Abstract
All-trans-retinoic acid (atRA) provides essential support to diverse biological systems and physiological processes. Epithelial differentiation and its relationship to cancer and embryogenesis have typified intense areas of interest into atRA function. Recently, however, interest in atRA action in the nervous system, the immune system, energy balance and obesity has increased considerably, especially concerning postnatal function. atRA action depends on atRA biosynthesis: defects in retinoid-dependent processes increasingly relate to defects in atRA biogenesis. Considerable evidence indicates that physiological atRA biosynthesis occurs via a regulated process, consisting of a complex interaction of retinoid binding-proteins and retinoid recognizing enzymes. An accrual of biochemical, physiological and genetic data have identified specific functional outcomes for the retinol dehydrogenases, RDH1, RDH10, and DHRS9, as physiological catalysts of the first step in atRA biosynthesis, and for the retinal dehydrogenases RALDH1, RALDH2, and RALDH3, as catalysts of the second and irreversible step. Each of these enzymes associates with explicit biological processes mediated by atRA. Redundancy occurs, but seems limited. Cumulative data supports a model of interactions among these enzymes with retinoid binding-proteins, with feedback regulation and/or control by atRA via modulating gene expression of multiple participants. The ratio apo-CRBP1/holo-CRBP1 participates by influencing retinol flux into and out of storage as retinyl esters, thereby modulating substrate to support atRA biosynthesis. atRA biosynthesis requires presence of both an RDH and an RALDH: conversely, absence of one isozyme of either step does not indicate lack of atRA biosynthesis at the site.
Keywords: retinol, retinoic acid, aldehyde dehydrogenase, cellular retinoic acid binding-protein, cellular retinol binding-protein, lecithin:retinol acyltransferase, retinol dehydrogenase, retinyl ester, retinal reductase, short-chain dehydrogenase/reductase
1. Introduction
Multiple functions of all-trans-retinoic acid (atRA) in diverse physiological systems have engendered interest beyond the traditional retinoid attention areas of cancer and development. Interest in atRA action has increased considerably concerning the nervous system, the immune system, energy balance and obesity, to name a few. Curiosity also has increased in the physiological biogeneration of atRA, because atRA biosynthesis contributes to regulating atRA action, and defects in atRA biosynthesis likely contribute to defects in retinoid action. Considerable evidence indicates that physiological atRA biosynthesis occurs via a regulated process, consisting of a complex interaction of retinoid binding-proteins and retinoid recognizing enzymes, quite distinct from the unregulated production of atRA pursuant to an excess of retinol, which is rarely observed in nature. Although vitamin A intake appears adequate in most (but not all) populations of developed countries, this likely represents an evolutionarily aberrant occurrence. As indicated by widespread deficiency in developing or underdeveloped countries, limited vitamin A intake seems the norm, resulting from nutritionally poor diets [1]. Thus, atRA generation must be considered in the context of the physiological forms of its precursors, retinol bound with cellular retinol binding-proteins, and the struggle to obtain and conserve an essential nutrient. The retinoid binding-proteins likely have multiple functions, including aiding in cellular uptake of retinol and protecting retinol from metabolism by xenobiotic clearing enzymes. In fact, at least two examples of retinoid knockouts are known that produced no phenotype when mice were fed a chow diet (contains copious vitamin A), but showed a distinctive retinoid-related phenotype when dietary vitamin A was lowered: CRBP2 and RDH1 [2, 3]. This indicates that these proteins evolved (in part) to help animals cope with restricted dietary vitamin A by maximizing its uptake and use, and shows that diets copious in vitamin A or pharmacological dosing with retinol eliminates a primary purpose for expressing these proteins. This review will focus on the biochemistry of atRA biosynthesis under physiological conditions, the evidence for involvement of retinoid binding-proteins in retinol metabolism, and the evidence for participation of specific enzymes. The data will be presented in context of the entire atRA generating “metabolon” and will consider regulatory nodes in an effort to present a model that integrates the functions of retinoid binding-proteins and enzymes that catalyze retinoid metabolism with their biochemistry and physiological functions.
