Abstract
Introduction
Crohn's disease (CD) and ulcerative colitis (UC) are the two major forms of inflammatory bowel disease (IBD). A high prevalence of Campylobacter concisus was previously detected in paediatric CD and adult UC. Currently, the prevalence of C. concisus in adult CD and the preferential colonization sites of Campylobacter species in the human intestine are unknown. In this study, we examined the prevalence of Campylobacter species in biopsies collected from multiple anatomic sites of adult patients with IBD and controls.
Methods
Three hundred and one biopsies collected from ileum, caecum, descending colon and rectum of 28 patients IBD (15 CD and 13 UC) and 33 controls were studied. Biopsies were used for DNA extraction and detection of Campylobacter species by PCR-sequencing and Campylobacter cultivation.
Results
A significantly higher prevalence of C. concisus in colonic biopsies of patients with CD (53%) was detected as compared with the controls (18%). Campylobacter genus-PCR positivity and C. concisus positivity in patients with UC were 85% and 77% respectively, being significantly higher than that in the controls (48% and 36%). C. concisus was more often detected in descending colonic and rectal biopsies from patients with IBD in comparison to the controls. C. concisus was isolated from patients with IBD.
Conclusion
The high intestinal prevalence of C. concisus in patients with IBD, particularly in the proximal large intestine, suggests that future studies are needed to investigate the possible involvement of C. concisus in a subgroup of human IBD. To our knowledge, this is the first report of the association between adult CD and C. concisus as well as the first study of the preferential colonization sites of C. concisus in the human intestine.
Introduction
Campylobacter species have been associated with various diseases in both animals and humans [1]. Campylobacter jejuni and Campylobacter coli are well established human pathogens, having been associated with a number of clinical conditions such as diarrhoea, abortion, septicaemia and Guillain-Barre syndrome [1]. Some other Campylobacter species including Campylobacter concisus have been considered as emerging human pathogens [2].
C. concisus is a curved Gram negative bacterium; with a single polar flagellum [3]. C. concisus was first isolated by Tanner et al in 1981 from human dental plague [4]. In a following-up study, Macuch and Tanner reported a higher isolation rate of C. concisus in patients at the initial stage of periodontitis in comparison to individuals with healthy gums [5].
Lately, C. concisus has been considered as an emerging human enteric pathogen [6]. Evidence that C. concisus may be an important human enteric pathogen has come from a number of recent studies reported that C. jejuni and C. concisus are the most commonly isolated Campylobacter species from diarrheal stool specimens [2], [7], [8], [9]. However, when Engberg et al compared the prevalence of C. concisus in 107 stool samples subjected to tests for enteric pathogens and in 107 age/sex matched healthy controls, they found that the prevalence of C. concisus in these two groups was not significantly different [9]. Furthermore, they found that C. concisus was more often isolated from children aged 0–9 years and individuals aged over 60 years as compared with other age groups. These results have led Engberg et al to conclude that C. concisus should be considered as a commensal bacterium and this bacterium may be an important opportunistic pathogen in individuals with compromised or immature immune systems [9].
In addition to periodontal and diarrheal diseases, recently C. concisus has been linked to inflammatory bowel disease (IBD). IBD is a chronic inflammatory condition of the gastrointestinal tract, with the two major forms being Crohn's disease (CD) and ulcerative colitis (UC). The inflammation in CD may occur anywhere along the gastrointestinal tract, however in UC the inflammation often occurs in colon and rectum [10]. The aetiology of IBD is currently unknown. It is understood that a complex interaction of a number of factors including host genetics, environment, immune system and intestinal microflora contributes to the development of IBD [10], [11], [12]. Despite strong evidence that the intestinal microbial flora plays a key role in the development of IBD, the exact causative agent (s) is still under investigation [11].
Previously, we detected a significantly higher prevalence of C. concisus by PCR in intestinal biopsies of children with CD (51%) as compared with the controls (2%) and isolated a C. concisus strain from intestinal biopsies of a child with CD [13]. In a later study, we detected high prevalence of C. concisus in stool samples of children with CD [14]. A recent study by Mukhopadhya et al reported a significantly higher prevalence of C. concisus detected by PCR in adult patients with UC as compared with the controls [15].
