Abstract
Chromosome 5p15.33 has been identified by genome-wide association studies as one of the regions that associate with lung cancer risk. A few single-nucleotide polymorphisms (SNPs) in the telomerase reverse transcriptase (TERT) and cleft lip and palate transmembrane 1-like (CLPTM1L) genes located in this region have shown consistent associations. We performed dense genotyping of SNPs in this region to refine the previously reported association signals for lung cancer risk. Two hundred and fifteen SNPs were genotyped on an Illumina iSelect panel, in a hospital-based case–control study of 1681 lung cancer cases and 1235 unaffected controls. Association was tested using unconditional logistic regression, while adjusting for age, sex and pack-years smoked. Furthermore, since many of the SNPs were in linkage disequilibrium (LD), haplotype blocks were constructed, from which tagging SNPs at an r2 threshold of ≥0.95 were included in a stepwise forward selection logistic regression model. Of the 215 SNPs, 69 were significant at P < 0.05 in univariate analysis; of these, 35 SNPs meeting the r2 threshold were included in the multiple logistic regression model. Two SNPs, rs370348 (odds ratio = 0.76, P = 1.6 × 10−6) and rs4975538 (odds ratio = 1.18, P = 0.005), significantly associated with risk in the overall sample. Among ever smokers, rs4975615 (odds ratio = 0.75, P = 1.2 × 10−4) and rs4975538 (odds ratio = 1.26, P = 0.002) were significant, whereas among never-smokers, rs451360 (odds ratio = 0.62, P = 7.6 × 10−5) was significant. We refined the consistent association signal in this region, allowing for the considerable LD between SNPs and identified four novel SNPs that were independently and significantly associated with lung cancer risk. Results of these analyses strongly suggest effects on risk from several loci in the TERT/CLPTM1L region.
Introduction
Genome-wide association studies (GWAS) have identified chromosome 5p15.33 as one of the regions that reproducibly associates with lung cancer risk and risk for several other cancers as well. 5p15.33 is one of the three regions, besides 15q25.1 and 6p21.33 with the strongest and most consistent association signals for lung cancer risk (1–3). Several studies have confirmed the association of single-nucleotide polymorphisms (SNPs) in the 5p15.33 region with lung cancer risk (4–8) suggesting that this region may have a role in lung cancer etiology. This 5p15.33 region comprises two candidate susceptibility genes, telomerase reverse transcriptase (TERT) and cleft lip and palate transmembrane 1-like (CLPTM1L). TERT encodes a catalytic subunit of telomerase that maintains telomere ends. Its overexpression leads to prolongation of the life span of the cell (9). Although not detectable in most normal tissues, it is overexpressed in cancer cells. CLPTM1L plays a role in apoptosis and has been found to be upregulated in cisplatin-resistant cell lines (10).
That the 5p15.33 region is important in susceptibility to lung cancer is further supported by replication of the association in diverse populations and in subgroups stratified by smoking history, histological subtype and sex. A few SNPs in the TERT and CLPTM1L genes have been replicated in African Americans (4) and Asians (7,11). Similarly, the rs2361000 SNP in the hTERT gene has been associated with lung cancer risk in smokers and shows stronger associations among never-smokers (8). Indeed, this SNP appears to strongly influence risk in those with adenocarcinoma (4,11), the most common form of lung cancer in never-smokers. Finally, tumor studies confirm the relevance of the 5p15.33 region in lung cancer etiology. Comparative genomic hybridization and fluorescence in situ hybridization studies of lung tumors by Kang et al. (12) have identified amplification in the 5p15.33 region as one of the most consistent alterations in early stage lung cancer. Likewise, Zienolddiny et al. (13) found that rs402710 (TERT–CLPTM1L locus) is associated with increased formation in the lungs of DNA adducts, which are possible precursor to lung carcinogenesis.
Since GWAS coverage is roughly one SNP per 10 000 bp and may not be adequate to refine the localization of the causal variants in this region, we conducted a dense analysis of 215 SNPs across the TERT and CLPTM1L genes to refine the association signal.
Materials and methods
Selection of cases and controls
The study participants for this case–control study were a consecutive series of newly diagnosed Caucasian lung cancer cases from an ongoing lung cancer study that has been accruing participants at The University of Texas MD Anderson Cancer Center since 1995. The controls were recruited from the Kelsey-Seybold Foundation, Houston’s largest multidisciplinary physician practice. The controls were frequency matched to the cases on age (±5 years), sex, smoking status and ethnicity (14). These study subjects were not included as participants in the GWAS for lung cancer conducted at MD Anderson that was reported recently (1). Only non-Hispanic white subjects (both cases and controls) were included in these analyses. All participants provided informed consent and the study was approved by the Institutional Review Board.
SNP selection
We used SNP browser version 4.0 (15) to identify SNPs for further study. The software was designed for selection of SNPs based on observed linkage disequilibrium (LD), including construction of metric LD maps and the selection of haplotype-tagging SNPs. SNP selection was based on the ethnic-specific LD patterns identified by HapMap Project http://hapmap.ncbi.nlm.nih.gov/.
