Abstract
Iron homeostasis is crucial to many biological functions in nearly all organisms, with roles ranging from oxygen transport to immune function. Disruption of iron homeostasis may result in iron overload or iron deficiency. Iron deficiency may have severe consequences, including anemia or changes in immune or neurotransmitter systems. Here, we report the variability phenotypic iron tissue loss and splenomegaly as well as the associated quantitative trait loci (QTL), polymorphic areas in the mouse genome which may contain one or more genes that play a role in spleen iron concentration or spleen weight under each dietary treatment. Mice from twenty-six BXD/Ty recombinant inbred strains, including the parent C57BL/6 and DBA/2 strains, were randomly assigned to adequate iron or iron deficient diets at weaning. After 120 days, splenomegaly was measured by spleen weight, and spleen iron was assessed using a modified spectrophotometry technique. QTL analyses and gene expression comparisons were then conducted using the WebQTL GenetNetwork. We observed wide, genetic-based variability in splenomegaly and spleen iron loss in BXD/Ty recombinant inbred strains fed an iron deficient diet. Moreover, we identified several suggestive QTL. Matching our QTL with gene expression data from the spleen revealed candidate genes. Our work shows that individual differences in splenomegaly response to iron deficiency are influenced at least partly by genetic constitution. We propose mechanistic hypotheses by which splenomegaly may result from iron deficiency.
Introduction
Nearly all organisms are dependent on iron for a variety of biological functions, including oxygen transport, metabolic pathways, and immune function (Aisen et al., 2001; Kuvibidila & Porretta, 2003; Omara & Blakley, 1994). Iron homeostasis is maintained through a large number of mechanisms regulating its absorption, transport, storage, and movement through cells. When iron homeostasis is disrupted, tissue iron overload or deficiency can be the result. Iron overload, often resulting from hemochromatosis and related to Parkinson’s disease, can result in many problems as a consequence of iron accumulation in tissues such as the brain, liver, heart, pancreas, and gonads (Aisen et al., 2001). Excess liver iron in particular increases the risk for cirrhosis, hepatoma, and other diseases.
Iron deficiency can also have adverse consequences, such as hypochromic microcytic anemia. Nearly 75% of all nutritional anemias are thought to be a direct consequence of iron deficiency (Ramakrishnan, 2001). When iron deficiency occurs early in development, pronounced and long-lasting behavioral and cognitive deficits can occur (Beard et al., 2003; Eriksonet al., 2000); however, little is known about the effects of early iron deficiency outside the central nervous system. Iron deficiency affects an estimated 15% of individuals worldwide, particularly pregnant women, women of childbearing age, elderly adults, and children (Ramakrishnan, 2001; Nelson et al., 1997).
Iron homeostasis is highly complex, involving more than twenty proteins and genes that can affect many aspects of iron metabolism, such as absorption or rate of loss (Aisen et al., 2001; Jones et al., 2007; Whitfield et al., 2000); however, we do not know yet exactly how many genes are involved. By understanding the genetics underlying iron homeostasis, we may better understand the biological mechanisms by which tissue-specific, optimal iron concentrations are maintained. Recent work on the hereditary cause of iron overload, hemochromatosis, suggests that even disruption of iron balance involves a multitude of genes that act as modifiers on the primary homozygous mutation of the HFE gene (Bensaid et al., 2004). Similarly, research on Australian twins, both identical and fraternal, displayed that nearly 23% and 31% of variance found in iron between men and women, respectively, could be attributed to genetic factors (Whitfield et al., 2000). Additionally, we applied the method of mapping genetic markers, i.e., quantitative trait loci (QTL) to show that, like many other important traits, the regulation of iron is under the influence of several genes. Despite recent progress in understanding the genetics of iron homeostasis, little is understood about the extent to which individuals vary in genetic susceptibility to iron deficiency and its resulting complications. This work aims to shed light on the topic, with a particular focus on variability in iron homeostasis in the spleen.