To date, the prevalence of C. concisus in adult patients with CD has not been investigated. Furthermore, no information is available regarding whether Campylobacter species preferentially colonize specific sites in the human intestine. In this study, we examined the prevalence of Campylobacter species in biopsies collected from four anatomic sites of intestines from adult individuals with normal intestinal histology and patients with IBD by PCR-sequencing and Campylobacter cultivation.
Materials and Methods
Ethics statement
Intestinal biopsies were obtained from colonoscopy procedures carried out at the Prince of Wales Hospital and the St George Hospital at Sydney, Australia. Ethics approval for this study was granted by the Ethics Committees of the University of New South Wales and the South East Sydney Area Health Service, Australia (HREC 09237/SESIAHS 09/078 and HREC08335/SESIAHS(CHN)07/48). Written informed consent was obtained from all subjects in this study.
Study subjects and biopsy collection
Sixty-one study subjects, including 28 patients with IBD and 33 controls, were recruited from the Prince of Wales Hospital and the St George Hospital at Sydney, Australia. Among the 28 patients with IBD (15 CD and 13 UC), ten patients (six CD and four UC) were relapsed cases and the remaining eighteen patients were newly diagnosed IBD. Disease location and severity were scored according to the Montreal criteria [16]. The controls, either presenting with gastrointestinal symptoms including abdominal pain and constipation or undertaking a screening colonoscopic examination due to previous history of polyps or a family history of colonic cancer, had no macroscopic or microscopic intestinal inflammation.
Five biopsies were collected from each individual. In the case where macroscopic inflammation was present, biopsies were taken from the edge of the inflamed areas. Of the five biopsies collected from each individual, four biopsies collected from each of the four anatomic sites (ileum, caecum, descending colon and rectum respectively) were used for DNA extraction and detection of Campylobacter species by PCR. The additional biopsy collected from caecum was used for Campylobacter cultivation.
DNA extraction from intestinal biopsies
Freshly collected intestinal biopsies were directly placed into cell lysis solution and DNA was extracted using the Puregene DNA Extraction kit (Gentra, Minneapolis, USA) according to the manufacturer's instructions.
Detection of Campylobacter species in intestinal biopsies by Campylobacter genus-PCR
To detect all Campylobacter species, DNA extracted from intestinal biopsies were subjected to a nested Campylobacter genus-PCR. Bacterial 16S rRNA gene was first amplified from 200 ng of DNA extracted from intestinal biopsies using universal primers F27 and R1496 [17]. The thermal cycling conditions were 94°C for 10 minutes, followed by 35 cycles of 94°C for 10 seconds, 53°C for 10 seconds and 72°C for 1 minute. The PCR reaction volume was 25 µl. The PCR product was then purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany). The purified PCR product (2 µl) was subjected to a Campylobacter genus-specific PCR using primers C418 and C1228 designed by Linton et al [18]. The thermal cycling conditions for the Campylobacter genus-specific PCR were 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds and 72°C for 30 seconds.
Campylobacter species identification
All positive PCR products were sequenced using the BigDyeTM terminator chemistry (Applied Biosystems, Foster City, USA) and the sequencing mixture was analysed on DNA sequence analyser ABI3720 (Applied Biosystems, Foster City, USA). The obtained sequences were compared to gene sequences of known bacterial identities available in GenBank through the National Centre for Biotechonology Information (NCBI website (http://www.ncbi.nlm.nih.gov).
C. concisus specific PCR
Three samples which had mixed sequences in the Campylobacter genus-PCR were subjected to a previously described C. concisus PCR to examine if C. concisus was present [14]. For the C. concisus PCR, DNA (50 ng) extracted from biopsies was subjected to the Campylobacter genus-PCR, then 1 µl of the Campylobacter genus-PCR product was subjected to C. concisus PCR as previously described [14].
Cultivation of Campylobacter species from intestinal biopsies
One caecal biopsy collected from each individual was subjected to Campylobacter cultivation. The biopsy was spread on agar plates prepared using blood agar base no 2 supplemented with 6% sterile defibrinated horse blood, trimethoprim (10 µg/ml), and vancomycin (10 µg/ml). The plates were incubated under microaerophilic conditions generated by a Campylobacter gas generating system (Oxoid Limited, Hampshire, United Kingdom) for two days. A bacterial suspension was prepared from the culture plates and filtered through a 0.6 µM filter membrane (Millipore, Billerica, USA) onto a fresh agar plate and further incubated for additional two days.
