance t-test, false discovery rate <1%.
Fig. 2.
Functional analysis of genes altered during the course of HCC development in c-Myc/Tgfα mice. (A) Dendrogram and heat-map overview of genes differentially expressed in c-Myc/Tgfα livers at various stages of HCC development [early dysplasia, 5 mice; late dysplasia, 6 mice; HCC and surrounding tissue (ST), 15 and 5 mice, respectively]. Data are presented in a matrix format in which rows and columns represent genes and samples, respectively. Gene expression values are the mean of two separate experiments (each RNA sample was analyzed twice using a dye-swap design). K-means algorithm organized significant genes in 10 clusters (C1–C10) and highlighted a progressive accumulation of transcriptional alterations during the course of HCC development in c-Myc/Tgfα mice. (B) Functional characterization of upregulated (C1–C4) and downregulated (C5–C8) genes by ingenuity pathway analysis. (C and D) Detailed time-course analysis of biological functions (C) and canonical pathways (D) enriched during hepatocarcinogenesis in c-Myc/Tgfα mice, using a progressive aggregation of deregulated gene clusters from early stage of dysplasia to fully developed HCC (C1 to C4 and C8 to C5 for up- and downregulated genes, respectively). P values were determined using the ingenuity scoring system and were judged significant at a 0.001 level (B–D, vertical dashed line).
From cluster organization to functional modules
An extensive functional analysis of the gene clusters was further performed using the Ingenuity Pathway Analysis tools. Validating our gene selection, this approach unambiguously correlated the transcriptional alterations with the histological changes observed during c-Myc/Tgfα hepatocarcinogenesis (8). Early stages were characterized by a sustained induction of genes linked to hyperproliferation and dysplasia, whereas the progressive induction of HCC-related genes was prevalent at the later stages (supplementary Figure 1 is available at Carcinogenesis Online). Induced genes (clusters C1–C4) were significantly associated with protein synthesis, cell proliferation and DNA repair, whereas repressed genes (clusters C5–C8) were related to immunological and metabolic disorders (Figure 2B–D). To further identify key molecular alterations that could be responsible for the rapid tumor initiation and progression in this model, we adopted a supervised approach based on the functional analysis of genes deregulated at each stage of HCC development.
Cell cycle and immune system abnormalities at an early stage of hepatocyte dysplasia
The first signs of hepatocyte dysplasia were obvious in c-Myc/Tgfα livers already at 3 weeks of age (Figure 1). Data mining of 197 differentially expressed genes (Table I) indicated a predominant induction of genes involved in cell proliferation and repression of immune-related genes as the first transcriptional changes at this early stage (Figure 2C–D; supplementary Table I is available at Carcinogenesis Online). The upregulation of genes involved in DNA replication (e.g. Mcm3-6, Rfc4) as well as the induction of survivin (Birc5), an inhibitor of apoptosis which expression is markedly elevated with the onset of DNA synthesis (14), was consistent with a high proliferation rate. The expression of key cell division regulators involved in mitotic spindle organization and chromosome segregation (e.g. Cenpa, Mad2l2, Stmn1, Pttg1) was simultaneously altered. The concomitant induction of DNA repair sensing genes (e.g. Ddb2, Mlh1, Msh2, Rad50, Rad51l1) suggested abnormalities of DNA replication and cell division well before the appearance of the first signs of focal growth.
Furthermore, numerous genes encoding innate immunity-related proteins such as acute-phase proteins (e.g. Ctsc, C9 complement component, Orm1-2, Saa1) were repressed at this stage (Figure 2D; supplementary Table I is available at Carcinogenesis Online). Genes involved in the MHC class I antigen presentation were also downregulated as exemplified by β2-microglobulin and H2-D1, -Q10 and -T9 genes. Clec2d gene, encoding a ligand for the NK cell inhibitory receptor Cd161, was induced.
