Abstract
The vitamin D receptor (VDR) mediates virtually all of the known biological actions of the hormonal ligand 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). These actions are directed toward the nucleus, where the VDR binds to the regulatory regions of target genes and modulates their transcriptional output. Recent technological advances have enabled the study of transcription factor binding on a genome-wide scale in cells and tissues that are major targets of vitamin D action. In this review, the results of several of these studies are discussed wherein overarching principles of gene regulation by the vitamin D hormone are beginning to emerge. In addition, several specific genes that are regulated by 1,25(OH)2D3 and which provide new insight into the increasingly complex mechanism whereby the receptor functions to modulate gene expression are considered. These studies suggest that while many of the principles that are now accepted regarding the regulation of gene expression by hormones and other regulatory factors are well grounded, others require extensive modification.
Keywords: transcription, ChIP-chip, ChIP-seq, VDR, RXR, enhancers, acetylation, CYP24A1, Tnfsf11/Rankl
1. Introduction
1,25-Dihydroxyvitamin D (1,25(OH)2D3), the active hormonal derivative of vitamin D, plays a central role in the regulation of mineral homeostasis through its control of diverse biological processes in the intestinal tract, kidney and in bone (DeLuca, 2004; Plum and DeLuca, 2010). More recently, it has become clear that the vitamin D hormone also functions to modulate a number of mature cell functions that are unrelated to mineral metabolism including intestinal regulation of xenobiotic degradation, modulation of specific components of the immune system, control of skin cell differentiation and regulation of endothelial cell and cardiovascular biology (Bouillon et al., 2008a; Bouillon et al., 2008b). This hormone also plays a more ubiquitous role in cellular proliferation, differentiation and survival, suggesting the possibility that it might reduce the risk of cancer or act as an anti-tumor agent in a variety of human cancers (Bouillon et al., 2006; Matthews et al., 2010; Zinser and Welsh, 2004).
The cellular actions of 1,25(OH)2D3 are mediated by the vitamin D receptor (VDR), an intracellular transcription factor whose functional activity in the nucleus of target cells is largely, although not exclusively, controlled by direct interaction with its cognate ligand (Haussler et al., 2008; Pike and Meyer, 2010; Pike et al., 2010). This activation process provokes heterodimer formation with nuclear retinoid X receptor (RXR) at selected DNA sequences that are located within enhancers and are termed vitamin D response elements (VDREs) (Kerner et al., 1989; Mangelsdorf and Evans, 1995; Ozono et al., 1990; Sone et al., 1991a; Sone et al., 1991b). These DNA segments function in cis with other cellular components to regulate the genes to which they are directly linked. Selective DNA binding by the VDR at these target genes results in the recruitment of coregulatory complexes that are capable of directing a number of key processes that are essential for altered gene output (Sutton and MacDonald, 2003). Although some of these principles are more relevant to the activation of genes by 1,25(OH)2D3, it is clear that chromatin modifications are also important to gene repression as well (Kato et al., 2007; Kim et al., 2009). Not surprisingly, while mechanisms of transcription factor activation differ, frequently through direct covalent factor modification rather that ligand activation, the general mechanisms of gene regulation downstream of these activation processes are similar.
Recently, a number of technological advances have been made that are directly relevant to the study of transcription. These advances are centered on the technique of chromatin immunoprecipitation (ChIP), wherein a protein factor can be cross-linked to its endogenous or induced site(s) of action on the genome and then utilized to precipitate small DNA segments to which it is associated (Wells and Farnham, 2002). While quantitation of precipitated DNA was assessed initially using primer sets directed toward the amplification of a predetermined region on the genome (PCR-ChIP analysis), this positional limitation was rapidly overcome by hybridizing the DNA segments on tiled microarrays (ChIP-chip analysis) (Kim et al., 2006; Kirmizis et al., 2004; Komashko et al., 2008) and more recently by using deep sequencing methodologies (ChIP-seq analysis) (Ernst and Kellis, 2010; Hawkins et al., 2010a; Hawkins et al., 2010b; Heintzman et al., 2009; Heintzman et al., 2007; Hon et al., 2009). A comparison of these three approaches to exploiting ChIP analysis has been documented recently (Pike, 2011). However, a major advantage of both ChIP-chip and particularly deep sequencing ChIP-seq methodologies is that they yield unbiased, genome-wide data sets that can provide detailed mechanistic information across complete genomes. Indeed, complex annotation of the genomes of both model organisms as well as humans with regard to structural, functional and regulatory features has grown almost exponentially over the past several years. This has both extended our understanding of the regulatory genome and uncovering entire new fields of exploratory biology. As one might imagine, these techniques have been applied to the vitamin D field with equal vigor.
In this review, we describe some of the advances that have been made in our understanding of the molecular actions of the vitamin D hormone and its nuclear receptor. We discuss first the results of several studies that have provided significant insight into the actions of 1,25(OH)2D3 on a genome-wide scale and then show how these principles are highlighted at the level of several genes of specific interest to the vitamin D field. It should come as no surprise that many of the ideas that have emerged over the past several decades using both limited gene data sets as well as the more traditional techniques have not been confirmed using these more recent approaches. Given the specific role of the vitamin D endocrine system in both the regulation of key physiological systems but also in a variety of disease processes, it is imperative that we identify gene networks whose activities in tissues are under vitamin D control and understand the genome-wide principles that govern their regulation by 1,25(OH)2D3.