Candidate colonies were subjected to microscopic examination of morphology, Gram staining, PCR targeting the 16S rRNA gene using primers F27 and R1649 and sequencing of the PCR products.
GenBank Sequence Submission
All 16S rRNA gene sequences of the PCR products were submitted to GenBank.
Statistical analysis
Fisher's exact test (two tailed) was used to compare the prevalence of Campylobacter species in patients with IBD and controls. Unpaired t test was used to compare the age of patients and controls. Statistical analysis was performed using GraphPad Prism 5 software (San Diego, CA).
Results
Clinical information of patients and controls
The average age of the patients with IBD and controls was 39±13 and 45±11 years old respectively. There were 12 male (43%) patients with IBD and 13 male in controls (39%). The age and sex between patients with IBD and controls were not statistically different.
A total of 301 biopsies (165 biopsies from 33 controls and 136 biopsies from 28 patients with IBD) were collected from four intestinal sites (ileum, caecum, descending colon and rectum) of patients with IBD and controls. Ileal biopsies were not available from two patients with CD, a caecal biopsy was not available from one patient with CD and a rectal biopsy was not available from an additional patient with CD. Both patients and controls did not receive antibiotics one month prior to colonoscopy.
All controls had normal intestinal histology. The Montreal classification of patients with IBD is summarized in Table 1.
Table 1. Montreal classification of patients with IBD.
| Montreal classification (CD) | CD (n = 15) |
| L1 | 7% (1/15) |
| L2 | 60% (9/15) |
| L3 | 33% (5/15) |
| Montreal classification (UC)-Extent | UC (n = 13) |
| Proctitis E1 | 8% (1/13) |
| Left sided UC E2 | 38% (5/13) |
| Extensive UC E3 | 54% (7/13) |
| Montreal classification (UC)-Severity | UC (n = 13) |
| Clinical remission S0 | 0 |
| Mild UC S1 | 54% (7/13) |
| Moderate UC S2 | 46% (6/13) |
| Severe UC S3 | 0 |
Prevalence of Campylobacter species in biopsies collected from different intestinal sites of individuals with normal intestinal histology
To examine the possible preferential colonization sites of Campylobacter species particularly C. concisus in the human intestine, DNA samples extracted from biopsies collected from four intestinal anatomic sites of 33 individuals with normal intestinal histology were subjected to Campylobacter genus-PCR. Among the 33 individuals examined, 48% (16/33) were positive for Campylobacter genus-PCR (an individual with at least one of the four intestinal biopsies collected from ileum, caecum, descending colon and rectum positive by the Campylobacter genus-PCR was considered Campylobacter genus-PCR positive). Of the 16 individuals who were positive by the Campylobacter genus-PCR, four individuals had one biopsy positive and 12 individuals had 2–4 biopsies positive by the PCR. Campylobacter genus-PCR positive rate in biopsies collected from ileum, caecum, colon and rectum were 27% (9/33), 30% (10/33), 27% (9/33), rectum 27% (9/33) respectively, with no statistical differences observed between sites. Campylobacter genus-PCR positive rate in male was 42% (5/12); with no statistical difference from that in females (52%, 11/21).
Sequencing of the positive PCR products yielded 503–766 bp sequences. The obtained sequences were used for identification of Campylobacter species. The similarities of 16S rRNA gene sequences between the Campylobacter genus-PCR products and the known Campylobacter species were 97%–100%. Five Campylobacter species were identified from biopsies collected from individuals with normal intestinal histology, including C. concisus, Campylobacter showae, Campylobacter hominis, Campylobacter ureolyticus and Campylobacter hyointestinalis. Among the 12 individuals who had multiple biopsies positive for the Campylobacter genus-PCR, single Campylobacter species was identified in eight individuals and two Campylobacter species were identified in the remaining four individuals.
Among the 33 individuals examined, 36% (12/33) of individuals were positive for C. concisus, 6% (2/33) of individuals were positive for C. showae, 9% (3/33) of individuals were positive for C. hominis, 6% (2/33) of individuals were positive for C. ureolyticus and 3% (1/33) of individuals were positive for C. hyointestinalis. For an individual to be classified as C. concisus positive, C. concisus had to be identified in at least one of the four biopsies collected. The same principle applied for the evaluation of the intestinal prevalence of the other Campylobacter species in this study.