Metabolic perturbations and immune surveillance disruption at the late stage of hepatocyte dysplasia
A widespread hepatocyte dysplasia preceding the appearance of preneoplastic foci is a hallmark feature of c-Myc/Tgfα hepatocarcinogenesis (Figure 1). At this stage, expression of 613 genes was altered when compared with the age-matched control animals (Table I). As observed previously, a sustained high proliferation rate in transgenic livers correlated with the induction of proliferation-associated markers, such as nucleolar protein 1 (Nol1) and numerous translation-related factors (e.g. Eif4b, Eef1b2, Eef2, Rpl4, Rpl12, Rps12) reflecting an increased protein synthesis (Figure 2; supplementary Table II is available at Carcinogenesis Online). At the same time, proapoptotic genes (e.g. Bmf, Bok, Tnfsf10/Trail) were repressed, and Birc6, an inhibitor of apoptosis, was induced.
Functional analysis of genes altered at this stage also pointed to a major metabolic disorder. As an example, genes involved in xenobiotic metabolism (e.g. aryl hydrocarbon receptor and Nrf2-mediated oxidative stress signaling pathways) were upregulated (Figure 2D) suggesting that transgenic hepatocytes were subjected to oxidative stress. Several heat shock proteins and Hsp-interacting proteins were also upregulated. One of the most prominent metabolic alterations involved genes associated with beta oxidation of fatty acids (e.g. Acadm, Ehhadh, Oxsm). A Cyp4a14 gene encoding a microsomal lipid peroxidizing enzyme involved in fatty acid omega oxidation was induced and the expression of several peroxisomal enzymes was altered (e.g. Acaa1b, Acox1, Acox3). The antioxidant enzyme peroxiredoxin 5, which reduces hydrogen peroxides and alkyl hydroperoxides, was downregulated. In the context of active proliferation associated with a chronic oxidative stress environment, an extensive DNA damage response signature was identified (Figure 2C–D; supplementary Table II is available at Carcinogenesis Online). Also, Pttg1 gene was strongly downregulated potentially further amplifying the genomic instability. Pttg1, encoding the homolog of the yeast securin which prevents separins from promoting sister chromatid separation, is known to be required for chromosomal stability (15).
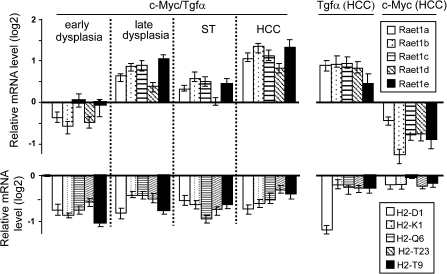
Most of the genes involved in innate immunity remained downregulated in the dysplastic livers (Figure 2C–D; supplementary Table II is available at Carcinogenesis Online), including components of the complement pathway (e.g. C6, C9, Mbl1), the aforementioned acute-phase proteins (e.g. Saa1-3, Orm1) and MHC-I-encoding genes (e.g. H-2K1, H-2D1). In contrast, a cluster of genes located at the 10A3 locus, and encoding retinoic acid early transcripts (Raet1a-e), was induced (Figure 3). Raet1 molecules are ligands for Nkg2d, an activating receptor expressed by immune cells such as NK and cytolytic T cells. Raet1s are thought to play an important role in mediating NK antitumor responses (16).
Fig. 3.
Raet1 and MHC-I messenger RNA levels in hepatic lesions derived from c-Myc/Tgfα, c-Myc and Tgfα mice. Relative messenger RNA level for Raet1a-e and MHC-I (H2-D1, -K1, -Q6, -T23, -T9) genes was determined at various stages of tumor development in c-Myc/Tgfα mice as well as in HCC from c-Myc and Tgfα single transgenic animals. In c-Myc/Tgfα mice, MHC-I genes were repressed in early dysplastic lesions and Raet1a-e genes were induced at a later stage. Similar profiles were observed in Tgfα-induced HCC. Expression values were expressed as log2 ratios using WT mice as a reference (bars, mean ± SD; 5–15 mice per group).