We analyzed SNPs located in the area between 1290 and 1450 kb on chromosome 5. Because we had a relatively large sample size and wanted as complete as possible coverage of the candidate region, we used liberal criteria for SNP selection: minor allele frequency ≥0.01, all non-synonomous SNPs were selected and no exclusion was initially proposed based on the distance from the neighboring SNP. The area includes four genes: SLC6A18, TERT, CLPTM1L and SLC6A3. A panel of 568 validated SNPs was generated.
Genotyping
A total of 221 SNPs in the 5p15.33 region were included as part of a custom-designed Illumina iSelect Genotyping Beadchip (San Diego, CA) that included 19 949 SNPs, a majority of which (11 930) were in inflammation pathways for a separate analysis. Of the 568 validated SNPs in the target region of interest, many SNPs that had been identified had to be removed because the Illumina iSelect chemistry cannot accommodate SNPs <50 bp from each other. SNPs were also excluded if other quality control metrics established by Illumina indicated that the SNP was likely to fail. Genotypes were called using the Beadstudio software. There were 64 expected duplicates and 4 unexpected duplicates (individuals who had DNA analyzed twice and were found to have the same DNA and are therefore either identical twins or the same person). The data were first filtered to remove any SNPs with <95% call rate and then individuals with <90% call rate across SNPs were removed. Among the remaining samples, the error rate obtained by comparing original to replicate samples was 0.0176%. The final data set consisted of 1681 Caucasian lung cancer cases and 1235 unaffected controls, with genotyped results for 215 SNPs.
Statistical analysis
Hardy–Weinberg equilibrium was tested for each of the SNPs using a Fisher’s exact test.
A three-step strategy was applied to perform the analyses (Figure 1fig1). First, association between each SNP and lung cancer was tested using unconditional logistic regression using STATA 10 (StataCorp LP, College Station, TX). Both allelic (additive or per allele) and genotypic (each genotype is compared with the other two by creating dummy variables) genetic models were tested. Only those SNPs significant at a χ2 P < 0.05 in univariate analysis were moved forward to the second stage of analysis. Next, to overcome problems of collinearity between SNPs significant in univariate analysis, haplotype blocks were constructed using Haploview v.4.1 (16) to identify sets of SNPs in LD. Then, to filter out highly correlated SNPs, Tagger (17), a program implemented within Haploview was used to select tagging SNPs from each block to represent SNPs that were in high LD with each other at an r2 threshold of ≥0.95. In this selection process, the SNP with the smallest P value (i.e. most significant SNP, rs370348) from the univariate analysis and two SNPs (rs401681 and rs31489) of the eight previously published SNPs that were not selected by Tagger were forced to be included so that all eight published SNPs were retained for analysis. This subset of SNPs was carried forward for further analysis by stepwise forward selection logistic regression to determine those SNPs that continued to show association (at P < 0.01) while adjusting for age (continuous), sex, pack-years smoked (continuous) and other SNPs in the model. Stepwise forward selection was repeated for subgroup analysis by sex, histological subtype and pack-years smoked.
Fig. 1.
Strategy for analysis of the densely genotyped SNPs in the 5p15.33 region.
In view of the strong correlation between smoking and lung cancer risk, we also performed logistic regression analyses stratified by smoking status and tested for SNP–smoking interaction by the Likelihood ratio test (testing the model with and without the SNP–smoking interaction term included in the model). In addition, we analyzed the SNP–lung cancer association with and without smoking (in pack-years) included as a covariate and examined differences in effect size in the smoking-adjusted and unadjusted logistic regression analyses. Finally, we examined the association of each SNP with smoking (ever smoker–never-smoker as the outcome variable) in cancer free controls, while adjusting for age and sex.
To control for type 1 error due to multiple testing, instead of applying the Bonferroni correction, we applied a method proposed by Li and Ji (18) that controls the error rate for correlated tests. We applied this method in view of the large number of highly correlated SNPs in our data. The Li and Ji method is based on calculating the effective number of independent tests performed—[M(eff)], first proposed by Cheverud (19). Li and Ji further developed the method to provide a more accurate estimate of the M(eff), to control the experiment-wise significance level. Using this method, the effective number of independent marker loci was calculated as 99.49 and the experiment-wide significance threshold required to keep the type I error rate at 5% was 0.0005.
We performed haplotype analysis for the SNPs that were significantly associated with lung cancer risk in the stepwise forward selection logistic regression analysis.
Imputation was performed to increase coverage of SNPs in the 5p15.33 region. An additional 136 SNPs were imputed using HapMap 3 reference data (release #2, February 2009) plus 1000 Genomes reference data (March 2010 genotypes; June 2010 haplotypes) that had predicted imputation r2 values ≥0.9. After imputation, the most likely genotype was used for analyses.