Campylobacter species detected in biopsies collected from the four intestinal anatomic sites of individuals with normal histology is shown in Table 2. Ileal, caecal and colonic biopsies showed similar C. concisus positive rates and the rectum had a lower C. concisus positive rate; however the difference was not statistically significant. Given the low positive rate for the remaining four Campylobacter species, no statistical analysis was applied to compare the prevalence of these Campylobacter species in different sites of the intestines (Table 2).
Table 2. Detection of Campylobacter species in biopsies collected from four intestinal anatomic sites of individuals (n = 33) with normal intestinal histology by Campylobacter genus-PCR and sequencing*.
| Ileum | Caecum | Colon | Rectum | |
| C. concisus | 21% (7/33) | 18% (6/33) | 18% (6/33) | 9% (3/33) |
| C. showae | 3% (1/33) | 3% (1/33) | 3% (1/33) | 6% (2/33) |
| C. hominis | 3% (1/33) | 6% (2/33) | 3% (1/33) | 9% (3/33) |
| C. ureolyticus | 0 | 0 | 3% (1/33) | 3% (1/33) |
| C. hyointestinalis | 0 | 3% (1/33) | 0 | 0 |
*Four biopsies, one each from ileum, caecum, descending colon and rectum of each individual, were examined. Identification of Campylobacter species was based on 97–100% similarity of the sequences of PCR products (503–766 bp) to the sequences of known Campylobacter species.
Comparison of intestinal prevalence of Campylobacter species in patients with IBD and controls
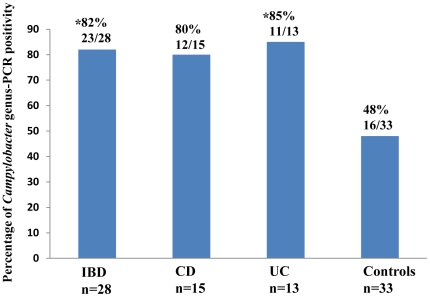
The above 33 individuals with normal intestinal histology were used as controls. Campylobacter genus-PCR positive rate in patients with IBD was 82% (23/28), which was significantly higher than that of the controls (48%, 16/33) (P<0.05). The Campylobacter genus-PCR positive rate was 80% (12/15) in patients with CD and 85% (11/13) in patients with UC; with the difference between UC group and the controls being statistically significant (P<0.05) and the difference between CD and controls being not statistically significant (Figure 1).
Figure 1. Campylobacter genus-PCR positivity in patients with IBD (CD and UC) and controls.
Four biopsies collected from each individual were examined. A Campylobacter genus-PCR positive individual is an individual who had at least one intestinal biopsy positive for Campylobacter genus-PCR. *Significantly different as compared with the controls (P<0.05).
The prevalence of different Campylobacter species in patients with IBD and controls is shown in Table 3. Eight Campylobacter species were identified in intestinal biopsies collected from patients with IBD. C. concisus was detected in 68% (19/28) patients with IBD, which was significantly higher as compared with the controls 36% (12/33) (P<0.05). The C. concisus positive rates in patients with CD and UC were 67% (10/15) and 69% (9/13) respectively; the C. concisus positive rate between UC and controls was statistically different (P<0.05) and the difference between CD and the controls was not significantly different. The prevalence of the remaining seven Campylobacter species in patients with IBD and controls was not statistically different (Table 3).
Table 3. Percentage of Campylobacter species positivity in patients with IBD (CD and UC) and controls@.
| IBD (n = 28) | CD (n = 15) | UC (n = 13) | Controls (n = 33) | |
| C. concisus | 68% (19/28)* | 67% (10/15) | 69% (9/13)* | 36% (12/33) |
| C. showae | 11% (3/28) | 7% (1/15) | 15% (2/13) | 6% (2/33) |
| C. hominis | 7% (2/28) | 7% (1/15) | 8% (1/13) | 9% (3/33) |
| C. ureolyticus | 10% (3/28) | 13% (2/15) | 8% (1/13) | 6% (2/33) |
| C. hyointestinalis | 4% (1/28) | 7% (1/15) | 0 | 3% (1/33) |
| C. rectus | 4% (1/28) | 0 | 8% (1/13) | 0 |
| C. jejuni | 4% (1/28) | 7% (1/15) | 0 | 0 |
| C. gracilis | 7% (2/28) | 7% (1/15) | 8% (1/13) | 0 |
@A specific Campylobacter species positive individual is an individual who has at least one biopsy positive for the Campylobacter species listed in Table 3, detected by Campylobacter genus-PCR and sequencing. *Significantly higher as compared with the controls (P<0.05).