A gene expression profile of dysplastic lesions at the HCC stage
By 9 months, 100% transgenic mice developed HCC in a background of severe hepatocyte dysplasia (Figure 1). Not surprisingly, the gene expression profiles of the dysplastic lesions at 3 and 9 months were highly similar. Actually, only 267 genes were differentially expressed between these two stages (Table I), and the differences were mostly quantitative (e.g. greater fold induction or repression). Thus, the genes which expression was altered in the surrounding non-tumorous tissues were enriched in the same functional modules which were identified in the livers of 3-month-old mice (Figure 2C–D). Genes involved in fatty acid β-oxidation (e.g. Acadm, Acox1, Ehhadh) were notably induced along with key enzymes involved in the detoxification of aldehydes arising from lipid peroxidation (e.g. Aldh1b1, Aldh3a2). Genes involved in detoxication pathways, including P450 cytochromes (e.g. Cyp2b10), uridine 5'-diphospho-glucuronosyltransferase glucuronosyltransferases (e.g. Ugt1a7c), glutathione-S-transferases (e.g. Gsta1) and sulfotransferases (e.g. Sult1c2), were similarly induced. These alterations coincided with the induction of Nr1i3/Car, a central regulator of xenobiotic metabolism known to control the transcription of these genes (17). As observed previously, immune-related genes remained downregulated, including components of complement pathway, MHC-I and numerous cytokines associated with their cognate receptors (e.g. Csf1/Csf1r, Il13/Il13ra1, Lif/Lifr).
Gene alterations in HCC
The most dramatic changes in terms of gene expression were found in tumors (Table I, Figure 2A). Alterations of genes involved in all functional categories described above, including protein synthesis, cell cycle/death, DNA repair, oxidative stress, detoxication, cytoskeleton organization, metabolic and immune disorders, culminated in HCC (Figure 2C–D; supplementary Table III is available at Carcinogenesis Online). For example, 15 translation initiation/elongation factors (e.g. Eif1a, Eef2), >55 ribosomal genes (e.g. Rpl3), aminoacyl-tRNA synthetases (e.g. Cars) and importins (e.g. Ipo4) were upregulated in HCC, indicating a global increase in protein synthesis and metabolic activity. An obvious oxidative stress signature was identified, which included the induction of key regulators of the Nrf2 pathway (Nfe2l2, Keap1, Mafk). In contrast, antioxidant enzymes were repressed (e.g. Gpx1, Prdx2). These results indicated that redox homeostasis was completely disrupted in transgenic livers. Master regulators of cell cycle checkpoint signaling pathways required for DNA damage response and genome stability were induced (e.g. Brca1, Rad21, Trp53). Finally, as observed at the earlier stages, tumor cells exhibited numerous alterations in the expression of genes involved in metabolism and immunity (Figure 2C–D, supplementary Table III is available at Carcinogenesis Online). Cross-comparison of gene expression profiles between HCCs from c-Myc/Tgfα mice and single transgenic counterparts (9) suggested that c-Myc and Tgfα mainly contributed to the deregulation of translation-associated and immune-related genes, respectively (supplementary Figure 2 is available at Carcinogenesis Online).
Disruption of immune microenvironment in the course of liver oncogenesis
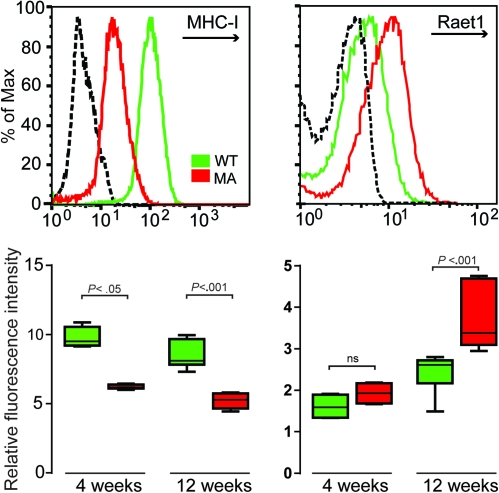
The deregulation of numerous immune-related genes at all stages of tumor development starting from early hepatocyte dysplasia suggested that the immune system may contribute to the rapid initiation and tumor progression in c-Myc/Tgfα transgenic mice. The network analysis revealed a significant enrichment of genes associated with the NK cell-mediated cytotoxicity pathway. NK cells activity is tightly controlled by a balance between inhibitory and activating signals, which are mediated through inhibitory (e.g. Ly49c/Klra3 in mouse) and activating (e.g. Nkg2d) receptors, respectively (18). Inhibitory receptors recognize MHC-I molecules, and activating receptors may eventually bind Raet1 ligands (18). Interestingly, the messenger RNA levels for MHC-I-associated genes (H-2Db/H-2Kd) were found to be decreased at the early stage of hepatocyte dysplasia (3 weeks), whereas the abundance of Raet1 messenger RNAs was increased at a later dysplastic stage (3 months) (Figure 3). Analysis of MHC-I and Raet1 expression in tumors derived from single transgenic mice suggested that Tgfα was the main contributor in modulating the expression of these genes (Figure 3). The expression of MHC-I and Raet1 was next evaluated on a protein level. Since microarray experiments were performed on RNA extracted from whole livers, including both hepatocytes and stromal cells, we performed fluorescence activated cell sorting analysis using purified hepatocytes. As shown in Figure 4, protein expression of MHC-1 and Raet1 closely recapitulated messenger RNA profiles from whole livers. Hepatocytes isolated from c-Myc/Tgfα transgenic mice exhibited a decrease in MHC-I expression starting at 4 weeks which was followed by increased Raet1 expression at 12 weeks (Figure 4). The fluorescence activated cell sorting results identified hepatocytes as the cell type responsible for the alterations in MHC-1 and Raet1 expression. These events, although representing an activating signal for NK cells (18), did not appear to slow down the rapid progression to malignancy suggesting that transformed hepatocytes are able to evade NK-mediated immune surveillance starting from the very early stage of disease.