2. An integrative model of physiological atRA homeostasis
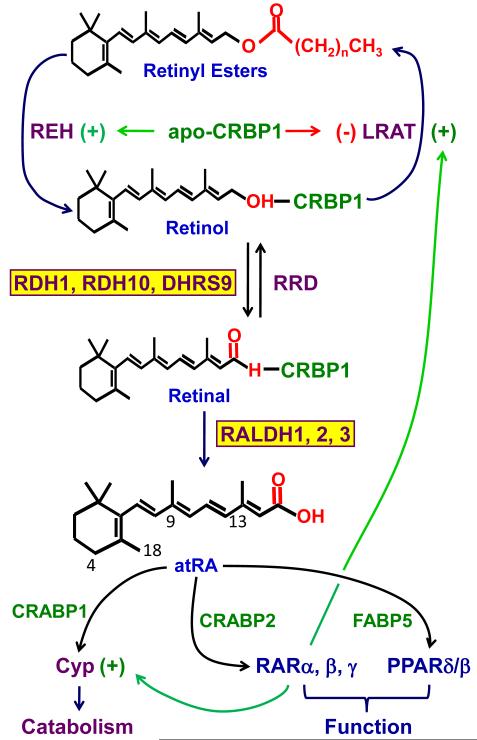
A problem with studying retinoid metabolism is the promiscuity and lability of retinoids in vitro, as well as the promiscuity of xenobiotic clearing enzymes, and even many enzymes assumed to be specific. Simply because a known enzyme has activity with retinoids in vitro does not indicate whether it has access in vivo. The overview in Figure 1 formulates a model developed largely via “de novo” approaches to delineate atRA homeostasis, i.e. by searching tissues for retinoid metabolizing enzymes with physiological amounts and forms of substrate.
Figure 1. An integrated model of atRA homeostasis.
This model accounts for the fact that the very high-affinity retinol binding-protein, holo-CRBP1, represents the major physiological form of intracellular retinol, and apo-CRBP1 interacts with enzymes to signal the intracellular concentration of retinol. The model presents the major enzymes that catalyze the rate-limiting step, retinol dehydrogenation, and the irreversible step, retinal dehydrogenation. These enzymes all have been validated biochemically, physiologically, and genetically, as contributing to atRA biosynthesis under physiological conditions. The model also postulates functions for the atRA binding proteins, CRABP1, CRABP2, and FABP5 in delivering atRA to distinct nuclear receptors or for catabolism. Finally, two of the autoregulatory nodes activated by atRA are shown, induction of CYP and LRAT to lower atRA and retinol concentrations, respectively.
3. Retinol homeostasis
Liver stores the largest amount of vitamin A, delivered on chylomicron remnants, as retinyl esters (RE), and releases retinol bound with the serum retinol binding-protein, RBP (encoded by Rbp4) into circulation [4-8]. RBP interacts with the extra-hepatic plasma membrane receptor STRA6, which mediates transfer of retinol into cells [9]. STRA6 enhances LRAT activity by delivering substrate [10]. The converse also is true: net transfer mediated by STRA6 occurs only if cells express either CRBP1 (encoded by Rbp1), and/or LRAT2. This insight complements observation of coupled retinol uptake and esterification, and of holo-CRBP1 (retinol bound with CRBP1) serving as a chaperone for retinol esterification [11-13]. This perspective also reflects holo-CRBP1 occurring as the most abundant form of intracellular non-esterified retinol, despite capacity for membranes to sequester far more retinol than occurs in cells during normal vitamin A nutriture [14]. The total CRBP1 concentration remains steady with varying vitamin A status, observed when feeding rats diets devoid of vitamin A vs. feeding diets containing ~2.5-fold more than the American Institute of Nutrition (AIN) recommended concentration for rodents (equivalent to 1.2 μg retinol = 4 IU/g diet) [15, 16]. The retinol concentration also remains within limited bounds with changes in dietary vitamin A, from a diet containing the 4 IU vitamin A/g diet recommended by the AIN, up to a single dosing with a copious amount of retinol (~20 mg/kg, >66,000 IU). In contrast, the amount of RE increases substantially with increasing dietary vitamin A [17]. The mouse recapitulates this phenomenon. As dietary vitamin A increases from marginal (0.6 IU vitamin A/g diet) to the AIN recommended amount, the liver retinol concentration increases 2-fold, but RE increase 6-fold [3]. Accumulation of RE in liver and other tissues allows for retinol storage in an innocuous form in lipid droplets, and for repetitive use of CRBP1, maximizing retinol uptake and storage, while minimizing need for expressing larger concentrations of CRBP1 (Figure 1).