Compared to other tissues, how the spleen manages iron appears to be unique. For example, we recently showed spleen iron levels do not correlate with other indices of peripheral iron status, such as liver iron, hematocrit, hemoglobin, total iron binding capacity, transferrin saturation, or plasma iron (Jones et al., 2007). The spleen also recycles iron stored in bone marrow and is essential for the removal of cell debris, cell creation, blood cell storage, and certain immune functions. Individuals who suffer from infectious diseases such as malaria, leishmaniasis, or trypanosomiasis may have permanently enlarged spleens, or splenomegaly (April, 1997). Interestingly, while studying iron deficiency, we noted striking variability in iron deficiency-induced splenomegaly among the strains in the BXD panel of recombinant inbred mice. While not considered to be a vital organ in adults, extremely large spleens may rupture and lead to massive hemorrhaging (April, 1997). Accordingly, understanding the mechanisms involved in splenomegaly may allow development of treatments for this condition.
In this article, we report our observations of genetic variability produced by iron deficiency and report QTL associated with both iron loss and hypertrophy in the spleen.
Materials and Methods
Animals
Twenty-six strains of the BXD/Ty mouse recombinant inbred (RI) strains, including the parental strains (C57BL/6 and DBA/2), provided the subjects for this experiment. The mice were housed in the Centralized Biological Laboratory located at the Pennsylvania State University at University Park, Pennsylvania. The mice were weaned at postnatal day 21 and housed in unisex groups of 2 – 5 mice per cage. The average number per strain × sex × diet group ranged between 10 and 12. Three groups had 2 animals and 3 groups had 3 animals.
Dietary treatment
Upon weaning, mice were separated into two groups: those fed an iron-adequate diet and those fed an iron deficient diet (Harlan-Teklad, Inc.). The iron-adequate diet was pelleted and contained approximately 240ppm of iron, and the iron deficient diet (also pelleted) contained approximately 3ppm of iron (Harland-Teklad, Inc.). All other nutritional factors were equal; both diets were based on the AIN-93G formulation (American Institute of Nutrition). Food and water (tap water, as Penn State tap water contains no detectable iron) were administered ad libitium. Housing conditions were controlled for temperature (23 ± 2 degrees Celsius) and humidity (40%). The lights in the colony were on a twelve hour cycle (on at 0600: off at 1800). All mice were weighed weekly to monitor weight gain and health. Mice were sacrificed at 120 days using carbon dioxide euthanasia. Cardiac puncture was used to obtain blood, and the liver and spleen were removed immediately. Additionally, the brain was removed and further dissected for another project. The Penn State Institutional Animal Care and Use Committee approved all experimental protocols.
Spleen weights and iron determination
Spleens were weighed and stored at −30°C until prepared for the analysis. Spleen tissue iron concentrations were determined using a modification of the Cook et al. protocol (1980). On day 1, spleens were removed and allowed to thaw. The spleens were weighed and placed in a glass test tube with protein precipitant proportional to spleen weight. For every 0.1 mg of spleen, 1 mL of protein precipitant was added. A marble was placed on the top of every test tube to prevent evaporation. The spleens were then allowed to sit in a sand bath of approximately 30°C over night. On day 2, the spleens were crushed with a glass rod and allowed to stay in the sand bath for an additional night. On day 3, the spleens were removed from the sand bath and allowed to cool. A standard curve of seven known iron concentrations (totaling 1 mL each) were made in similar test tubes and 100 μL of protein precipitant were added to each standard. 2 mL of chromagen were added to each standard, leaving 3 mL of liquid in each test tube. These standards were vortexed and allowed to develop for 10 minutes. A reagent blank was made by adding 100 μL of protein precipitant to 3 mL of chromagen. The reagent blank was also vortexed and allowed to sit for 10 minutes. New sample test tubes were labeled in duplicate, and 3 mL of chromagen were placed in each sample. Increments of spleen sample were then added identically to both sample test tubes containing chromagen. Increments often started at approximately 25 μL and were added until the sample appeared visually to contain the same amount of iron (same shade of purple) as the middle standards of the standard curve. Both sample test tubes were vortexed after every addition of spleen sample. All samples were allowed to develop for 10 minutes. The samples were then read using a spectrophotometer, set at a wavelength of 535 nm. Calculations against the standard curve were then performed to determine the iron concentration of spleen tissue in each mouse (μg iron/g tissue).