Fig. 4.
Loss of MHC-I and gain of Raet1 protein expression in c-Myc/Tgfα dysplastic hepatocytes. At 4 and 12 weeks, hepatocytes were isolated from the liver of WT and c-Myc/Tgfα (MA) transgenic mice and analyzed by flow cytometry for the cell surface expression of MHC-I and Raet1 proteins. Upper two panels: representative flow cytometry profiles obtained at 12 weeks showing a loss of MHC-I and a gain of Raet1 expression in hepatocytes isolated from transgenic mice (red histogram) compared with WT animals (green histogram). Dashed black histogram, WT hepatocytes stained with the appropriate isotype controls. The results are representative of six independent experiments using individual mice. Lower two panels: quantitative analysis of MHC-I (left) and Raet1 (right) protein expression in WT (green box) and c-Myc/Tgfα (red box) at 4 and 12 weeks (four to six mice per each group). The results were obtained from four to six independent experiments. P values were determined using a non-parametric two-tailed Mann–Whitney test.
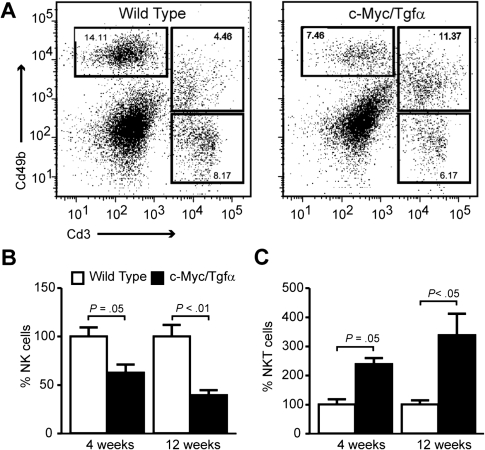
To evaluate the contribution of NK cells, intrahepatic mononuclear cells were isolated and analyzed by fluorescence activated cell sorting using anti-Cd49b and anti-CD3 antibodies. Although the cumulative frequency of Cd49b+ cells, which included both NK and NKT cells, was comparable in transgenic and WT animals, the relative contribution of hepatic NK (Cd49b+CD3−) and NKT (Cd49b+CD3+) cells was strikingly different (Figure 5A). The proportion of NK was significantly reduced, whereas the abundance of NKT cells was increased in c-Myc/Tgfα dysplastic livers as compared with WT controls (Figure 5B). The role of hepatic NKT cells in tumor rejection is complex since NKT cells may both promote or inhibit antitumor immunity (19,20). NKT cells can recognize tumor-derived phospholipids and glycolipids which are present in the context of Cd1d1, a non-classical MHC molecule (19). Interestingly, we have found that Cd1d1 expression was repressed in HCC developed in c-Myc/Tgfα transgenic mice (supplementary Figure 3 is available at Carcinogenesis Online).
Fig. 5.