We also evaluated whether or not rare variants in the region associated with lung cancer risk using the WHaIT: weighted haplotype and imputation-based tests program (20). In this analysis, we restricted the study to only include SNPs that had a minor allele frequency of ≤0.05 and then evaluated whether, in aggregate, there was evidence that SNPs showed an association with lung cancer risk, upweighting according to the rarity of alleles.
Results
The analyses included 1681 lung cancer case subjects and 1235 unaffected controls. Although frequency matching was performed by age (±5 years), sex and smoking status, the matching was incomplete; the cases were on average 6 years older than the controls, the proportion of affected males was higher than the affected females and smokers were over-represented in the case group (Table I).
Table I.
Descriptive characteristics
| Total | Cases | Controls | P* | |
| Sex, n (%) | ||||
| Male | 1424 (48.8) | 847 (59.5) | 577 (40.5) | 0.05 |
| Female | 1492 (51.2) | 834 (55.9) | 658 (44.1) | |
| Smokera, n (%) | ||||
| Never | 970 (33.3) | 462 (47.6) | 508 (52.4) | <0.001 |
| Former | 1144 (39.2) | 801 (70.0) | 343 (30.0) | |
| Current | 802 (27.5) | 418 (52.1) | 384 (47.9) | |
| Number of cigarettes | ||||
| Mean (SD) | 26.1 (13.8) | 27.4 (13.3) | 23.7 (14.4) | <0.001 |
| Years smoked | ||||
| Mean (SD) | 34.2 (13.6) | 37.0 (12.9) | 29.5 (13.5) | <0.001 |
| Pack-years of smoking | ||||
| Mean (SD) | 46.8 (32.4) | 52.3 (32.7) | 37.3 (29.6) | <0.001 |
| Age, years | ||||
| Mean (SD) | 60.8 (12.4) | 63.5 (11) | 57.2 (13.2) | <0.001 |
| Cancer type, n (%) | ||||
| Adenocarcinoma | 953 (56.7) | |||
| Squamous cell | 323 (19.2) | |||
| Small cell | 133 (7.9) | |||
| Mixed | 262 (15.6) | |||
| Unknown or missing | 10 (0.6) | |||
Smoker definitions: those who smoked fewer than 100 cigarettes over a lifetime were defined as never-smokers; those who quit smoking 1 year before diagnosis (cases) or before interview date (controls) were defined as former smokers and those who smoked or quit within the past 12 months were defined as current smokers.
*P values based on χ2 test for categorical variables and Wilcoxon test for continuous variables.
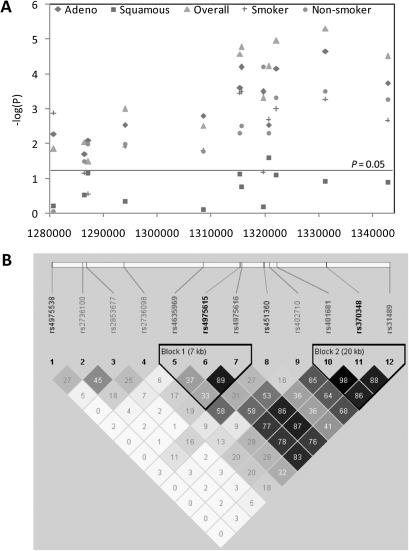
Of the 215 SNPs genotyped, 69 were statistically significant at P < 0.05 in univariate analysis in either an additive (per allele) or a genotypic model. Of these 69 SNPs, four SNPs (rs2735940, rs2853672, rs37002 and rs402710) were out of Hardy–Weinberg equilibrium at P < 0.05, but we did not exclude these from the analysis. All eight previously reported SNPs in the 5p15.33 region (3–8,11,21) (Supplementary Table 1 is available at Carcinogenesis Online) were significantly associated with lung cancer risk at P < 0.05 in our data at this stage of the analysis. To filter out highly correlated SNPs to overcome collinearity in the data, 35 tag SNPs were selected from haplotype blocks to represent the 69 significant SNPs using Tagger in Haploview. These 35 SNPs that were retained for further analysis included all the eight published SNPs and the most significant SNP (rs370348) in the univariate analysis. The adjusted odds ratios, 95% confidence intervals and P values for the 35 selected SNPs are presented in Table II. The results of the association analysis for the 35 SNPs are presented in Figure 2fig2, along with the observed correlation (r2) between the SNPs. The results of the eight previously reported SNPs in this region are indicated by square boxes in Figure 2A; all eight were significant at P < 0.05 in our data.
Table II.