2. A Genome-Wide Perspective on Gene Regulation by 1,25(OH)2D3
Several studies have been conducted recently that provide a genome-wide perspective on the actions of vitamin D and its receptor. Not surprisingly, these studies have focused upon the actions of 1,25(OH)2D3 in osteoblasts, a particularly relevant target of vitamin D action in view of the potential bone anabolic activity inherent to this important calcemic hormone (Meyer et al., 2010a). Studies have also focused on intestinal/colon cells, an equally important vitamin D target tissue (Meyer et al., 2011). As the cells used in this latter study represent a colorectal cancer cell line, the focus here was not restricted to genes involved in calcium and phosphorus homeostasis alone but also to xenobiotic metabolism and growth regulation as well. A recent study of vitamin D action was also conducted in EB virus transformed lymphoblastoid cell lines (Ramagopalan et al., 2010). While this cell type does not a highly studied vitamin D target cell type, the rationale for the examination of these lines appears to be the availability of large sets of ancillary genomic data that have emerged as a result of the ENCODE Project (Bernstein et al., 2010). The following section describes the results of these three studies in some detail.
2.1. Genome-wide Studies of VDR/RXR Heterodimer Binding in Osteoblasts
2.1.1 ChIP-chip and Chip-seq Analysis in Osteoblasts
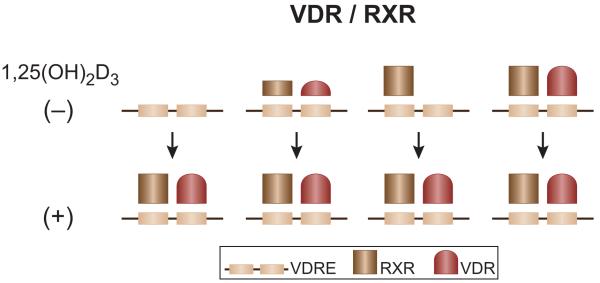
ChIP-chip and subsequent ChIP-seq analysis in osteoblasts has revealed that the VDR is bound to approximately 1200 sites across the mouse genome under basal conditions and to approximately 8000 sites following activation by 1,25(OH)2D3 (Meyer et al., 2010a). These sites therefore comprise the VDR cistrome, a term which refers to the genome-wide collection of all binding sites for a particular factor in a given cell type under a specific set of conditions. A large portion of these basal VDR binding sites overlap those observed in the presence of 1,25(OH)2D3. These data suggest that while VDR DNA binding is highly regulated by 1,25(OH)2D3 at most enhancers, selective occupancy by the VDR at a subset of these sites does not require ligand activation (Figure 1). This feature differs from a number of other nuclear receptors of this class where residual DNA binding appears to be a predominant characteristic (Lefterova et al., 2008). Although ligand-independent VDR enhancer binding activity is both gene- and cell type-specific, the mechanism that defines this selectivity and its functional consequence, if any, is unknown. ChIP-chip analysis also revealed that while RXR binding is widely distributed at distinct sites across the genome, it is also ubiquitously present at almost all VDR DNA binding sites as well as following activation by 1,25(OH)2D3. Interestingly, RXR is often pre-bound at sites that become occupied by the VDR following ligand activation (Figure 1). A classic example of this features is observed at the osteopontin gene (Meyer et al., 2010a). While this observation suggests that RXR may point to sites of potential VDR action, the nature of RXR binding under these conditions is unknown. Regardless, these observations strongly support the original hypothesis that RXR represents a heterodimeric partner necessary for VDR DNA binding and gene activation.
Figure 1.
Relative VDR and RXR binding activities across mouse and human genomes in the absence and presence of 1,25(OH)2D3.
The identification of large numbers of VDR/RXR binding sites across the osteoblast genome enabled a de novo search for sequence motifs that represent direct binding sites for the VDR/RXR heterodimer. As illustrated in Figure 2, the most frequent DNA sequence identified at these sites was comprised of two directly repeated half-sites of the consensus sequence AGGTCA separated by three base pairs, a sequence first identified in the human osteocalcin gene (Kerner et al., 1989; Ozono et al., 1990) and subsequently in most other vitamin D target genes as well (Haussler et al., 2010; Pike and Meyer, 2010). In many cases, more than one VDRE was identified at sites of VDR/RXR interaction on the genome. Not surprisingly, a biased bioinformatic search for specific VDRE-like sequences at these sites also resulted in the identification of at least one plausible VDRE at almost every site. It is worth noting, however, that close inspection of both the VDREs identified within this genome-wide VDR/RXR binding data set as well as those identified over the past decade in specific genes suggests that VDREs are actually comprised of a single conserved consensus half-site paired with a second half-site that is highly degenerate. The implications for this configuration remain unclear, however, because the polarity of the heterodimer relative to VDR and RXR binding at these sites and the impact of this orientation on gene expression has not been rigorously explored. Despite this uncertainty regarding this feature of VDR/RXR binding, it is clear that genome-scale identification of DNA binding sites for the heterodimer confirms the structure of the first VDRE originally identified over two decades ago (Ozono et al., 1990).
Figure 2.
Position weight matrix motif (VDRE) derived from 168 VDR/RXR binding sites in human colorectal cancer cells.