The C. concisus positive rate in relapsed CD was 67% (4/6), which was not significantly different as compared with the newly diagnosed CD cases (67%, 6/9). The C. concisus positive rate in relapsed UC was 75% (3/4), which was not significantly different from that of the new cases (67%, 6/9).
Of the three biopsies which showed mixed sequences by Campylobacter genus-PCR; two samples were positive and one was negative by C. concisus PCR.
Comparison of prevalence of C. concisus in biopsies collected from different anatomic intestinal sites of patients with IBD and controls
Given that C. concisus was the only Campylobacter species showing statistical difference between patients with IBD and the controls in this study (Table 3), the prevalence of C. concisus in biopsies collected from ileum, caecum, colon and rectum of patients with IBD and controls was further compared and the results are shown in Table 4. Ileal biopsies collected from patients with IBD and controls showed similar C. concisus positivity. Caecal biopsies of patients with UC had a low C. concisus positive rate; however it was not statistically different from the other groups. C. concisus positive rate of colonic biopsies of patients with IBD was 43% (12/27), which was significantly higher compared to the controls (P<0.05). The C. concisus positivity in colonic biopsies of patients with CD and UC was 53% (8/15) and 31% (4/13) respectively; with patients with CD showing a statistically significant difference when compared to the controls (P<0.05). The C. concisus positivity in rectal biopsies of patients with IBD was higher than that of the controls; however it was not statistically different (Table 4).
Table 4. Detection of C. concisus in biopsies collected from four intestinal anatomic sites of patients with IBD (CD and UC) and controls@.
| IBD n = 28 | CD n = 15 | UC n = 13 | Control n = 33 | |
| Ileum | 23% (6/26) | 23% (3/13) | 23% (3/13) | 21% (7/33) |
| Caecum | 15% (4/27) | 21% (3/14) | 9% (1/13) | 18% (6/33) |
| Colon | 43% (12/28)* | 53% (8/15)* | 31% (4/13) | 18% (6/33) |
| Rectum | 26% (7/27) | 21% (3/14) | 31% (4/13) | 9% (3/33) |
@ Identification of C. concisus was based on Campylobacter genus-PCR and sequencing of the positive PCR products, except for three biopsy samples. The three biopsy samples showed mixed sequences by Campylobacter genus-PCR, therefore were further subjected to C. concisus PCR to examine the presence of C. concisus.
Biopsies collected from four intestinal anatomic sites were examined; ileal biopsies were not available from two patients with CD, caecal biopsy was not available from one patient with CD and rectal biopsy was not available from one patient with CD.
*Significantly higher in patients with IBD as compared with the controls (P<0.05).
Prevalence of C. concisus in relation to Montreal classification of IBD
No significant differences were noted between the prevalence of C. concisus in patients with different Montreal classifications.
Isolation of C. concisus from intestinal biopsies of patients with IBD and controls
C. concisus was isolated from intestinal biopsies of two patients with IBD, one patient with CD and one patient with UC. The identity of the C. concisus isolates was confirmed by bacterial morphology (small curved and spiral rods), Gram stain (Gram negative) and sequence of 1200 bp 16S rRNA gene (100% similarity to the known C. concisus in GenBank).
Sequences accession numbers
The accession numbers of the sequences of the PCR products submitted to GenBank were JN544934-JN545008.
Discussion
This study aimed to investigate the prevalence of C. concisus in adult patients with CD and the possible preferential colonization sites of Campylobacter species in the human intestine; by examining the presence of Campylobacter species in 301 intestinal biopsies collected from 28 patients with IBD and 33 controls using PCR-sequencing and Campylobacter cultivation.
The high positive rate of Campylobacter genus-PCR and intestinal prevalence of C. concisus in adult patients with CD and UC observed in this study are consistent with our previous findings in paediatric CD and the findings by Mukhopadhya et al in adult UC [13], [14], [15]. A further finding of this study is the increased prevalence of C. concisus in the proximal large intestines (descending colon and rectum) of patients with IBD as compared with the controls (Table 4). In adult patients with CD, only biopsies collected from the descending colon showed a significantly higher prevalence of C. concisus as compared with the controls (Table 4).