Gene expression in spleen
Spleens were taken from 6 to 8 week old male mice. RNA was extracted and prepared for hybridization to Illumina Ref-8 arrays according to the manufacturer’s instructions. Normalized data was entered into the GeneNetwork database site at the University of Western Australia for WebQTL systems genetics analyses (Morahan et al., 2003; Morahan & Williams, 2007).
Statistical analysis
The raw data were evaluated by analysis of variance with strain, sex, and diet as between-subject variables. Correlational analyses were determined by Pearson’s r using the BXD published phenotypes database within WebQTL (http://www.genenetwork.org). For the SNP marker data, differences between means were evaluated by Student’s t-tests. QTL analysis was also performed using WebQTL (http://www.genenetwork). The likelihood of the odds (LOD) ratio was selected to identify the significance of identified QTL. Narrow-sense heritability values were estimated for each treatment condition separately by ANOVA as SSstrain/SStotal (Belknap, 1993). Outliers of greater than two standard deviations from the mean were removed before any statistical analyses.
For each QTL marker, we performed a search for nearby genes whose expression in the spleen was cis-regulated, i.e, with significant QTL for expression close to the encoding region. Combining genetic analysis (QTL) with genomic analysis (gene expression) has been proposed by others (Hitzemann, Reed, et al., 2004) as an efficient means to identify candidate genes. For those QTL markers lying near cis-regulated genes, we separated the strains carrying alleles from DBA/2 and C57BL/6 parental strains and calculated Student t-tests using strain means for spleen iron concentration or spleen weights as indices.
Results
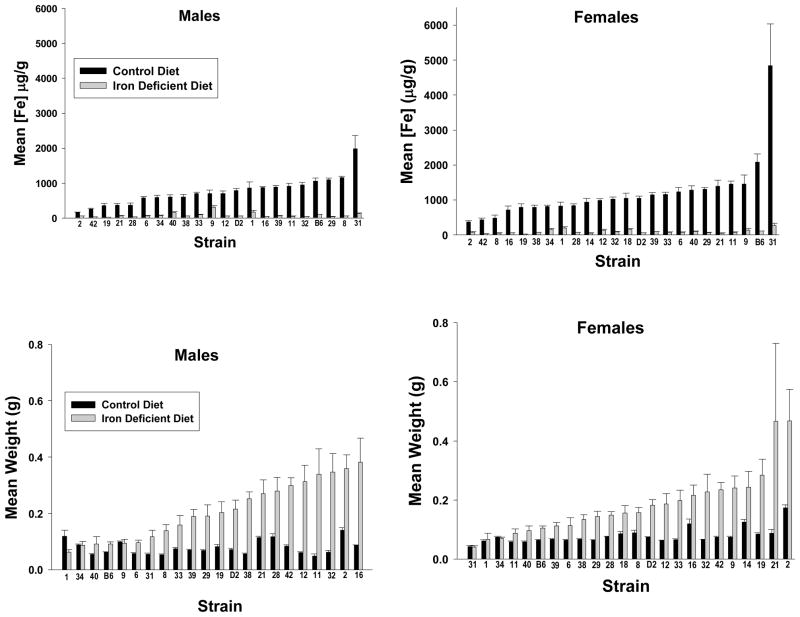
Table 1 displays the summary statistics for spleen weight and iron concentration for both adequate diet and iron deficient mice. For spleen iron concentration, there was a significant effect of strain (F25,906=6.140, p < 0.001), strain by sex interaction (F22,906=2.199, p<0.001), strain by diet interaction (F23,906=6.673, p<0.001), and sex by strain by diet interaction (F21,906=2.553, p<0.001). There was no significant main effect of sex or diet. Figure 1 (top panel) illustrates the effects of iron deficient diet versus iron-adequate diet on spleen iron concentrations and spleen weights by strain. The spleen iron concentrations for males found in this study correlated significantly with spleen iron concentrations from previous work in our lab on mice (sexes combined) fed an iron-adequate diet (r=0.505, p<0.02) (Jones, Beard, et al., 2007).