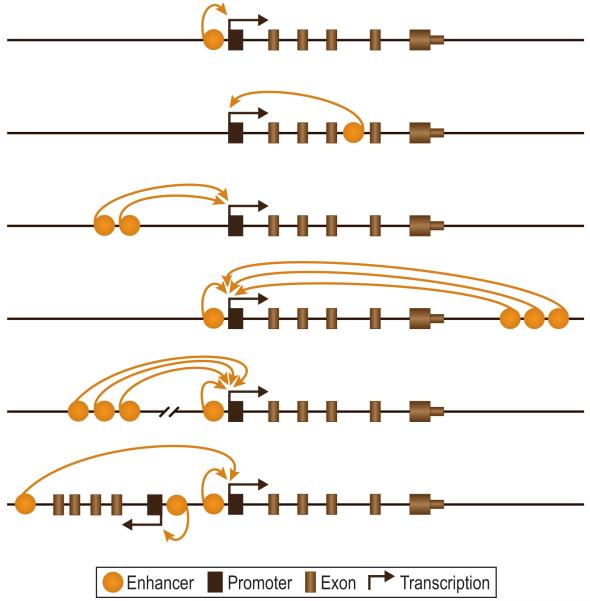
ChIP-chip and ChIP-seq assessment of VDR/RXR binding sites in the osteoblast genome coupled with nearest gene neighbor analysis indicates that while regulatory elements for this receptor pair can be found near the promoters for target genes, they are found more frequently distal to promoters in either intergenic regions that surround the transcription units or within introns (Figure 3). The data also suggest that genes are frequently regulated by multiple enhancers and that these elements are often located in clusters. This finding for both the VDR as well as other transcription factors is somewhat unexpected and clearly diverges from previous findings that suggest that the regulation of eukaryotic genes is controlled by enhancers located a few kilobases from gene promoters. Examples of genes that display these regulatory organizations are listed in the legend in Figure 3. It is now clear from this study of the VDR as well as studies of other transcription factors that multiple distal regulatory elements routinely control the expression of genes and that many of the earlier conclusions that were drawn using cellular transfection of plasmids were not correct. This modification to our understanding of gene regulation represents a significant paradigm shift and has broad implications.
Figure 3.
Possible locations of regulatory enhancers across mammalian genomes. Promoters, enhancers, exons, and the direction of transcription is shown. Examples of genes that display the indicated configuration from the top to the botton panel are as follow: 1) CYP24A1, 2) VDR, 3) TRPV6 and c-FOS, 4) CYP24A1, 5) RANKL, and 6) c-MYC and PAX6.
2.1.2. Measuring the Consequences of VDR/RXR Binding at Target Genes
ChIP-seq analysis has also revealed that VDR/RXR binding across the genome results in cofactor recruitment, changes in the levels of histone H3 and H4 lysine acetylation and changes in RNA polymerase II (RNA pol II) occupancy at promoters that are regulated by 1,25(OH)2D3. While the patterns of response across the genome are exceedingly complex, certain conclusions can be drawn regarding the function of enhancers and their role in modulating the output of genes in osteoblasts such as osteocalcin, osteopontin, alkaline phosphatase, bone sialoprotein, and others as well. For example, ligand-induced VDR/RXR occupancy at most sites across the genome is correlated with increased recruitment of coactivators such as SRC1, a histone acetyltransferase that functions to increase histone acetylation and to enhance transcription factor access via chromatin decondensation (Smith and O’Malley, 2004). Indeed, a striking increase in H3 and H4 acetylation is directly correlated with the appearance of the VDR/RXR heterodimer and coregulators such as SRC1. These changes occur at not only enhancers bound directly by the VDR, but other regions as well including promoters. The broad nature of this modification suggests the capacity of this covalent activity to spread across the gene locus (Hawkins et al., 2010a; Heintzman et al., 2009; Hon et al., 2009).
VDR binding at enhancers also correlates with the recruitment of coregulators such as the MED1 component of Mediator, a complex that functions, at least in part, to increase RNA pol II density at target gene promoters (Rachez et al., 1999; Yuan et al., 1998). Surprisingly, the recruitment of MED1 and RNA pol II as well as other components of the basal machinery are not limited to the promoters of regulated genes, but rather also occur at many of the enhancers to which the VDR/RXR is bound. Thus, it is possible that in addition to their roles in providing increased access to key chromatin regions, enhancers may also function as RNA pol II recruitment centers, thereby increasing the concentration of the primary enzyme complex required for transcript production (Szutorisz et al., 2005). While this hypothesis remains currently viable, the presence of these components at enhancers together with the discovery that much of the genome is actively transcribing RNA has prompted examination of the production of RNA from these regions as well (Wang et al., 2008). The results suggest that indeed these enhancer regions are frequently producing RNA transcript of several types. Although the function of most of these RNAs is not clear, it is possible that active transcription may play a role in the gene-regulating activities of enhancers. As a result of these unbiased genome-wide studies, however, it is now certain that new paradigms are emerging to describe the regulation of gene expression by 1,25(OH)2D3. These include the selective binding of the VDR and its RXR partner at multiple sites across the genome that are frequently located many kilobases for target genes and that are capable of modulating chromatin architecture, and the recruitment of RNA pol II to these regions where they may function to synthesize unexpected RNA transcripts capable of also modulating target gene transcription. Possible mechanisms through which RNA transcripts might regulate gene expression have been recently reviewed (Ong and Corces, 2011). Most of these concepts are a direct result of the application of the new technologies of ChIP-chip and ChIP-seq analysis.
2.2. Genome-wide Studies of VDR/RXR Heterodimer Binding in Intestinal/colonic Cells
ChIP-chip and ChIP-seq analysis have also been utilized to explore VDR/RXR binding sites across the human intestinal LS180 cell line (Meyer et al., 2011). This line is derived from a colorectal adenocarcinoma and therefore could also provide insight into mechanisms whereby 1,25(OH)2D3 inhibits uncontrolled cell growth. Genome-wide analysis of DNA binding sites for the VDR and RXR in the absence and presence of 1,25(OH)2D3 suggest that while the total number of sites is reduced, the patterns are similar to those observed in the mouse osteoblasts. Thus, while both VDR and RXR was found to be localized to a restricted set of targets in the absence of 1,25(OH)2D3, the overall number for both receptors was increased substantially in the ligand’s presence. As anticipated, RXR was present at most sites that containing the ligand-activated VDR. As with the VDR/RXR cistrome in osteoblasts, a nearest gene neighbor analysis indicated that the majority of the sites were located not at promoters, but rather at either distal intergenic regions upstream or downstream of transcription units or within introns. Finally, de novo motif finding analysis also revealed the presence of one or more classic VDREs within each of these VDR/RXR-bound activity peaks. Less than half of these de novo identified sites were comprised of two half-sites separated by 3 base pairs, however, suggesting that the VDR bindings to other configurations as well. These studies suggest that while the quantitative nature of the VDR cistrome differs as a result of cell type or species, the fundamental nature of both the cis and trans elements are similar.