Early disruption of NK and NKT cell abundance in the liver of c-Myc/Tgfα mice. Mononuclear cells were isolated from the liver of WT and c-Myc/Tgfα mice at 4 and 12 weeks. Cells (1–13 × 106) were stained using Cd3 and Cd49b monoclonal antibodies and analyzed by flow cytometry to evaluate the abundance of NK (Cd49b+CD3−) and NKT (Cd49b+Cd3+) cells. (A) Representative profiles of stained mononuclear cells isolated from livers of 12-week-old mice showing a decrease in NK cells and an increase in NKT cells in c-Myc/Tgfα mice compared with WT. The results are representative of four independent experiments using individual mice. (B and C) Quantitative analysis of NK (B) and NKT (C) cells in the liver of WT (white bars) and c-Myc/Tgfα (black bars) mice at 4 and 12 weeks (three to five individuals mice per each group). The results were obtained from three to five independent experiments. P values were determined using a non-parametric two-tailed Mann–Whitney test.
Discussion
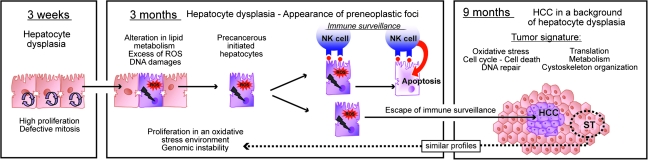
The conceptual basis for these studies was our earlier work that established c-Myc/Tgfα mice as the best-fit model to mimic human liver cancer with bad prognosis (5). Indeed, tumors developed in c-Myc/Tgfα mice recapitulated human HCC characterized by increased proliferation, decreased apoptosis and reduced patient survival. Here, applying a functional genomics approach under well-controlled experimental conditions, we identified and defined the exact sequence of underlying cellular and molecular mechanisms involved in tumor onset and progression in c-Myc/Tgfα mice (Figure 6). We demonstrated that the hepatic coexpression of c-Myc and Tgfα caused a synchronized activation of the stage-specific transcriptional programs from the early stages of hepatocyte dysplasia to fully developed HCC.
Fig. 6.
Proposed model for a multistage hepatocarcinogenesis in c-Myc/Tgfα mice.
The early transcriptome profiles remarkably corroborated the phenotypical and biochemical changes observed at the dysplastic stage, including abnormal production of reactive oxygen species, increased lipid peroxidation and high genomic instability (21–23). Although each category of biological functions and canonical pathways included upregulated and downregulated genes, we observed a general shift toward either up- or downregulation, suggesting a global activation or repression of given pathways. The c-Myc/Tgfα hyperproliferative phenotype correlated with the early deregulation of specific genes and signaling pathways that control cell cycling, execution of mitosis and DNA damage response. Among the notable genes at this stage was survivin, a member of the apoptosis inhibitor family. Survivin is highly expressed in most human tumors and has been shown to play a contributing role in regulation of cell-cycle entry and progression, particularly in G2/M phase (24). Constitutive activation of c-Myc and Tgfα also increased expression of genes involved in faithful chromosome segregation and spindle organization/biogenesis, such as Cenpa and Stmn1 (25,26). In addition, we found downregulation of Pttg1, encoding a homolog of yeast securin proteins preventing separins from promoting sister chromatid separation (20), as well as Mad2l2, a component of the mitotic spindle assembly checkpoint that controls a proper alignment of chromosomes at the anaphase onset (27). An early imbalance of these important regulators may provide an explanation for high genomic instability observed in c-Myc/Tgfα transgenic mice (22,23). Deregulation of genes found at 3 months persisted in the non-tumorous livers at 9 months, suggesting that the key alterations involved in the transformation were initiated at 3 months. Supporting this notion, we have shown previously that the transplantation of early dysplastic liver tissue into nude mice yielded rapid HCC growth (8).