Table 1.
Summary Statistics for Adequate Iron and Iron Deficient Mouse Spleen Parameters.
| AD* Spl. Wt. (g) | ID† Spl. Wt. (g) | AD Spl. Fe (μg/g) | ID Spl. Fe (μg/g) | |
|---|---|---|---|---|
| Males | ||||
| mean | 0.1282 | 0.1593 | 516.965 | 473.493 |
| SD | 0.1276 | 0.1300 | 580.540 | 562.859 |
| min | 0.0206 | 0.0300 | 12.739 | 10.772 |
| max | 0.9544 | 0.7403 | 3869.388 | 3891.667 |
| Females | ||||
| mean | 0.1178 | 0.1219 | 592.450 | 543.846 |
| SD | 0.1372 | 0.0921 | 603.035 | 1057.022 |
| min | 0.0146 | 0.0310 | 8.194 | 17.767 |
| max | 1.011 | 0.6035 | 3511.572 | 10415.254 |
AD = Adequate Diet,
ID = Iron Deficient
Figure 1.
Results for Adequate Diet (AD) and Iron Deficient (ID) Mice by Strain. The top panel displays spleen iron concentrations for males (left panel) and females (right panel) by strain. The bottom panel displays spleen weights for males (left panel) and females (right panel) by strain. All animals were fed their respective diets from weaning at 21 days of age until 120 days of age and then sacrificed for tissue iron concentrations and measurements. The data presented are means ± s.e.m.
For spleen weights, (Figure 1, bottom panel) there was a significant effect of strain (F25,965=10.546, p<0.001), diet (F1,965=257.874, p<0.001), sex (F1,965=5.836, p<0.02), sex by strain interaction (F24,965=2.145, p<0.001), diet by strain interaction (F23,965=5.490, p<0.001), and diet by strain by sex interaction (F21,965=2.550, p<0.001).
Heritability estimates
For spleen weights, the narrow sense heritability estimate was 0.45 for the iron-adequate diet and 0.25 for the iron deficient condition. For spleen iron, the values were 0.26 and 0.20, respectively.
Genetic correlational analysis between measures
Using strain means, there was a significant correlation between spleen weight and spleen iron concentration for iron-adequate diet males (r= −0.541, p<0.01), iron deficient males (r=−0.589, p<0.005), iron-adequate females (r=−0.465, p<0.03), and iron deficient females (r=−0.428, p<0.04). Similarly, there was a significant correlation between adequate diet and iron deficient females for spleen weight (r=0.710, p<0.001) and spleen iron concentration (r=0.614, p<0.001).
Genetic correlations between sex by measure
The correlation between males and females was significant under the iron-adequate condition for both spleen weight (r=0.634, p<0.001) and spleen iron concentration (r=0.755, p<0.001). Likewise, the correlation between males and females was significant under the iron deficient condition for both spleen weight (r=0.519, p<0.02) and spleen iron concentration (r=0.434, p<0.04).
Simple marker-based quantitative trait loci analysis
Table 2 summarizes QTL markers identified for iron parameters by sex.
Table 2.
QTL Identified by Phenotype.
| Phenotype | Diet | Sex | Chr(Mb) | Marker | LOD |
|---|---|---|---|---|---|
| [Fe] | AD | M | 10 (118.38) | mCV25264026 | 2.39 |
| ID | F | 2 (103.12) | Rs3143810 | 2.03 | |
| ID | F | 17 (16.40) | rs3675740 | 2.23 | |
| Spleen Wt | AD | M | 15 (3.23) | rs13459176 | 2.99 |
| ID | M | 2 (103.12) | rs3143810 | 2.60 | |
| AD | F | 10(64.17) | rs13480625 | 2.30 | |
| ID | F | 11(83.52) | rs13481128 | 2.04 |
Spleen iron concentration
For spleen iron concentration, three suggestive QTL were defined depending on condition. For iron-adequate males, one suggestive QTL was located on chromosome 10 near 118 Mb (marker, mCV25264026, LOD=2.39). For iron-deficient females, two suggestive QTLs were identified; one on chromosome 2 near 103 Mb (rs3143810, LOD=2.03) and the other on chromosome 17 near 16 Mb (rs3675740, LOD=2.23).