Additional ChIP-seq analysis was also conducted in LS180 cells to identify the genome-wide distribution of coregulatory factors such as SRC1, CBP, Med1 and NCoR and to assess the impact of 1,25(OH)2D3 on this distribution. The results suggest that while these factors are bound to numerous regulatory sites across the genome in the basal state, as would be expected of general transcriptional regulators, the number of these sites increased substantially upon 1,25(OH)2D3 stimulation. Interestingly, tag density analysis suggested that sites that contained both the VDR/RXR heterodimer as well as one or more of the coregulators were highly correlated to genes that were shown to be regulated by 1,25(OH)2D3 as well. These include genes involved in calcium and phosphate transport, xenobiotic transport and metabolism, and other cellular functions. The identification of regulatory regions confirmed sites on genes previously identified and frequently defined new sites of action as well. These analyses support both the recruitment by the DNA-bound VDR/RXR heterodimer of coregulators and the capacity of these coregulators to influence the expression of genes located nearby.
Although the structures of gene enhancers are unknown, most studies have suggested that they are modular in nature, capable of selectively binding multiple transcription factors activated via numerous signaling pathways. This functional feature of enhancers enabled us to search for binding motifs within the enhancers which could bind the VDR/RXR heterodimer and to identify potential sites for other factors that might influence vitamin D action as well. This analysis revealed a series of motifs that were present with varying frequencies within the peaks of VDR/RXR binding activity and included potential sites for the factors AP1, ET1, C/EBPβ and CDX2. Indeed, subsequent ChIP-seq analysis of two of these regulators (CDX2 and C/EBPβ) demonstrated that they were frequently presence at the enhancers for 1,25(OH)2D3. C/EBPβ represents a remodeling transcription factor functionally active in the intestine whereas CDX2 represents a homeobox factor essential for embryonic development of the intestine, for stem cell maturation and for mature epithelial cell activity (Heinz et al., 2010; Lin et al., 2010; Suh and Traber, 1996). We speculate that these factors may be essential to the formation and regulation of VDR/RXR enhancer function.
2.3. Genome-wide Studies of VDR Binding in Epstein-Bar Virus B Cells
A recent ChIP-seq analysis of the VDR together with gene expression data has been reported in several EB-virus transformed lymphoblastoid cell lines (Ramagopalan et al., 2010). These cell lines have been used as analytical targets within the ENCODE Project and as a result, their genomes have been extensively explored and heavily annotated for digital DNAse1 hypersensitivity (DHS), DNA methylation, histone variants, histone methylation and acetylation marks, and for a multitude of transcription factor binding activities as well (Bernstein et al., 2010). Thus, the identification of both basal and ligand-induced VDR binding sites was placed in this valuable context. VDR binding properties were found to be similar to those identified in our studies. Thus, the VDR bound to 623 sites in the absence of 1,25(OH)2D3 and to 2776 sites following hormonal treatment. Among these sites, the authors confirmed our finding of the presence of the VDR at sites within the VDR gene (Zella et al., 2010; Zella et al., 2006), although they did not confirm previous findings of VDR binding at the ALOX5 gene (Seuter et al., 2007). The authors observed a much stronger bias for promoter localization of these sites irrespective of the state of activation. Consistent with this observation, VDR binding sites were enriched in regions that exhibited increased DNAse1 hypersensitivity and manifested higher levels of H3K4me3 and H3K427ac activity (histone signatures that are more frequently found at promoters and within transcription units) relative to H3K4 me1 activity (a histone signature found more frequently at distal enhancers) (Heintzman et al., 2007). Thus, these data support an association in these cell lines of the VDR with more promoter proximal rather than promoter distal sites of actions. Discounting VDR sites located near active promoters, however, the authors also determined that the median distance of a VDR enhancer to the nearest 1,25(OH)2D3 regulated gene was 66.6 kb, thereby supporting the idea that a significant number of VDR enhancers are located at sites that are remote relative to their target genes. Regardless of the location of these sites, however, further study using MEME analysis revealed the frequent presence of one or more consensus VDRE-like motifs comprised of the sequence AGGTCA XXG AGTTCA, thereby confirming the structural nature of typical VDREs. The frequency of the above half-site was somewhat higher than that observed in intestinal cells (Meyer et al.,2011). Many of the targets of vitamin D action, as defined through this particular study using both gene expression and ChIP-seq analyses, highlight both previous as well as potentially new targets of 1,25(OH)2D3 activity. Many of these target genes appeared to be involved in disease. It is worth noting, however, that most of these results are correlative in nature. Thus, the authors provide no evidence that the regulatory regions they define de novo are either active or are linked to the disease genes they find located nearby. Given our fundamental understanding that enhancers can be locate a many different sites across the genome, a direct functional link between an enhancer and a gene needs to be established for virtually all enhancers that are found using ChIP-seq analysis. Despite this caveat, this study provides valuable fundamental insight into mechanisms and potential targets of vitamin D action.
3. Functional Examination of Specific Gene Targets of 1,25(OH)2D3 Action
As referred to earlier, research over the past several decades has identified a number of genes that represent direct targets of 1,25(OH)2D3 and its receptor. These included human osteocalcin, the first target identified (Kerner et al., 1989), as well as subsequent targets that include osteopontin (SPP1), BGLAP, the calbindins D9K and D28K, Cyp24a1, Cyp27b1, PTH, and others (Haussler et al., 1998). Accordingly, both the location of the regulatory regions and the VDRE(s) within the regions were identified. While many of these analyses were successful in defining site of regulatory action, however, examination of others known also to be regulated by 1,25(OH)2D3 were not as successful. These failures suggested that either an alternative mechanisms was at play or that the regulatory elements were located elsewhere. The question of whether regulatory regions identified near the promoters of the genes examined provided the exclusive means by which 1,25(OH)2D3 functioned to modulate their expression was also unresolved. Clearly, the genome-wide analyses summarized above suggest that regulatory elements may be located far from the promoters they control and that both proximal and distal elements also may be involved. In the following sections, we describe our recent work on the Cyp24a1 and the Tnfsf11 (receptor activator of NF-κB ligand; Rankl) genes that highlight these principles.