Adjusted logistic regression analysisa of lung cancer risk by selectedb SNPs in the 5p15.33 region overall and by smoking status
| Overall |
Smoker |
Never-smoker |
|||||||||
| SNP | Chromosomal position | Alleles | HWc P | MAF (cases, controls) | Adjusted ORd (95% CI) | P | Adjusted ORe (95% CI) | P | Adjusted ORf (95% CI) | P | Pintg |
| rs370348g | 1331219 | A:G | 0.15 | 0.390, 0.453 | 0.77 (0.69–0.86) | 4.9 × 10−6 | 0.79 (0.69–0.9) | 5.4 × 10−4 | 0.70 (0.58–0.85) | 3.1 × 10−4 | 0.31 |
| rs401681h | 1322087 | G:A | 0.06 | 0.392, 0.453 | 0.78 (0.70–0.87) | 1.1 × 10−5 | 0.80 (0.70–0.91) | 1.0 × 10−3 | 0.71 (0.59–0.86) | 4.7 × 10−4 | 0.21 |
| rs3816659 | 1317820 | G:A | 0.17 | 0.372, 0.432 | 0.78 (0.70–0.87) | 1.5 × 10−5 | 0.79 (0.69–0.90) | 5.5 × 10−4 | 0.73 (0.61–0.89) | 1.3 × 10−3 | 0.64 |
| rs4975616h | 1315660 | A:G | 0.71 | 0.374, 0.434 | 0.78 (0.70–0.87) | 1.7 × 10−5 | 0.78 (0.68–0.89) | 3.2 × 10−4 | 0.75 (0.62–0.90) | 2.8 × 10−3 | 0.83 |
| rs4975615g | 1315343 | A:G | 0.56 | 0.352, 0.411 | 0.78 (0.70–0.89) | 2.7 × 10−5 | 0.78 (0.68–0.89) | 3.5 × 10−4 | 0.76 (0.63–0.92) | 4.7 × 10−3 | 0.90 |
| rs31489h | 1342714 | C:A | 0.13 | 0.362, 0.421 | 0.79 (0.70–0.88) | 3.1 × 10−5 | 0.81 (0.70–0.92) | 2.1 × 10−3 | 0.71 (0.59–0.86) | 5.5 × 10−4 | 0.46 |
| rs27996 | 1345474 | A:G | 0.11 | 0.394, 0.454 | 0.8 0(0.71–0.89) | 5.9 × 10−5 | 0.79 (0.69–0.91) | 8.3 × 10−4 | 0.75 (0.62–0.90) | 2.6 × 10−3 | 0.85 |
| rs402710h | 1320722 | G:A | 0.04 | 0.301, 0.352 | 0.79 (0.70–0.88) | 6.0 × 10−5 | 0.80 (0.70–0.92) | 2.0 × 10−3 | 0.75 (0.62–0.92) | 4.9 × 10−3 | 0.84 |
| rs37011 | 1348798 | T:A | 0.23 | 0.392, 0.448 | 0.81 (0.73–0.91) | 2.3 × 10−4 | 0.82 (0.72–0.94) | 5.1 × 10−3 | 0.74 (0.62–0.90) | 1.9 × 10−3 | 0.64 |
| rs37008 | 1351538 | G:A | 0.43 | 0.389, 0.444 | 0.81 (0.73–0.91) | 2.8 × 10−4 | 0.81 (0.71–0.93) | 3.3 × 10−3 | 0.76 (0.63–0.92) | 4.7 × 10−3 | 0.73 |
| rs451360g | 1319680 | C:A | 0.54 | 0.195, 0.233 | 0.78 (0.68–0.90) | 4.9 × 10−4 | 0.86 (0.73–1.01) | 0.066 | 0.61 (0.48–0.78) | 6.3 × 10−5 | 0.06 |
| rs2736098h | 1294086 | G:A | 0.10 | 0.307, 0.269 | 1.23 (1.09–1.39) | 0.001 | 1.21 (1.04–1.41) | 0.012 | 1.30 (1.06–1.59) | 0.010 | 0.15 |
| rs37002 | 1356944 | G:A | 0.04 | 0.462, 0.510 | 0.84 (0.75–0.94) | 0.002 | 0.85 (0.74–0.97) | 0.013 | 0.78 (0.64–0.93) | 0.007 | 0.65 |
| rs27064 | 1359938 | G:A | 0.23 | 0.154, 0.122 | 1.29 (1.10–1.51) | 0.002 | 1.26 (1.03–1.53) | 0.021 | 1.40 (1.07–1.83) | 0.013 | 0.39 |
| rs2736108 | 1297488 | G:A | 0.81 | 0.333, 0.297 | 1.20 (1.07–1.35) | 0.003 | 1.15 (0.99–1.33) | 0.062 | 1.34 (1.10–1.63) | 0.004 | 0.11 |
| rs4635969h | 1308552 | G:A | 0.21 | 0.176, 0.211 | 0.81 (0.70–0.93) | 0.003 | 0.81 (0.68–0.96) | 0.016 | 0.75 (0.59–0.95) | 0.017 | 0.83 |
| rs410805 | 1323196 | G:A | 0.59 | 0.203, 0.168 | 1.24 (1.08–1.44) | 0.003 | 1.32 (1.06–1.58) | 0.002 | 1.19 (0.94–1.51) | 0.140 | 0.59 |
| rs27071 | 1346081 | A:G | 0.82 | 0.242, 0.282 | 0.83 (0.73–0.94) | 0.004 | 0.86 (0.74–1.00) | 0.055 | 0.72 (0.58–0.89) | 0.002 | 0.10 |