Spleen weight
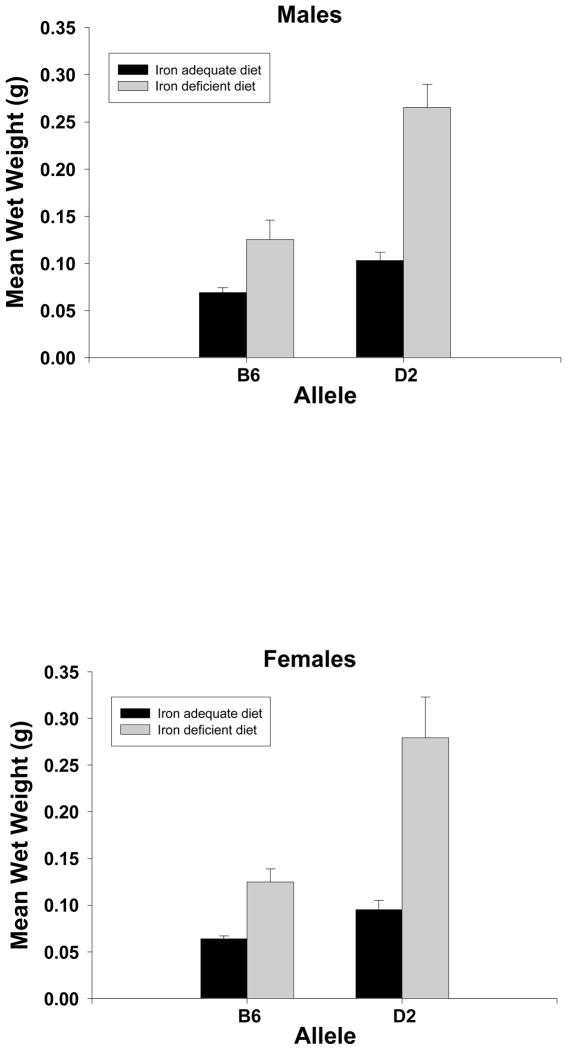
For spleen weight, four suggestive QTL were identified, one for each diet by sex combination. For iron-adequate diet males, the marker, rs13459176 is located on chromosome 15 at 3.229128 Mb. For iron-deficient males, the marker, rs3143810 is located on chromosome 2 at 103.12449 Mb. For iron-adequate females, the marker, rs13480625 is located on chromosome 10 at 64.173094 Mb. For iron-deficient females, an area on chromosome 11 from approximately 82 Mb to 90 Mb contained nine QTL; however, bootstrap analysis supported only one, rs13481128, located at 83.521935 Mb. Statistical analysis by Student t-test by marker (illustrated in Figure 2) showed that, for the iron adequate males, the D2 allele was associated with higher mean spleen weight than the B6 allele (t21=3.81, p<0.001); this was also true for the iron-deficient diet (t21=3.96, p<0.001). The females showed the same pattern with the iron-adequate diet (t22=3.28, p<0.004) and iron-deficient diet (t22=3.83, p<0.001).
Figure 2.
Spleen weights by allele, diet and sex. The bars are the mean and s.e.m from several strains carrying alleles from either the C57BL/6J strain (B6) or DBA/2J strain (D2). For the males the marker for the iron adequate diet is rs13459176 located on chromosome 15 at 3.229128 Mb and for the iron deficient diet, rs3143810 located on chromosome 2 at 103.124490 Mb. For the females the marker for the iron adequate diet is rs13480625, located on chromosome 10 at 64.173094 Mb and for the iron deficient diet, rs13481128 located on chromosome 11 at 83.521935 Mb. There were no significant differences between the sexes; within each sex, all pairwise comparisons between means are significant at at least p<0.01. The number of strains per diet: M/AD, B allele = 16, D allele =7; M/ID B allele = 9, D allele =14; F/AD, B allele = 13, D allele = 11; F/ID, B allele = 14, D allele = 10.