3.1. Regulation of Cyp24a1 Expression by 1,25(OH)2D3
Cyp24a1 is a quintessential vitamin D target gene (Prosser and Jones, 2004). Its striking upregulation by 1,25(OH)2D3 in all cells that contain the VDR serves as a negative control mechanism to curb the activity of 1,25(OH)2D3 through direct metabolic degradation of the hormone. As a consequence, the ambient activity of 1,25(OH)2D3 in tissues is a direct consequence of the amount of 1,25(OH)2D3 that enters cells from the circulation, the amount of 1,25(OH)2D3 that is produced locally or within the cell, and the rate at which intracellular 1,25(OH)2D3 is degraded by Cyp24a1. Mechanistically, the cloning of the Cyp24a1 gene provided the opportunity to explore the underlying mechanism responsible for the regulation of this gene by 1,25(OH)2D3 (Ohyama et al., 1993; Ohyama et al., 1991). This effort led to the identification of two VDREs located within the first 300 bp of the promoter for Cyp24a1 (Ohyama et al., 1996; Zierold et al., 1995). Although subsequent work has provided additional insight into features of Cyp24a1 regulation by 1,25(OH)2D3 (Kim et al., 2005), the general conclusion from all of these studies has been that the Cyp24a1 gene is regulated through activities that occur at the gene’s proximal promoter.
3.1.1. Regulation of Cyp24a1 expression involves both the promoter and a downstream intergenic cluster of regulatory elements
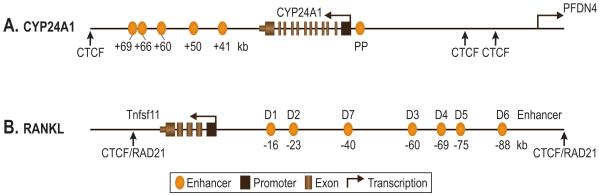
To test the hypothesis that the regulation of Cyp24a1 expression by 1,25(OH)2D3 was exclusively proximal in nature, we conducted unbiased ChIP-chip and ChIP-seq analyses of VDR and RXR binding across an extended segment of human chromosome 20 and mouse chromosome 2 which contained the Cyp24a1 genes using intestinal and bone osteoblast cell lines (Meyer et al., 2010b; Meyer et al., 2007). In both cell types, ChIP-chip analysis confirmed both the absence of VDR and RXR at the mouse and human Cyp24a1 proximal promoters under basal conditions and the co-localization of these factors to this region of the two genes in response to 1,25(OH)2D3, thereby confirming previous studies (Figure 4A). Surprisingly, however, both VDR and RXR occupancy was also strongly induced by 1,25(OH)2D3 at a cluster of intergenic sites located downstream of the Cyp24a1 gene some +35-40 kb in mouse and +40-66 kb in human cells. Interestingly, while VDR and RXR binding was dependent upon the presence of 1,25(OH)2D3 at almost all the sites, significant ligand-independent VDR and RXR binding was observed at one particular site at +50 kb and the level of the receptors were only upregulated (2-fold) following activation by the hormone. Further dissection revealed the presence of one or more functional VDREs in several of these enhancers.
Figure 4.
Locations of active regulatory enhancers for the (A) human CYP24a1 and (B) mouse Tnfsf11 (Rankl) genes. The individual transcription units and their associated exons and introns are shown with the arrow indicating the promoter and the direction of transcription. The positions of CTCF/RAD21 binding sites are designated with arrows. The Tnfsf11 enhancers D1-D7 and their positions relative to the gene’s transcriptional start site (TSS) are documented. The TSS, promoter proximal and downstream enhancer cluster for the CYP24A1 gene are indicated.
To explore this downstream cluster of VDR/RXR binding sites further, we conducted ChIP-chip analysis using antibodies to the coregulators SRC1, MED1, NCoR and SMRT. These studies revealed that SRC1, MED1 and SMRT, but not NCoR, were recruited in a ligand-dependent manner not only to the proximal promoter, but to most of the downstream regulatory regions as well. Interestingly, measureable levels of all of the cofactors, including NCoR were evident under basal as well as ligand-induced conditions at the +50 kb site that contained prebound VDR and RXR. We also observed that while histone H4 acetylation levels across the Cyp24a1 locus were unremarkable under basal conditions, a striking increase occurred at both the promoter and across the entire downstream enhancer regions from +40 to +69 kb as a result of 1,25(OH)2D3. These findings suggest that VDR/RXR binding at the promoter and at this downstream cluster is coincident with coregulator recruitment and chromatin modification, both of which appear necessary for the regulation of Cyp24a1 expression by 1,25(OH)2D3.