Kinetic analysis reveals that LRAT (in its native association with microsomal membranes) accesses retinol bound to CRBP1 [13, 18]. RE formation in vitro occurs in the presence of holo-CRBP1 at a rate faster than allowed by the low concentration of “free” retinol in solution that would equilibrate with holo-CRBP1, assuming any substantial retinol actually can occur in a aqueous solution. The latter is highly unlikely—retinol solubility in aqueous medium does not exceed ~60 nM in the absence of CRBP1 [19]. In the presence of μM holo-CRBP1 with CRBP1 in excess, the law of mass action predicts nM retinol concentrations. The rate of RE formation in the presence of holo-CRBP1 very much exceeds the rate that would be supported if unbound retinol were the only substrate, and the relationship between holo-CRBP1 and RE formation displays Michaelis-Menten kinetics.
Although LRAT catalyzes RE biosynthesis in most tissues, an alternative activity, ARAT, occurs in some tissues, notably in the mammary gland and skin [20-23]. In skin, the enzyme DGAT1 (diacylglyceracyltransferase) accounts for ARAT activity [24]. The physiological advantage of requiring a second retinol esterifying enzyme has not been determined.
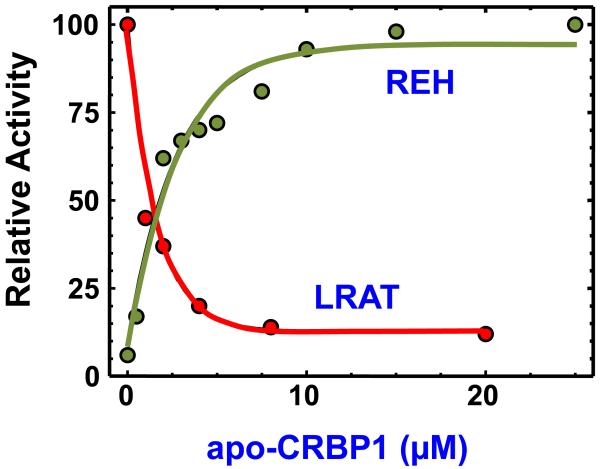
Stored RE require mobilization into retinol to support vitamin A function. Several classes of membrane-associated lipases convert RE into retinol, and each may be specific for a specific phase of vitamin A distribution, such as hydrolysis of chylomicron or chylomicron remnant-containing RE or endosomal packaged RE [25, 26]. A bile-salt independent neutral REH seems physiologically relevant for generating retinol from intracellular stores [27-31]. This deduction is supported by the microsomal localization of the hydrolase, i.e. intracellular, and the ability of apo-CRBP1 to stimulate hydrolysis of endogenous microsomal RE via the bile salt independent REH [32]. Alternative proteins that can bind retinol, such as albumin and β-lactoglobulin, and another member of the FABP gene family that binds atRA but not retinol, CRABP1, do not have the same effect. Thus, RE hydrolysis reacts to apo-CRBP1 specifically. Stimulation of RE hydrolysis by apo-CRBP1 conforms to Michaelis-Menten kinetics: it is saturable and 2.6 μM apo-CRBP1, a concentration in the range of the physiological concentration in liver, produces half-maximal stimulation (Figure 2).