Gene expression QTL (eQTL) analysis
For iron-adequate males for the marker, rs13459176, there were no nearby cis-regulated genes. For SNP rs13478434 in iron-deficient males, one cis-regulated gene, catalase, is located at 103.29 Mb and the marker for its expression has an associated LOD score of 6.44. There is also a significant eQTL for its expression with a LOD score of 5.83 located on chromosome 9 at 40 Mb. For iron-adequate females, as with the iron-adequate males, we found no cis-regulated genes near the marker, rs13480625. For iron-deficient females, we found three cis-regulated genes, Slf2, Schlafen gene 2, Mmp28, matrix metallopeptidase 28 (epilysin), and Acaca, acetyl-Coenzyme A carboxylase. Slf2 is also trans-regulated with its marker located on chromosome 8 at approximately 40. In addition to performing eQTL analysis on the Illumina microarray database described above, we cross-checked with the recently available Affymetrix gene expression data from UTHSC Affymetrix MoGene 1.0 ST Spleen (Dec10) RMA Genenetwork.org, accession number: GN283. As for the iron-adequate males and females, there were no significant cis-regulated genes identified near the markers. For the iron-deficient males, catalase gene expression failed to reach significance in the Affymetrix data. For the iron-deficient females, however, there was agreement between the platforms for Slfn2 and Mmp28. In addition, in the Affymetrix assay, eQTL analysis indicates Slfn9 as possible candidate.
Discussion
In this work, we explored an interesting finding that many of the mice fed an iron deficient diet developed pronounced splenomegaly. Additionally, we conducted QTL mapping to identify specific areas of the mouse genome that contained genes related to our measures, and we also reported cis-regulated genes in the spleen as potential candidate genes for these QTLs.
Concerning natural spleen iron concentration, there was wide variability among strains. All mice fed an iron deficient diet, regardless of strain, displayed a significant decrease in spleen iron concentration, with much lower strain-related variability, probably due to scale restriction of the measure. Whether there is a minimum below which spleen iron concentration does not drop is an interesting possibility that awaits further study. Among all of the tissues that we have examined (unpublished observations), the spleen appears to be one of the most sensitive to nutritional iron deficiency.
For spleen weight, we again observed a significant effect of both strain and diet. Unlike spleen iron, we observed greater strain-related variability in both sexes in spleen weights under the iron deficient condition compared to control.
There was high variability within some strains as well. In many of the strains, a clear bimodal trend was observed in which the mice either had spleen weights that were low/more similar to same-strain iron-adequate mice or spleen weights that were very high. Such variability in these particular strains is interesting, considering that these animals are more than 99% genetically identical within strain. It may be that these strains have high-genetically based environmental lability – i.e., sensitivity to even very small changes in care, housing, and husbandry.
The narrow-sense heritability for spleen iron concentration and spleen weight for iron adequate and iron deficient diets were moderate and indicate that genetic background is not the only driving factor for these parameters. Despite this, a number of suggestive QTL were identified; however, it is important to note that these may have reached genome-wide significance levels if more strains were available to us. These suggestive QTL may be confirmed by further studies.
For spleen iron concentration, we identified a suggestive QTL on chromosome 10. There are no obvious candidates as no cis-regulated genes were found in the vicinity. For iron-deficient females, one of our suggestive QTL on chromosome 2 points to the same cis-regulated gene as for iron-deficient male spleen weight.