3.1.2. Linking the Activity of the Downstream Enhancer Cluster to Cyp24a1 Expression
While the above studies suggest that downstream elements may participate in the regulation of Cyp24a1 expression, they do not provide direct evidence of such activity. As a consequence, we created a series of large mouse and human bacterial artificial chromosome (BAC) minigene reporters that contained not only the Cyp24a1 transcription unit but associated regulatory regions as well. This BAC clone collection was comprised of not only an unmodified version of the locus, but altered versions that contained mutations which blocked activity at the proximal Cyp24a1 promoters, mutations that removed the downstream cluster of putative regulatory enhancers, or the combination. These BAC clones were then utilized to create cell lines that contained stably integrated copies of these minigenes and the cell lines explored for basal and 1,25(OH)2D3 inducible reporter activity. While the native Cyp24a1 BAC clones were fully activated by 1,25(OH)2D3, independent mutation of the proximal VDREs or deletion of the downstream enhancer cluster reduced but did not eliminate the ability of 1,25(OH)2D3 to induce Cyp24a1 reporter function. However, when both the proximal mutations and the downstream deletion were combined within the Cyp24a1 BAC clone, all 1,25(OH)2D3 inducible activity was lost (Meyer et al., 2010b). These results provide clear evidence that both the proximal VDREs defined in the mid 90’s as well as the downstream regulatory elements identified in this study contribute to the regulation of Cyp24a1 expression by 1,25(OH)2D3. Studies using chromosome conformation capture (3C) analysis, which provides a measure of the relative proximity of two discontinuous DNA segments (Dekker et al., 2002), also revealed that the downstream DNA segment that contained the VDR enhancer cluster was indeed located near the Cyp24a1 promoter in three dimensional terms (Meyer et al., 2010b), providing additional support for this emerging feature of the Cyp24a1 gene. These findings highlight the increased complexity of gene regulation at a gene that has served as a prototypic vitamin D target for almost three decades.
3.2. Regulation of Tnfsf11 Gene Expression by 1,25(OH)2D3
Early studies by Suda and colleagues suggested that 1,25(OH)2D3 and PTH could induce the formation of osteoclasts through an indirect mechanisms that involved the regulation of a factor produced in osteoblasts (Suda et al., 1992; Suda et al., 1999). This factor was identified in 1999 as Rankl, a membrane-bound TNF-like molecule that not only induced the formation of osteoclasts, but stimulated their activation and survival as well (Burgess et al., 1999; Kong et al., 1999; Lacey et al., 1998). Currently, Rankl is known to be expression in not only osteoblasts but in other cell types as well and plays an essential role in bone remodeling; its upregulation following menopause as well as in a variety of disease state is central to bone loss and osteoporosis (Leibbrandt and Penninger, 2008; Theill et al., 2002). Despite this insight, early attempts to identify a mechanism whereby 1,25(OH)2D3 could induce Rankl (Tnfsf11) gene expression were less successful. Thus, while Kitazawa and colleagues demonstrated that the proximal regions of both the mouse and human Rankl gene promoters contained elements that mediated 1,25(OH)2D3 response, these elements diverged to some extent from typical VDREs and where capable of only modest activity in response to 1,25(OH)2D3 (Kitazawa et al., 2008; Kitazawa et al., 2003) Efforts by others to replicate these responses met with limited success (Aguilera et al., 2007; O’Brien, 2010).
3.2.1 Rankl Gene Expression is Regulated by 6 Upstream Distal Enhancers
Given this above background, we initiated an effort to determine whether regulation of Rankl was indeed mediated by 1,25(OH)2D3 through the promoter or whether it involved regulatory elements located elsewhere within the gene locus. To this end, we conducted a ChIP-chip analysis of 1,25(OH)2D3 induced VDR and RXR binding that centered on the Rankl gene yet spanned over 500 kb of DNA surrounding the gene itself (Kim et al., 2006; Nerenz et al., 2008). This analysis revealed the presence of at least 5 intergenic sites to which the VDR/RXR heterodimer bound following 1,25(OH)2D3-treatment that were located upstream of the Rankl promoter at −16, −23, −60, −69 and −75/76 kb (Figure 4B). In this analysis, however, neither VDR nor RXR were observed at the Rankl promoter. Further studies identified the VDREs located within several of these regions, and a particularly robust action mediated by the regulatory region located at −75 kb. These results suggest the possibility that the induction of Rankl by 1,25(OH)2D3 in this cell type was mediated not through proximal elements, but rather through a complex set of regulatory element located far upstream. O’Brien and colleagues, using an alternative approach involving large BAC clones as described above, arrived at a similar conclusion that the distal region located at −75/76 kb is capable of mediating the actions of both 1,25(OH)2D3 and PTH (Fu et al., 2006). The dissection of this region revealed the presence of several CREB sites responsible for the actions of PTH as well (Kim et al., 2007). Further studies demonstrated that both 1,25(OH)2D3 and PTH induced not only VDR/RXR and CREB binding, respectively, at these sites, but also induced the recruitment of several coregulatory complexes and an upregulation of histone H4 acetylation as observed at the Cyp24a1 gene locus. An additional enhancer at −88 kb was also identified more recently that mediates the induction of the Rankl gene by the cytokines OSM and IL-6 via the transcription factor STAT3 (Bishop et al., 2009). These studies have been instrumental in defining how the Rankl gene is regulated by 1,25(OH)2D3 and other inducers such as PTH and highlight the frequent role of multiple elements located distal to promoters in the regulation of gene expression.
3.2.2 Validation of the Role of Rankl Gene Enhancers in 1,25(OH)2D3 and PTH Action in Vivo
As with the Cyp24a1 gene, linking the activities of enhancers located far upstream of Rankl to the expression of the gene itself requires direct evidence. While the use of natural and mutant BAC clones provided preliminary evidence of this linkage, the apparent central role of the enhancer located at −75/−76 kb prompted O’Brien and colleagues to delete this segment of DNA in the mouse genome and to explore the consequence of this maneuver on the basal as well as 1,25(OH)2D3 and PTH induced expression of Rankl in vivo (Galli et al., 2008). Basal levels of Rankl expression were found to be suppressed in this mouse not only in bone but in the spleen as well suggesting that the −75/−76 kb enhancer was involved in basal Rankl expression in both bone and T cells. Perhaps more interestingly, treatment of these enhancer knockout mice with either 1,25(OH)2D3 or PTH revealed a defect in regulation such that neither of the two hormones were as effective at inducing Rankl as was observed in wildtype mice. Further examination of the biologic phenotype of these enhancer-deleted mice suggests that these mice developed mild osteopetrosis as a result of the decrease in Rankl expression. Collectively, the studies support the idea that at least one enhancer located a significant distance for the Rankl gene promoter is fundamental to the regulation of the Rankl gene. Additional deletion studies are ongoing to further define the roles of each of the other enhancers that were identified in the Rankl upstream control region.