| rs428499 | 1322663 | G:A | 0.08 | 0.219, 0.187 | 1.22 (1.06–1.41) | 0.005 | 1.25 (1.05–1.49) | 0.011 | 1.24 (0.98–1.56) | 0.067 | 0.63 |
| rs7725218 | 1282414 | G:A | 0.73 | 0.365, 0.333 | 1.17 (1.04–1.31) | 0.009 | 1.24 (1.07–1.43) | 0.004 | 1.09 (0.90–1.31) | 0.400 | 0.54 |
| rs2736100h | 1286516 | C:A | 0.46 | 0.466, 0.498 | 0.86 (0.77–0.96) | 0.009 | 0.88 (0.77–1.00) | 0.069 | 0.82 (0.68–0.98) | 0.032 | 0.19 |
| rs2735940 | 1296486 | A:G | 0.02 | 0.461, 0.496 | 0.87 (0.78–0.97) | 0.011 | 0.89 (0.78–1.02) | 0.092 | 0.80 (0.67–0.96) | 0.016 | 0.12 |
| rs2736103 | 1300401 | A:G | 0.59 | 0.448, 0.413 | 1.15 (1.03–1.29) | 0.011 | 1.12 (0.98–1.29) | 0.093 | 1.25 (1.04–1.50) | 0.018 | 0.67 |
| rs4975538g | 1280830 | G:C | 0.57 | 0.371, 0.343 | 1.16 (1.03–1.30) | 0.014 | 1.27 (1.10–1.47) | 0.001 | 1.01 (0.84–1.22) | 0.880 | 0.09 |
| rs2735946 | 1300429 | C:A | 0.94 | 0.261, 0.291 | 0.86 (0.76–0.97) | 0.018 | 0.83 (0.72–0.97) | 0.019 | 0.88 (0.71–1.08) | 0.210 | 0.76 |
| rs7734992 | 1280128 | A:G | 0.37 | 0.434, 0.407 | 1.14 (1.02–1.27) | 0.025 | 1.22 (1.06–1.40) | 0.005 | 1.03 (0.86–1.24) | 0.720 | 0.34 |
| rs35387865 | 1255844 | G:A | 0.97 | 0.019, 0.010 | 1.76 (1.07–2.88) | 0.026 | 1.79 (0.98–3.26) | 0.056 | 1.83 (0.82–4.07) | 0.140 | 0.89 |
| rs2735845 | 1300584 | C:G | 0.91 | 0.217, 0.190 | 1.17 (1.02–1.34) | 0.027 | 1.26 (1.06–1.49) | 0.008 | 1.08 (0.86–1.36) | 0.480 | 0.49 |
| rs7727912 | 1318960 | A:G | 0.85 | 0.102, 0.087 | 1.23 (1.02–1.49) | 0.032 | 1.26 (0.99–1.60) | 0.050 | 1.14 (0.83–1.56) | 0.410 | 0.02 |
| rs2853677h | 1287194 | A:G | 0.31 | 0.453, 0.423 | 1.13 (1.01–1.26) | 0.033 | 1.08 (0.94–1.23) | 0.280 | 1.27 (1.06–1.53) | 0.010 | 0.21 |
| rs2736109 | 1296759 | G:A | 0.69 | 0.430, 0.405 | 1.12 (1.00–1.25) | 0.044 | 1.11 (0.97–1.27) | 0.140 | 1.20 (1.00–1.45) | 0.050 | 0.47 |
| rs6889886 | 1358822 | G:A | 0.99 | 0.262, 0.280 | 0.88 (0.78–0.99) | 0.048 | 0.98 (0.85–1.14) | 0.830 | 0.67 (0.54–0.84) | 4.3 × 10−4 | 0.03 |
| rs2735944 | 1304432 | G:A | 0.84 | 0.103, 0.121 | 0.85 (0.71–1.01) | 0.066 | 0.85 (0.69–1.05) | 0.140 | 0.75 (0.56–1.02) | 0.066 | 0.69 |
| rs35033501 | 1253918 | G:A | 0.46 | 0.019, 0.027 | 0.73 (0.50–1.06) | 0.099 | 0.70 (0.44–1.11) | 0.130 | 0.81 (0.44–1.49) | 0.490 | 0.74 |
| rs27065 | 1359255 | G:A | 0.86 | 0.491, 0.502 | 0.98 (0.88–1.10) | 0.733 | 0.87 (0.76–0.99) | 0.044 | 1.20 (0.99–1.45) | 0.050 | 0.03 |
MAF, minor allele frequency.
Additive model where the log risk increments by the number of minor alleles.
35 SNPs selected at threshold of r2 ≥ 0.95 by Tagger in Haploview with the most significant SNP, rs370348, and eight previously published SNPs included.
HW = Hardy–Weinberg exact P.
Adjusted for age, sex and pack-years smoked.
Adjusted for age and sex.
Likelihood ratio test P value for interaction, testing the model with and without the SNP × smoking interaction terms included in the model.
SNPs significant in forward selection logistic regression.
SNPs significantly associated with lung cancer risk from previously reported GWAS.