For spleen weight, we identified suggestive QTL on chromosomes 2, 10, 11 and 15. QTL mapping combined with gene expression data for nearby genes can prove to be an efficient way to identify candidate genes (Hitzemann et al., 2004). For iron deficient females, three markers for iron-deficient spleen weights on chromosome 11 are near cis-regulated genes ccl12 and ccl9, chemokine (C-C motif) ligands, and acaca, acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase). More distally on chromosome 11 for the same phenotype in females are appbp2 and ppm1d, risc, encoding amyloid beta precursor protein (cytoplasmic tail) binding protein 2, protein phosphatase 1D magnesium-dependent, delta isoform, and retinoid-inducible serine caroboxypetidase respectively. Whether any or all of the genes on chromosome 11 are viable candidates requires further investigation, as they are not separated by recombination in the panel of the BXD/Ty RI strains having been derived from an F2 intercross. As with any QTL study, the biological function of the candidate genes if true, becomes an important issue. For catalase, its major role is to convert hydrogen peroxide to oxygen and water. Exactly how this relates to iron-induced splenomegaly is not clear at this time. For matrix metalopeptidase 28, this enzyme is involved in extracellular matrix remodeling, as is probably the case in splenomegaly. Schlafen 2 is important in T-cell quiescence and it is quite likely that spleen iron status has a major impact on T-cell turnover. Ccl12 is highly expressed in macrophaghes and again, how its function is related to iron-related splenomegaly is yet to be explored.
In a recent article, Wang and colleagues (2007) reported a highly significant QTL for spleen non-heme iron on mouse chromosome 9 based on a F2 intercross between C57BL/10J and SWR mice. The QTL was close to Mon1a and appeared to be related to a missense mutation in this gene. The allele was carried by the B10 strain and was associated with low non-heme iron loading in splenic macrophages. The B6 strain also shows low non-heme iron content in the spleen (likely based on the same mutation in Mon1a) and liver, and the spleen iron content also showed a QTL on chromosome 9 near the transferrin gene, which is about 4 Mb upstream from Mon1a (Grant, et al., 2006). In our work, we measured total iron content in the spleen and did not see a QTL on chromosome 9. In fact, in total iron content when fed the 270 ppm Fe diet, B6 was above the median across all strains. We also saw no correlation between the expression of Mon1a in the spleen and spleen iron content.
We suggest two possible mechanisms by which the splenomegaly may occur in response to iron deficiency. The first is that severe iron deficiency likely influences immune function (Kuvibidila & Porretta, 2003). As discussed earlier, spleens can enlarge as a secondary result of large changes in immune function (April, 1997); therefore, it is possible that each individual has a particular “threshold” by which changes in immune function, which could be induced by iron deficiency, will cause the spleen to enlarge. Potentially, this “threshold” is established genetically, thereby causing a susceptibility to splenomegaly as a result of iron deficiency. Not all mice of a susceptible strain may reach this threshold, however, thus leading to the large variability that we see in some of the strains. The second potential mechanism is by way of hemoglobin reduction in anemia. Considering that nearly all of our iron deficient mice established a baseline or minimum iron concentration, regardless of strain, it is likely that all of our mice, exhibiting various degrees of anemia, were bringing compensatory mechanisms into play. We have unpublished data showing that the change in hemoglobin during iron deficiency is also highly variable with some strains showing a more than 60% decrease and others less than 20%. Because the spleen is involved in maintaining hemoglobin, it is possible that this change, too, could cause the spleen in some strains to be overworked in order to process the necessary amount of hemoglobin, resulting in enlargement. Again, it is possible that there is a genetically established “threshold” for how much iron deficiency the spleen can handle before enlarging, leading to susceptibility to splenomegaly.
Future research should establish a mechanism by which splenomegaly is triggered as a result of iron deficiency, more clarification about the biological (pathological) significance, and how genetic susceptibility plays a role in either buffering against or promoting spleen enlargement.
In conclusion, we have demonstrated that spleen enlargement can result from iron deficiency, that there is a genetic component to this susceptibility, and moreover, that susceptibility is most likely mediated by several genes. Finally, we offered two potential mechanisms that may trigger splenomegaly as a result of iron deficiency, including changes to immune function or hemoglobin.