4. Final Conclusions and Future Perspectives
The observations summarized in this article provide new insight into the mechanisms through which 1,25(OH)2D3 and its receptor regulate the expression of vitamin D target genes. Although studies over the past several decades have defined a number of general guiding principles of vitamin D action, including definition of key components in addition to the VDR/RXR heterodimer and identification of the structural organization of the VDREs themselves, more recent studies using newly devised technologies capable of providing a genome-wide perspective suggest that many of these principles require significant modification. Among those is the idea that regulatory regions are located near promoters. Clearly, the evidence presented above suggests that although genes do contain these proximal elements, they are more frequently located distal to transcriptional start sites and often configured in regulatory clusters. This distal nature makes it imperative that enhancers identified through ChIP-chip and ChIP-seq analysis be evaluated directly for their functional contribution to the regulation of specific target genes using genome-deletion in vivo, recombineered BAC clones, chromosome conformation capture or other types of analyses as discussed above. Indeed, this may be most difficult consequence of the results provided by these powerful genomic analyses.
Future studies are likely to focus on the mechanisms whereby enhancers regulate gene expression. Thus, while binding of regulatory factors initiate coregulator recruitment and provoke epigenetic changes to the covalent states of histones, the mechanism(s) through which enhancers function to modify the expression of target genes remains unclear. An emerging concept is that enhancers may direct the synthesis of RNA transcripts that may participate in some way in the regulation of target genes, as discussed earlier in this article (Wang et al., 2008). Indeed, considerable evidence now suggests that at least some of these RNAs may function in a regulatory mode. Additional efforts are also likely to focus on the structural composition of enhancers and the identification of factors that are involved in the creation of these regulatory regions as well (Lin et al., 2010). Clearly, the phenotype of a cell is determined by the genes which are expressed, and the ability of these genes to be expressed in one cell type and not another is determined by the presence of appropriately placed yet dynamically regulated enhancers. Focused studies are just beginning to delineate the role of lineage-determining factors in the creation and maintenance of enhancers that are essential to the expression of cell specific gene networks. These and additional studies are likely to provide new insight into the cell- and tissue-selective actions of the vitamin D hormone.
Acknowledgments
The author gratefully acknowledges the contributions of each of the members of the Pike laboratory to the work discussed and the artistic skills of Laura Vanderploeg to the figures presented.
This work was supported by National Institute of Heath Grants DK-072281, DK-073995, DK-074993 and AR-045173
Footnotes
Disclosure Statement: The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera O, Peña C, García JM, Larriba MJ, Ordóñez-Morán P, Navarro D, Barbáchano A, López de Silanes I, Ballestar E, Fraga MF, et al. The Wnt antagonist DICKKOPF-1 gene is induced by 1alpha,25-dihydroxyvitamin D3 associated to the differentiation of human colon cancer cells. Carcinogenesis. 2007;28:1877–1884. doi: 10.1093/carcin/bgm094. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKl gene expression. Mol Endocrinol. 2009;23:2095–2110. doi: 10.1210/me.2009-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillon R, Bischoff-Ferrari H, Willett W. Vitamin D and health: perspectives from mice and man. J Bone Miner Res. 2008a;23:974–979. doi: 10.1359/jbmr.080420. [DOI] [PubMed] [Google Scholar]
- Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, Lieben L, Mathieu C, Demay M. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008b;29:726–776. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. Vitamin D and cancer. J Steroid Biochem Mol Biol. 2006;102:156–162. doi: 10.1016/j.jsbmb.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Burgess T, Qian Y, Kaufman S, Ring B, Van G, Capparelli C, Kelley M, Hsu H, Boyle W, Dunstan C, et al. The ligand for osteoprotegerin (OPGL) directly activates mature osteoclasts. J Cell Biol. 1999;145:527–538. doi: 10.1083/jcb.145.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26:6453–6468. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C, Zella LA, Fretz JA, Fu Q, Pike JW, Weinstein RS, Manolagas SC, O’Brien CA. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Haussler CA, Bartik L, Whitfield GK, Hsieh JC, Slater S, Jurutka PW. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutr Rev. 2008;66:S98–112. doi: 10.1111/j.1753-4887.2008.00093.x. [DOI] [PubMed] [Google Scholar]
- Haussler MR, Haussler CA, Whitfield GK, Hsieh JC, Thompson PD, Barthel TK, Bartik L, Egan JB, Wu Y, Kubicek JL, et al. The nuclear vitamin D receptor controls the expression of genes encoding factors which feed the “Fountain of Youth” to mediate healthful aging. J Steroid Biochem Mol Biol. 2010;121:88–97. doi: 10.1016/j.jsbmb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13:325–349. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010a;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. In Nat Rev Genet. 2010b:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, Cheng J, Murre C, Singh H, Glass C. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Fujiki R, Kim MS, Kitagawa H. Ligand-induced transrepressive function of VDR requires a chromatin remodeling complex, WINAC. J Steroid Biochem Mol Biol. 2007;103:372–380. doi: 10.1016/j.jsbmb.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Kerner SA, Scott RA, Pike JW. Sequence elements in the human osteocalcin gene confer basal activation and inducible response to hormonal vitamin D3. Proc Natl Acad Sci U S A. 1989;86:4455–4459. doi: 10.1073/pnas.86.12.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kondo T, Takada I, Youn M, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde NK, Pike JW. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol. 2007;21:197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa R, Mori K, Yamaguchi A, Kondo T, Kitazawa S. Modulation of mouse RANKL gene expression by Runx2 and vitamin D3. J Cell Biochem. 2008;105:1289–1297. doi: 10.1002/jcb.21929. [DOI] [PubMed] [Google Scholar]