Fig. 2.
Association of 35 selected SNPs on chromosome 5p15.33 with lung cancer. (A) Association of SNPs with lung cancer, adjusted for sex, age and number of pack-years smoked. Empty squares indicate SNPs significantly associated with lung cancer risk in previously published GWAS (rs2736100: the most consistently replicated SNP). Chromosomal position is on the x-axis and negative logarithm to the base 10 of the P values from logistic regression analysis is on the y-axis. (B) Genes in the analyzed region and patterns of LD, denoted by r2 values and shading. Higher r2 values and darker shading indicate greater correlation between the SNPs.
In the stepwise forward selection logistic regression analysis, two SNPs, rs370348 and rs4975538, were significantly associated with lung cancer (Table III). However, only rs370348 met the multiple testing corrected threshold for significance at P < 0.0005. Among smokers, rs4975615 (but not rs370348) and rs4975538 were significant, whereas among never-smokers, rs451360 was the only SNP significantly related to lung cancer risk (Table III, Figure 3). In stratified analysis by sex, rs370348 was significantly associated with lung cancer risk in men, reaching genome-wide significance (P = 4.9 × 10−8), but none of the SNPs reached statistical significance in women. In analysis by histological subtype, results for adenocarcinoma were consistent with results of the overall analyses. Detailed results by histological subtype are presented in Figure 3.
Table III.
SNPs significantly associated with lung cancer risk in the most parsimonious stepwise multiple logistic regression models overall and in subgroups
| rs370348 |
rs4975538 |
rs4975615 |
rs451360 |
|||||||||
| ORa | 95% CI | P | ORa | 95% CI | P | ORa | 95% CI | P | ORa | 95% CI | P | |
| Overall | 0.76 | 0.68–0.85 | 1.6 × 10−6 | 1.18 | 1.05–1.33 | 0.005 | ||||||
| Smokers | 1.26 | 1.09–1.47 | 0.002 | 0.75 | 0.65–0.87 | 1.2 × 10−4 | ||||||
| Never-smokers | 0.62 | 0.48–0.78 | 7.6 × 10−5 | |||||||||
| Menb | 0.65 | 0.55–0.76 | 4.9 × 10−8 | |||||||||
| Adenocarcinoma | 0.76 | 0.67–0.86 | 1.8 × 10−5 | 1.2 | 1.05–1.37 | 0.007 | ||||||
| NSCLC | 0.76 | 0.68–0.86 | 1.0 × 10−5 | |||||||||
NSCLC, non-small cell lung cancer, which includes adenocarcinoma and squamous cell carcinoma.
Adjusted for age, sex and pack-years smoked.
No significant results found in women.
Fig. 3.
Association of selected SNPs on chromosome 5p15.33 with lung cancer risk overall and stratified by histological subtype and smoking status. (A) The panel includes four SNPs selected by stepwise forward selection logistic regression analysis and eight previously published GWAS SNPs. Association of SNPs with lung cancer, adjusted for sex, age and number of pack-years smoked in the overall analysis and analysis by histological subtype. Analysis by smoking status adjusted for age and sex. Chromosomal position is on the x-axis and negative logarithm to the base 10 of the P values from logistic regression analysis is on the y-axis. (B) Patterns of LD, denoted by r2 values and shading.
We also performed analyses to determine whether the genes identified in the 5p15.33 region are possibly associated with smoking and not lung cancer. In stratified analysis by smoking status (presented in Table II), the top SNPs in overall analyses continued to be the top SNPs in both smokers and non-smokers, although the P values were attenuated due to stratification. The exception was rs6889886, a SNP that was not significant in ever smokers but was statistically significant in never-smokers (P = 4.3 × 10−4). This SNP was also statistically significant for interaction with smoking (P = 0.03). However, this SNP was not independently associated with lung cancer in the forward selection logistic regression analysis overall or by smoking subgroups. Two other SNPs, rs7727912 and rs27065, were significant at P < 0.05 in the test for multiplicative interaction but the main effects for both these SNPs were only marginally significant. Positive tests for interactions may reflect the large number of tests and should be viewed with some skepticism. We also performed logistic regression analysis with and without adjusting for smoking (pack-years smoked) and found that the effect size did not change by >10% for any of the SNPs (Supplementary Table 2 is available at Carcinogenesis Online). Finally, in the logistic regression analysis of the SNP–smoking association in unaffected controls (with ever–never smoked as the outcome), none of the SNPs was statistically significant (P > 0.05; results not shown). These results indicate that the association detected for the SNPs in the 5p15.33 region is with lung cancer risk and not with smoking.
In the haplotype analysis (results not presented), none of the haplotypes was more significant than the most significant underlying SNP tests. Hence, haplotype analysis did not identify a haplotype that fit the data better than individual SNP studies did. The omnibus test was also not better than the individual SNP tests.