References
- Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Bio. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- April EW. National Medical Series for Independent Study: Clinical Anatomy. Philadelphia: Williams and Wilkins; 1997. [Google Scholar]
- Beard JL, Wiesinger JA, Connor JR. Pre- and postweaning iron deficiency alters myelination in sprague-dawley rats. Dev Neurosci. 2003;25:308–315. doi: 10.1159/000073507. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genetc. 1993;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Bensaid M, Fruchon S, Mazeres C, Bahram S, Roth MP, Coppin H. Multigenetic control of hepatic iron loading in a murine model of hemochromatosis. Gastroenterology. 2004;126:1400–1408. doi: 10.1053/j.gastro.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Cook GA, King MT, Veech RL. Changes in liver inorganic pyrophosphate content during ethanol metabolism. Adv Exp Med Biol. 1980;132:433–440. doi: 10.1007/978-1-4757-1419-7_43. [DOI] [PubMed] [Google Scholar]
- Erikson KM, Jones BC, Beard JL. Iron deficiency alters dopamine transporter functioning in rat striatum. J Nutr. 2000;130:2831–2837. doi: 10.1093/jn/130.11.2831. [DOI] [PubMed] [Google Scholar]
- Grant GR, Robinson SW, Edwards RE, Clothier B, Davies R, Judah DJ, Broman KW, Smith AG. Multiple polymorphic loci determine basal hepatic and splenic iron status in mice. Hepatology. 2006;44:174–185. doi: 10.1002/hep.21233. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, Phillips T, Crabbe J. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004;28:1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Jones BC, Beard JL, Gibson JN, Unger EL, Allen RP, McCarthy KA, Earley CJ. Systems genetic analysis of peripheral iron parameters in the mouse. Am J Physiol Regul Integr Comp Physiol. 2007;293:R116–R124. doi: 10.1152/ajpregu.00608.2006. [DOI] [PubMed] [Google Scholar]
- Kuvibidila SR, Porretta C. Iron deficiency and in vitro chelation reduce the expression of cluster of differentiation molecule (CD)28 but not CD3 receptors on murine thymocytes and spleen cells. Br J Nutr. 2003;90:179–189. doi: 10.1079/bjn2003864. [DOI] [PubMed] [Google Scholar]
- Morahan G, Peeva V, Mehta M, Williams R. Systems genetics can provide new insights into immune regulation and autoimmunity. J Autoimmun. 2003;3:233–236. doi: 10.1016/j.jaut.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Morahan G, Williams RW. Systems genetics: the next generation in genetics research? Novartis Found Symp. 2007;281:181–188. doi: 10.1002/9780470062128.ch15. [DOI] [PubMed] [Google Scholar]
- Nelson C, Erikson K, Pinero DJ, Beard JL. In vivo dopamine metabolism is altered in iron-deficiency anemic rats. J Nutr. 1997;127:2282–2288. doi: 10.1093/jn/127.12.2282. [DOI] [PubMed] [Google Scholar]
- Omara FO, Blakley BR. The effects of iron deficiency and iron overload on cell-mediated immunity in the mouse. Br J Nutr. 1994;72:899–909. doi: 10.1079/bjn19940094. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan U. Nutritional Anemias. Boca Raton: CRC Press LLC; 2001. [Google Scholar]
- Wang F, Paradkar PN, Custodio AO, Ward DM, Fleming MD, Campagna D, Roberts KA, Boyartchuk V, Dietrich WF, Kaplan J, Andrews NC. Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nature Genet. 2007;39:1025–1032. doi: 10.1038/ng2059. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Cullen LM, Jazwinska EC, Powell LW, Heath AC, Zhu G, Duffy DL, Martin NG. Effects of HFE C282Y and H63D polymorphisms and polygenic background on iron stores in a large community of sample twins. Am J Hum Genet. 2000;66:1246–1258. doi: 10.1086/302862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick M, Pinggera W, Lehmann P. Iron Metabolism, Anemias, Diagnosis, and Therapy. Vol. 4. New York: SpingerWien; 2000. [Google Scholar]