Beyond abnormalities in cell cycle regulation and critical metabolic alterations promoting oxidative stress and genomic instability, our study emphasizes the disruption of immune microenvironment as an important early event in the carcinogenic process induced by c-Myc and Tgfα. The liver is known to play important functions both in innate and adaptive immunity and therefore is considered as an immunological organ per se (28,29). It exhibits unique features, such as a selective enrichment for immune cells, including macrophages, NK and NKT cells (28,29), implicated in the clearance of pathogens and tumor immunity. Cancer immune surveillance is a complex mechanism that notably involves CD8+ cytotoxic T lymphocytes, NK and NKT cells (30). Functional analysis of genes altered in the early dysplastic livers suggested that switches in the expression of specific ligands and receptors may allow transformed hepatocytes to escape immune surveillance. For example, early downregulation of MHC-I may represent a first mechanism by which transformed hepatocytes escape elimination by cytotoxic T lymphocyte, which recognition and killing activity are restricted to MHC-I molecules (30). NK cells also play an important role in tumor immunity through expression of activating and inhibitory receptors. Early and sustained induction of Clec2d gene during the course of oncogenesis in c-Myc/Tgfα transgenic mice may explain how transformed hepatocytes offset the premature loss of MHC-I that represents an activating signal for NK cells. Indeed, Clec2d is a ligand for the inhibitory receptor Cd161 expressed by NK cells as well as subsets of T and NKT cells (31). Importantly, the engagement of Cd161 with Clec2d expressed by target cells has been shown to inhibit NK cell-mediated cytotoxicity and interferon γ secretion (31). In addition, Raet1 genes which encode activating ligands for NK cells were not expressed in hepatocytes at 3 weeks thus contributing to prevent the killing activity by NK cells. Overexpression of Raet1 proteins occurred at the later stage (3 months). Raet1 genes are normally not expressed in adult tissues but their induction is considered to be a hallmark of stressed, infected or transformed cells (32), reinforcing the notion that hepatocytes acquired a transformed phenotype within the first 3 months in c-Myc/Tgfα livers. Strikingly, a hepatocyte-mediated increase in Raet1 expression occurred coincidentally with a decrease in the number of liver NK cells. Therefore, despite the correct expression of cell surface markers required for NK cells activation, initiated hepatocytes could escape immune surveillance through a mechanism limiting the number of NK cells, either by promoting apoptosis or by secreting factors inhibiting their recruitment. This mechanism may be particularly relevant in the context of high proliferation characteristic for the early stage of hepatocyte dysplasia in c-Myc/Tgfα mice (33). Besides the decrease in the number of NK cells, the functional relevance of the increased NKT abundance at early stages of hepatocarcinogenesis remains to be determined. Although the role of NK cells in mediating tumor immunity is well documented, the antitumor activity of NKT cells is more complex (34). Indeed, evidence suggests the existence of functionally distinct NKT cell subsets that exhibit either immunosuppressive activity promoting tumor formation or cell-mediated immunity against tumor cells (35–37). Altogether, our results suggest that transformed hepatocytes may evade immune surveillance by a dual mechanism involving a defect in recruiting NK cells into the liver at an early stage followed by a decrease in Cd1d1-restricted activation of NKT cells at later stages. Further experiments will be needed to validate this hypothesis, as an example by monitoring tumor progression in response to NK or specific NKT subset depletion or dysfunction.
In conclusion, our study provides a comprehensive characterization of hepatocarcinogenesis in c-Myc/Tgfα mice and emphasizes the critical role of immune microenvironment at the early stages of liver cancer development. By reproducing the broad spectrum of pathological and genetic changes characteristic of human HCC, c-Myc/Tgfα mouse model may offer novel opportunities to (i) functionally test and validate new candidate cancer genes, (ii) study the cross-talk between immune and tumor cells in vivo and (iii) design innovative immune-based therapies against liver cancer.
Supplementary material
Supplementary Figures 1–3 and Tables I–III can be found at http://carcin.oxfordjournals.org.
Funding
This research was supported by the intramural research program of the NIH/CCR/NCI, USA. C.C. is a recipient of a fellowship from NIH and Association pour la Recherche sur le Cancer, France.