Different Campylobacter species may have preferable intestinal colonization sites in their hosts. For example, a study from Inglis et al examining the colonization of C. jejuni and Campylobacter lanienae in asymptomatic beef cattle, C. jejuni was found to colonize the proximal small intestine whereas C. lanienae was detected primarily in the caecum, descending colon and rectum [19].
In individuals without intestinal inflammation, biopsies collected from ileum, caecum, descending colon had a similar C. concisus positive rate and the rectal biopsies had a lower C. concisus positive rate (Table 2). However, in patients with IBD, a higher prevalence of C. concisus in the proximal large intestine (descending colon and rectum) was detected (Table 4). It is not entirely clear why C. concisus was more prevalent in the proximal large intestine of patients with IBD, particularly in descending colon of patients with CD (Table 4). It is possible that this may relate to the fact that C. concisus requires hydrogen enriched microaerophilic atmosphere for growth [2]. In the human intestine, hydrogen is produced by bacterial flora through fermentation of unabsorbed carbohydrates; previous studies showing that 99% of hydrogen in the intestine is produced in the colon [20]. The amount of hydrogen produced in the intestine is affected by food type and intestinal bacterial composition [20], [21]. It may be that the microenvironment of proximal large intestine in some individuals is more suitable for C. concisus growth. Whether the high prevalence of C. concisus in the proximal large intestine of patients with IBD is a primary event or secondary to the disease is not known. The finding in this study that the prevalence of C. concisus in newly diagnosed patients is similar to that of the relapsed cases suggests that the high prevalence of C. concisus in patients with IBD is likely a primary event.
The finding that C. concisus has a preferable intestinal colonization site (the proximal large intestine) in patients with IBD suggests that different bacterial species may be associated with IBD occurring at different parts of the gastrointestinal tract. Other evidence from both human and animal studies supports this view. For example, in human studies adherent and invasive Escherichia coli has been found to be associated with ileal CD only [22]. Furthermore, antibiotics used to treat patients with IBD were effective only in a subgroup of patients [23]. In animal studies, IL-10 -/- mice developed caecal inflammation when monoassociated with E. coli but distal colitis when colonized with Enterococcus faecalis [24].
Whether C. concisus detected in patients with IBD has contributed to the pathogenesis of the disease requires further investigation. C. concisus is a bacterium with great diversity; which has been demonstrated by various research groups using different methodologies [25], [26], [27], [28], [29]. Intestinal C. concisus strains have been shown to be able to induce production of IL-8 in HT-29 cells and some C. concisus strains were invasive to Caco2 cells [30], [31]. In addition, the presence of bacterial virulence factors such as phospholipase A2 and a cytolethal distending toxin (CDT)-like toxin in some C. concisus strains suggest that some C. concisus strains may have the enteric pathogenic potential [32], [33].
Examination of prevalence of C. concisus in patients with gum disease by Macuch and Tanner has revealed an interesting relationship between C. concisus and oral mucosal inflammation [5]. In their study, Macuch and Tanner found that the isolation of C. concisus from subgingival plaque samples of patients with initial periodontitis was greatly higher than the controls. However the isolation rate of C. concisus in patients with established periodontitis was greatly reduced in comparison with the healthy controls [5]. These results suggest that C. concisus may be only associated with mild oral mucosal inflammation. A more severe inflammatory microenvironment such as the established periodontitis is certainly no longer a favourable environment for C. concisus. We have observed a similar phenomenon in patients with CD. In our previous study in a paediatric population, we found that biopsies taken from macroscopic normal area near the inflamed area had higher C. concisus detection than biopsies taken from the centre of the severely inflamed area [13]. Patients with UC included in this study had mild to moderate disease severities (Table 1); we therefore were unable to examine the prevalence of C. concisus in patients with severe UC.
These data suggest that the role of C. concisus in the pathogenesis of IBD, if there is any, would be most likely to facilitate the establishment of the inflammation in the early stage of the disease or to promote inflammation from a mild form to a more severe form. Recently, we found that C. concisus has the ability to modulate the gut mucosal immune system through upregulation of the intestinal epithelial expression of Toll-like receptor (TLR)-4 (unpublished data). The low level intestinal epithelial expression of TLR-4 is one of the mechanisms allowing gut mucosal system to maintain its tolerance to commensal intestinal bacteria flora [34]. Accumulated evidence suggests that some intestinal commensal bacterial species are involved in the pathogenesis of IBD [11]. Perhaps the increased intestinal expression of TLR-4 induced by C. concisus has upregulated responses of the gut mucosal immune system to some intestinal commensal bacterial species otherwise it would tolerate. This hypothesis requires further investigation.