Other than rs370348 and rs4975615 (r2 = 0.78), there was low correlation between the other significant SNPs (Figure 3). LD mapping with the eight previously reported SNPs showed that two of the significant SNPs identified, rs4975538 (maximum r2 = 0.27 with rs2736100) and rs451360 (maximum r2 = 0.58 with rs4635969) were largely independent of the other SNPs (Figure 3).
Results of the imputed SNPs are presented in Supplementary Figure 1 is available at Carcinogenesis Online along with the results of the genotyped SNPs in this region. None of the imputed SNPs reached a significance level greater than the genotyped SNPs so the imputed SNPs were not analyzed further.
Analysis of rare variants was not contributory and the P value derived from the rare variant analysis program WHaIT (20) did not identify an excess of rare variants in the case or control populations (P = 0.52 for ever smokers and P = 0.8 for never-smokers).
Discussion
In this fine mapping analysis of genetic variants in the 5p15.33 region, we identified four novel SNPs associated with lung cancer risk, one of which was specific to smokers and one was specific to non-smokers. None of the SNPs was in protein-coding region or a promoter or splice site variant; rs4975538 is an intronic SNP in TERT, rs451360 and rs370348 are intronic SNPs in CLPTM1L and rs4975615 is in the intergenic region between the two genes. Although none of these SNPs is in a putative functional region, our findings confirm that the TERT–CLPTM1L region is related to lung cancer risk. Interestingly, there are very few common variants in the exonic protein-coding regions in TERT or CLPTM1L and other than rs2736098 (corresponding to A305A); all are rare variants with a <5% minor allele frequency.
The GWAS by McKay et al. (5) first suggested two genes, TERT and CLPTM1L, in the 5p15.33 region that could have a role in lung cancer susceptibility. Their GWAS identified four SNPs in this region, rs402710, rs2736100, rs401681 and rs31489, of which the first two SNPs were replicated in an independent sample of cases and controls. The studies that followed confirmed the association of these and other SNPs in the TERT–CLPTM1L region (Supplementary Table 1 is available at Carcinogenesis Online). Furthermore, lung cancer risk for SNPs in the TERT–CLPTM1L region was also reported by other authors in specific subgroups, including never-smokers (8,11), people with adenocarcinoma (4), women (11,21), African Americans (21) and Asians (7,11,22). SNPs in the TERT–CLPTM1L region that reached genome-wide significance in these studies are listed in Supplementary Table 1 (available at Carcinogenesis Online). In particular, rs2736100 was found to be significant across different studies and subgroups, which suggest that this or another SNP in LD with it is probably to be causally related to lung cancer risk. In comparison, although all the eight previously reported SNPs were nominally significant at P < 0.05 in our study (Table II and Figure 2A), rs2736100 was not one of the most significant SNPs (P = 0.009).
There is evidence that the 5p15.33 region may be important in susceptibility to other cancers as well. Rafnar et al. examined rs401681 and rs2736098, two SNPs in the 5p15.33 region for their association with risk for many different types of cancer and found a significant association for rs401681 with several cancers including basal cell carcinoma, lung, bladder, prostate and cervical cancers (6). Another study confirmed the association of variants in this region with bladder cancer (23) and associations were also determined for rs401681 with squamous cell carcinoma of the head and neck (24). Furthermore, in a GWAS for pancreatic cancer, rs401681 was identified as one of the susceptibility loci (25). Interestingly, rs401681 was the second most significant SNP in our study (P = 1.1 × 10−5). Rafnar et al. also examined the association between rs401681 and rs2736098 and telomere length in DNA from whole blood as telomere shortening is a possible mechanism of carcinogenesis related to the 5p15.33 region. Their results suggested that the variants may lead to a gradual shortening over time, although this effect was only apparent in older women (6). However, these results were not confirmed by Pooley et al., who found that rs401681, which is located in intron 13 of CLPTM1L was not associated with mean telomere length (26).
Both TERT and CLPTM1L at the 5p15.33 susceptibility locus are attractive candidate genes for lung cancer as they have both been plausibly linked with carcinogenesis. TERT encodes the catalytic subunit of telomerase, an enzyme that maintains telomere ends by adding the telomere repeat TTAGGG. It has been shown that telomerase expression is high in progenitor and cancer cells and absent or low in normal somatic cells (27). Telomere length is linked to aging and anti-apoptosis, and mouse studies have shown that dysregulation of telomerase expression may be involved in oncogenesis (28). Similarly, CLPTM1L encodes an enzyme—cleft lip and palate transmembrane 1-like that is upregulated in cisplatin-resistant cell lines and may be associated with apoptosis (10). Furthermore, the risk allele of rs402710 within the CLPTM1L gene has also been found to be associated with a higher accumulation of DNA damage measured by bulky aromatic/hydrophobic DNA adducts, which may be an early step in lung carcinogenesis (13).