Supplementary Material
Acknowledgments
We thank Anita Ton for her experimental help with isolation of mononuclear cells and Daekwan Seo for his help with GEO submission of microarray data.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- MHC
major histocompatibility complex
- NK
natural killer
- NKT
natural killer T
- WT
wild-type
References
- 1.El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, et al. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Lee JS, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 6.Murakami H, et al. Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis. Cancer Res. 1993;53:1719–1723. [PubMed] [Google Scholar]
- 7.Jhappan C, et al. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61:1137–1146. doi: 10.1016/0092-8674(90)90076-q. [DOI] [PubMed] [Google Scholar]
- 8.Santoni-Rugiu E, et al. Evolution of neoplastic development in the liver of transgenic mice co-expressing c-myc and transforming growth factor-alpha. Am. J. Pathol. 1996;149:407–428. [PMC free article] [PubMed] [Google Scholar]
- 9.Coulouarn C, et al. Oncogene-specific gene expression signatures at preneoplastic stage in mice define distinct mechanisms of hepatocarcinogenesis. Hepatology. 2006;44:1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- 10.Coulouarn C, et al. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong F, et al. Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J. Clin. Invest. 2002;110:1503–1513. doi: 10.1172/JCI15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon R, et al. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 13.Santoni-Rugiu E, et al. Acceleration of c-myc-induced hepatocarcinogenesis by co-expression of transforming growth factor (TGF)-alpha in transgenic mice is associated with TGF-beta1 signaling disruption. Am. J. Pathol. 1999;154:1693–1700. doi: 10.1016/s0002-9440(10)65425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deguchi M, et al. Expression of survivin during liver regeneration. Biochem. Biophys. Res. Commun. 2002;297:59–64. doi: 10.1016/s0006-291x(02)02128-9. [DOI] [PubMed] [Google Scholar]
- 15.Jallepalli PV, et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 16.Diefenbach A, et al. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, et al. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- 18.Waldhauer I, et al. NK cells and cancer immunosurveillance. Oncogene. 2008;27:5932–5943. doi: 10.1038/onc.2008.267. [DOI] [PubMed] [Google Scholar]
- 19.Berzofsky JA, et al. The contrasting roles of NKT cells in tumor immunity. Curr. Mol. Med. 2009;9:667–672. doi: 10.2174/156652409788970706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swain MG. Hepatic NKT cells: friend or foe? Clin. Sci. (Lond) 2008;114:457–466. doi: 10.1042/CS20070328. [DOI] [PubMed] [Google Scholar]
- 21.Factor VM, et al. Disruption of redox homeostasis in the transforming growth factor-alpha/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. J. Biol. Chem. 1998;273:15846–15853. doi: 10.1074/jbc.273.25.15846. [DOI] [PubMed] [Google Scholar]
- 22.Sargent LM, et al. Ploidy and karyotypic alterations associated with early events in the development of hepatocarcinogenesis in transgenic mice harboring c-myc and transforming growth factor alpha transgenes. Cancer Res. 1996;56:2137–2142. [PubMed] [Google Scholar]
- 23.Sargent LM, et al. Nonrandom cytogenetic alterations in hepatocellular carcinoma from transgenic mice overexpressing c-Myc and transforming growth factor-alpha in the liver. Am. J. Pathol. 1999;154:1047–1055. doi: 10.1016/s0002-9440(10)65357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki A, et al. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–3234. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 25.Belmont LD, et al. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 26.Valdivia M, et al. CENPA a genomic marker for centromere activity and human diseases. Curr. Genomics. 2009;10:326–335. doi: 10.2174/138920209788920985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, et al. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 2001;15:1765–1770. doi: 10.1101/gad.898701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao B, et al. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 29.Racanelli V, et al. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 30.Zitvogel L, et al. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat. Rev. Immunol. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 31.Aldemir H, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J. Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 32.Nausch N, et al. NKG2D ligands in tumor immunity. Oncogene. 2008;27:5944–5958. doi: 10.1038/onc.2008.272. [DOI] [PubMed] [Google Scholar]
- 33.Santoni-Rugiu E, et al. Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor alpha. Cancer Res. 1998;58:123–134. [PubMed] [Google Scholar]
- 34.Subleski JJ, et al. Application of tissue-specific NK and NKT cell activity for tumor immunotherapy. J. Autoimmun. 2009;33:275–281. doi: 10.1016/j.jaut.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terabe M, et al. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat. Immunol. 2000;1:515–520. doi: 10.1038/82771. [DOI] [PubMed] [Google Scholar]
- 36.Subleski JJ, et al. Enhanced antitumor response by divergent modulation of natural killer and natural killer T cells in the liver. Cancer Res. 2006;66:11005–11012. doi: 10.1158/0008-5472.CAN-06-0811. [DOI] [PubMed] [Google Scholar]
- 37.Crowe NY, et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J. Exp. Med. 2005;202:1279–1288. doi: 10.1084/jem.20050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.