Despite the high prevalence of C. concisus detected in patients with IBD, the amount of C. concisus DNA in the intestinal biopsies was generally low. An initial examination of 20 biopsies collected from 5 patients with CD using direct Campylobacter genus-PCR revealed low positivity. Given this, we decided to use a nested PCR method to amplify the 16S rRNA gene of universal bacteria and then use Campylobacter genus-PCR. The nested PCR has greatly increased the detection rate of Campylobacter species in biopsy samples. It is likely that the preparation procedure for colonoscopy, which involves induction of severe diarrhoea, may have contributed to the low number of C. concisus in the biopsies.
In addition to detection of C. concisus from intestinal biopsies by PCR, we have isolated C. concisus from intestinal biopsies of one patient with CD and one patient with UC.
Some other Campylobacter species detected in this study have been shown to be clinically important. However, the prevalence of these Campylobacter species in patients with IBD was low and not significantly different from that in the controls (Table 3).
In summary, in this study we detected a significantly higher prevalence of C. concisus in colonic biopsies of adult patients with CD as compared with the controls and isolated C. concisus from intestinal biopsies of adult patients with IBD. Furthermore, we found that C. concisus preferentially colonizes the proximal large intestine of patients with IBD. These results suggest that future studies are needed to investigate the possible involvement of C. concisus in a subgroup of human IBD. To our knowledge, this is the first report of the association between adult CD and C. concisus; the first study of the preferential colonization sites of C. concisus in the human intestine; and the first isolation of C. concisus from intestinal biopsies of adult patients with IBD.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Broad Medical Research Program of the Broad Foundation (Grant No: IBD0273-R). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Moore JE, Corcoran D, Dooley JSG, Fanning S, Lucey B, et al. Campylobacter. Vet Res. 2005;36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 2.Lastovica AJ. Emerging Campylobacter spp.: the Tip of the Iceberg. Clin Microbiol Newsl. 2006;28:7. [Google Scholar]
- 3.Vandamme P, Dewhirst FE, Paster BJ, On SLW. Genus I. Campylobacter. . In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2 ed. New York: Springer; 2005. pp. 1147–1160. [Google Scholar]
- 4.Tanner ACR, Badger S, Lai CH, Listgarten MA, Visconti RA, et al. Wolinella gen-nov, Wolinella-succinogenes (vibrio-succinogenes-wolinet-al) Comb-nov, and description of Bacteruides-gracilis sp-nov, Wolinella-recta sp-nov, Campylobacter-concisus sp-nov and Eikenella-corrodens from humans with periodontal-disease. Int J Syst Bacteriol. 1981;31:432–445. [Google Scholar]
- 5.Macuch PJ, Tanner ACR. Campylobacter species in health, gingivitis, and periodontitis. J Dent Res. 2000;79:785–792. doi: 10.1177/00220345000790021301. [DOI] [PubMed] [Google Scholar]
- 6.Newell DG. Campylobacter concisus: an emerging pathogen? Eur J Gastroen Hepat. 2005;17:1013–1014. doi: 10.1097/00042737-200510000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Aabenhus R, On SLW, Siemer BL, Permin H, Andersen LP. Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J Clin Microbiol. 2005;43:5091–5096. doi: 10.1128/JCM.43.10.5091-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijders F, Kuijper EJ, deWever B, vanderHoek L, Danner SA, et al. Prevalence of Campylobacter-associated diarrhea among patients infected with human immunodeficiency virus. Clin Infect Dis. 1997;24:1107–1113. doi: 10.1086/513643. [DOI] [PubMed] [Google Scholar]
- 9.Engberg J, On SLW, Harrington CS, Gerner-Smidt P. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol. 2000;38:286–291. doi: 10.1128/jcm.38.1.286-291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podolsky DK. Inflammatory bowel disease. New Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 11.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 12.Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182–205. doi: 10.1016/s0016-5085(98)70381-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Man SM, Day AS, Leach ST, Lemberg DA, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn's disease. J Clin Microbiol. 2009;47:453–455. doi: 10.1128/JCM.01949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Man SM, Zhang L, Day AS, Leach ST, Lemberg DA, et al. Campylobacterconcisus and Other Campylobacter Species in Children with Newly Diagnosed Crohn's Disease. Inflamm Bowel Dis. 2010;16:1008–1016. doi: 10.1002/ibd.21157. [DOI] [PubMed] [Google Scholar]