One of the lingering questions about the GWAS hits identified for lung cancer is whether genes identified are associated with smoking and not lung cancer. We tested for gene–smoking interaction to see if smoking modified the SNP–lung cancer association. However, other than for rs6889886, our results did not show that smoking modified the SNP–lung cancer association for any of the SNPs. Even for rs6889886, which is an intergenic SNP between CLPTM1L and SLC6A3 genes, evidence for an interaction with smoking could reflect type 1 error, given the number of tests performed. We also examined smoking (as pack-years smoked) as a confounder of the SNP–lung cancer association and did not find a significant change in the effect sizes when we compared the results of the unadjusted and adjusted analyses. Finally, we examined the SNP–smoking association in the unaffected controls and found that none of the SNPs was associated with smoking. Our findings clearly suggest that the SNPs in the 5p15.33 region are strongly associated with lung cancer and not smoking.
In summary, in this analysis, we used a fine mapping approach to evaluate additional, possibly causal SNPs in the 5p15.33 GWAS-identified lung cancer susceptibility locus. We used multiple logistic regression according to haplotype blocks to identify independent variants associated with lung cancer risk. We identified rs370348 and rs4975538 as novel SNPs associated with lung cancer risk and two additional SNPs that may be susceptibility markers for lung cancer risk in smokers (rs4975615) and non-smokers (rs451360). Our results show that after fine mapping, the 5p15.33 locus that has repeatedly been identified as a strong susceptibility locus for lung cancer, there appears to be several distinct loci influencing disease risk. None of the SNPs we identified were obvious functional SNPs, that is, in exonic, splice site or promoter regions. A limitation of this study was incorporation of a limited number of SNPs on the SNP array of the total number of SNPs identified in the SNP selection process. Future analyses using sequencing approaches may help to identify all causal variants in this region and animal and cell models may be needed to establish mechanisms of cancer risk.
Supplementary material
Supplementary Tables 1–2 and Figure 1 can be found at http://carcin.oxfordjournals.org.
Funding
This research was supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant (CA016672); research grants (CA55769, CA127219, R01 CA121197, 1P50 CA70907, U19 CA148127); Cancer Prevention & Research Institute of Texas grant (RP100443).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contribution of Emily Lu for help with data imputation and analysis, Huifeng Zhang for genotyping and Stephanie Deming for scientific editing.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- SNPs
single-nucleotide polymorphisms
- GWAS
genome-wide association studies
- TERT
telomerase reverse transcriptase
- CLPTM1L
cleft lip and palate transmembrane 1-like
References
- 1.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat.Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat.Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay JD, et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truong T, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl Cancer Inst. 2010;102:959–971. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis. 2010;31:234–238. doi: 10.1093/carcin/bgp287. [DOI] [PubMed] [Google Scholar]
- 9.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto K, et al. A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem. Biophys. Res. Commun. 2001;280:1148–1154. doi: 10.1006/bbrc.2001.4250. [DOI] [PubMed] [Google Scholar]
- 11.Hsiung CA, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:e1001051. doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang JU, et al. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer Genet. Cytogenet. 2008;182:1–11. doi: 10.1016/j.cancergencyto.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Zienolddiny S, et al. The TERT-CLPTM1L lung cancer susceptibility variant associates with higher DNA adduct formation in the lung. Carcinogenesis. 2009;30:1368–1371. doi: 10.1093/carcin/bgp131. [DOI] [PubMed] [Google Scholar]
- 14.Hudmon KS, et al. Identifying and recruiting healthy control subjects from a managed care organization: a methodology for molecular epidemiological case-control studies of cancer. Cancer Epidemiol. Biomarkers Prev. 1997;6:565–571. [PubMed] [Google Scholar]
- 15.De La Vega FM, et al. A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns. Pac. Symp. Biocomput. 2006;11:487–498. [PubMed] [Google Scholar]
- 16.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.de Bakker PI, et al. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 18.Li J, et al. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 19.Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, et al. To identify associations with rare variants, just WHaIT: weighted haplotype and imputation-based tests. Am. J. Hum. Genet. 2010;87:728–735. doi: 10.1016/j.ajhg.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dyke AL, et al. Chromosome 5p region SNPs are associated with risk of NSCLC among women. J. Cancer Epidemiol. 2009;2009:242151. doi: 10.1155/2009/242151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin G, et al. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis. 2009;30:987–990. doi: 10.1093/carcin/bgp090. [DOI] [PubMed] [Google Scholar]
- 23.Gago-Dominguez M, et al. Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis. 2011;32:197–202. doi: 10.1093/carcin/bgq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, et al. Genetic variations in TERT-CLPTM1L genes and risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2010;31:1977–1981. doi: 10.1093/carcin/bgq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen GM, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pooley KA, et al. No association between TERT-CLPTM1L single nucleotide polymorphism rs401681 and mean telomere length or cancer risk. Cancer Epidemiol. Biomarkers Prev. 2010;19:1862–1865. doi: 10.1158/1055-9965.EPI-10-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wai LK. Telomeres, telomerase, and tumorigenesis—a review. MedGenMed. 2004;6:19. [PMC free article] [PubMed] [Google Scholar]
- 28.Calado RT, et al. Telomerase: not just for the elongation of telomeres. Bioessays. 2006;28:109–112. doi: 10.1002/bies.20365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.