- Kitazawa S, Kajimoto K, Kondo T, Kitazawa R. Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J Cell Biochem. 2003;89:771–777. doi: 10.1002/jcb.10567. [DOI] [PubMed] [Google Scholar]
- Komashko VM, Acevedo LG, Squazzo SL, Iyengar SS, Rabinovich A, O’Geen H, Green R, Farnham PJ. Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 2008;18:521–532. doi: 10.1101/gr.074609.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- Lefterova M, Zhang Y, Steger D, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Liu X, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibbrandt A, Penninger JM. RANK/RANKL: regulators of immune responses and bone physiology. Ann N Y Acad Sci. 2008;1143:123–150. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Matthews D, LaPorta E, Zinser GM, Narvaez CJ, Welsh J. Genomic vitamin D signaling in breast cancer: Insights from animal models and human cells. J Steroid Biochem Mol Biol. 2010;121:362–367. doi: 10.1016/j.jsbmb.2010.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch P, Pike J. Overlapping VDR/RXR and TCF4/b-catenin cistromes in colorectal cancer cells: Impact on c-FOS and c-MYC regulation. submitted to Genome Sciences. 2011 [Google Scholar]
- Meyer M, Goetsch P, Pike J. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J Steroid Biochem Mol Biol. 2010a;121:136–141. doi: 10.1016/j.jsbmb.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 Expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010b;285:15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Zella LA, Nerenz RD, Pike JW. Characterizing early events associated with the activation of target genes by 1,25-dihydroxyvitamin D3 in mouse kidney and intestine in vivo. J Biol Chem. 2007;282:22344–22352. doi: 10.1074/jbc.M703475200. [DOI] [PubMed] [Google Scholar]
- Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2008;22:1044–1056. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA. Control of RANKL gene expression. Bone. 2010;46:911–919. doi: 10.1016/j.bone.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama Y, Noshiro M, Eggertsen G, Gotoh O, Kato Y, Björkhem I, Okuda K. Structural characterization of the gene encoding rat 25-hydroxyvitamin D3 24-hydroxylase. Biochemistry. 1993;32:76–82. doi: 10.1021/bi00052a011. [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Noshiro M, Okuda K. Cloning and expression of cDNA encoding 25-hydroxyvitamin D3 24-hydroxylase. FEBS Lett. 1991;278:195–198. doi: 10.1016/0014-5793(91)80115-j. [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Ozono K, Uchida M, Yoshimura M, Shinki T, Suda T, Yamamoto O. Functional Assessment of Two Vitamin D-responsive Elements in the Rat 25-Hydroxyvitamin D3 24-Hydroxylase Gene. J Biol Chem. 1996;271:30381–30385. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono K, Liao J, Kerner SA, Scott RA, Pike JW. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J Biol Chem. 1990;265:21881–21888. [PubMed] [Google Scholar]
- Pike JW. Genome-scale technique highlight the epigenome and redefine fundamental principles of gene regulation. Journal of Bone and Mineral Reseach. 2011 doi: 10.1002/jbmr.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3. Endocrinol Metab Clin North Am. 2010;39:255–269. doi: 10.1016/j.ecl.2010.02.007. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Meyer MB, Martowicz ML, Bishop KA, Lee SM, Nerenz RD, Goetsch PD. Emerging regulatory paradigms for control of gene expression by 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2010;121:130–135. doi: 10.1016/j.jsbmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum LA, DeLuca HF. Vitamin D, disease and therapeutic opportunities. Nat Rev Drug Discov. 2010;9:941–955. doi: 10.1038/nrd3318. [DOI] [PubMed] [Google Scholar]
- Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, Handunnetthi L, Handel AE, Disanto G, Orton SM, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuter S, Väisänen S, Rådmark O, Carlberg C, Steinhilber D. Functional characterization of vitamin D responding regions in the human 5-Lipoxygenase gene. Biochim Biophys Acta. 2007;1771:864–872. doi: 10.1016/j.bbalip.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Sone T, Kerner S, Pike JW. Vitamin D receptor interaction with specific DNA. Association as a 1,25-dihydroxyvitamin D3-modulated heterodimer. J Biol Chem. 1991a;266:23296–23305. [PubMed] [Google Scholar]
- Sone T, Ozono K, Pike JW. A 55-kilodalton accessory factor facilitates vitamin D receptor DNA binding. Mol Endocrinol. 1991b;5:1578–1586. doi: 10.1210/mend-5-11-1578. [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Martin TJ. Modulation of osteoclast differentiation. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie M, Martin T. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- Suh E, Traber P. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton AL, MacDonald PN. Vitamin D: more than a “bone-a-fide” hormone. Mol Endocrinol. 2003;17:777–791. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N, Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Theill LE, Boyle WJ, Penninger JM. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev Immunol. 2002;20:795–823. doi: 10.1146/annurev.immunol.20.100301.064753. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Yuan C, Ito M, Fondell J, Fu Z, Roeder R. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zella L, Meyer M, Nerenz R, Lee S, Martowicz M, Pike J. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 2010;24:128–147. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zella LA, Kim S, Shevde NK, Pike JW. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2006;20:1231–1247. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- Zierold C, Darwish HM, DeLuca HF. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J Biol Chem. 1995;270:1675–1678. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]
- Zinser G, Welsh J. Effect of Vitamin D3 receptor ablation on murine mammary gland development and tumorigenesis. J Steroid Biochem Mol Biol. 2004;89-90:433–436. doi: 10.1016/j.jsbmb.2004.03.012. [DOI] [PubMed] [Google Scholar]