- 15.Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, et al. Detection of Campylobacter concisus and Other Campylobacter Species in Colonic Biopsies from Adults with Ulcerative Colitis. Plos One. 2011;6:e21490. doi: 10.1371/journal.pone.0021490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverberg MS, Satsangi J, Ahmad T, Arnott IDR, Bernstein CN, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19:5A–36A. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 17.Lane DJ. Chichester. Vol. 115. Wiley J & Sons Ltd.; 1991. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. p. 175. [Google Scholar]
- 18.Linton D, Owen RJ, Stanley J. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol. 1996;147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 19.Inglis GD, Kalischuk LD, Busz HW, Kastelic JP. Colonization of cattle intestines by Campylobacter jejuni and Campylobacter lanienae. Appl Environ Microbiol. 2005;71:5145–5153. doi: 10.1128/AEM.71.9.5145-5153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levitt MD. Production and excretion of hydrogen gas in man. New Engl J Med. 1969;281:122–127. doi: 10.1056/NEJM196907172810303. [DOI] [PubMed] [Google Scholar]
- 21.Cummings JH. Fermentation in the human large-intestine-evidence and implications for healthy. Lancet. 1983;1:1206–1209. doi: 10.1016/s0140-6736(83)92478-9. [DOI] [PubMed] [Google Scholar]
- 22.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 23.Guslandi M. Antibiotics for inflammatory bowel disease: do they work? Eur J Gastroenterol Hepat. 2005;17:145–147. doi: 10.1097/00042737-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, et al. Variablephenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Vandamme P, Falsen E, Pot B, Hoste B, Kersters K, et al. Identification of EF group-22 Campylobacters from gastroenteritis cases as Campylobacter-concisus. J Clin Microbiol. 1989;27:1775–1781. doi: 10.1128/jcm.27.8.1775-1781.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsheka MI, Lastovica AJ, Elisha BG. Molecular identification of Campylobacter concisus. J Clin Microbiol. 2001;39:3684–3689. doi: 10.1128/JCM.39.10.3684-3689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastyns K, Chapelle S, Vandamme P, Goossens H, Dewachter R. Specific detection of Campylobacter concisus by PCR amplification of 23S rDNA areas. Mol Cell Probe. 1995;9:247–250. doi: 10.1016/s0890-8508(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 28.Aabenhus R, Permin H, On SLW, Andersen LP. Prevalence of Campylobacter concisus in diarrhoea of immunocompromised patients. Scand J Infect Dis. 2002;34:248–252. doi: 10.1080/00365540110080566. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, et al. Isolation and Detection of Campylobacter concisus from Saliva of Healthy Individuals and Patients with Inflammatory Bowel Disease. J Clin Microbiol. 2010;48:2965–2967. doi: 10.1128/JCM.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Man SM, Kaakoush NO, Leach ST, Nahidi L, Lu HK, et al. Host Attachment, Invasion, and Stimulation of Proinflammatory Cytokines by Campylobacter concisus and Other Non-Campylobacter jejuni Campylobacter Species. J Infect Dis. 2010;202:1855–1865. doi: 10.1086/657316. [DOI] [PubMed] [Google Scholar]
- 31.Kalischuk LD, Inglis GD. Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. Bmc Microbiology. 2011;11:53–66. doi: 10.1186/1471-2180-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Istivan TS, Coloe PJ, Fry BN, Ward P, Smith SC. Characterization of a haemolytic phospholipase A(2) activity in clinical isolates of Campylobacter concisus. J Med Microbiol. 2004;53:483–493. doi: 10.1099/jmm.0.45554-0. [DOI] [PubMed] [Google Scholar]
- 33.Engberg J, Bang DD, Aabenhus R, Aarestrup FM, Fussing V, et al. Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin Microbiol Infect. 2005;11:288–295. doi: 10.1111/j.1469-0691.2005.01111.x. [DOI] [PubMed] [Google Scholar]
- 34.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–143. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]