Figure 2. Concentration effects of apo-CRBP1 in the stimulation of retinyl ester hydrolase (REH) and inhibition of RE biosynthesis.
Apo-CRBP1 (apoCRBP) activates hydrolysis of endogenous RE in microsomes by a cholate-independent REH [32]. Half-maximum stimulation occurs ~2.6 μM. apo-CRBP1 also serves as a potent competitive inhibitor of LRAT, with 50% inhibition occurring ~2 μM [33]. Thus, the ratio apo-CRBP1/holo-CRBP1 would serve as a signal of intracellular retinol status to influence the relative rates of RE biosynthesis vs. RE hydrolysis.
In contrast to stimulation of RE hydrolysis, apo-CRBP1 inhibits retinol esterification by LRAT with a inhibitory constant of ~0.2 μM [33]. This suggests that the ratio holo-CRBP1/apo-CRBP1 serves as a signal of retinol status, which directs flux of retinol into and out of storage as RE, while maintaining a steady-state concentration of holo-CRBP1 to support atRA biosynthesis. As mentioned above, RE represent the major disposition of retinol upon entering the liver. As the cell accumulates retinol, the RE concentration increases far more than the retinol concentration. Ability of apo-CRBP1 to inhibit LRAT activity would prevent all retinol from converting (or re-converting) into RE. RE production would continue as long as retinol continued to enter the liver. But as retinol presentation diminishes, LRAT action would deplete holo-CRBP1 of retinol, generating apo-CRBP1 and limiting the rate of esterification, while prompting mobilization of RE. This seems an elegant mechanism to insure that not all retinol ends up as RE, to provide substrate for atRA biosynthesis, and to prevent futile cycling of retinol back and forth into and out of RE. Presumably, the ratio apo-CRBP1/holo-CRBP1 functions extrahepatically, as it seems to function in liver. Retinol concentrations in mouse tissues other than liver range from 0.2 to 0.6 μM, whereas RE concentrations range from 0.2 to 2 μM, in mice fed the recommended amounts of vitamin A [34]. As a rule of thumb, RE concentrations exceed those of retinol in these tissues, but the differences are not as large as in liver.
4. CRBP1 as a chaperone
Although affinity of CRBP1 for retinol had been estimated as ~16 nM, this value has been re-determined as 3 nM, by availability of a more sensitive fluorometer [35-37]. Both values likely represent an upper limit rather than a true value, because fluorometer sensitivity has limited the generation of apparent kd values for retinoid binding-proteins. Estimates of the true value approach 0.1 nM, based on competition experiments, rather than fluorescence quenching [38]. Sequestration of retinol inside a high-affinity binding-protein draws retinol into the cell and protects it from unfettered metabolism, but clearly allows RE formation [36, 39]. Tight binding and a direct path of retinol from RBP through STRA6 via CRBP1 and LRAT to form RE would seem to preclude atRA formation. On the other hand, a chaperone might have limited function, if retinol could easily escape its influence. The ability of holo-CRBP1 to serve as substrate for retinol metabolism addresses these issues. This concept has sometimes been misunderstood as suggesting that select enzymes require holo-CRBP1 to function, or “engulf” holo-CRBP1 as a substrate, rather than the existence of a microenvironment in which holo-CRBP1 interacts with enzymes in context of their membranes (LRAT and RDH). This would allow transfer of retinol from CRBP1 to select enzymes without requiring diffusion throughout the cell. In other words, select enzymes participate in a microenvironment with holo-CRBP1 and can “seize” retinol as CRBP1 undergoes the dynamic motions that partially expose but don’t release retinol. Put another way, the chaperone model postulates that CRBP1 safeguards efficient retinol metabolism; not that retinol metabolism has an absolute requirement for CRBP1. Reactions that occur in the presence of a chaperone, also may occur in the absence of a chaperone. In the absence of a chaperone, however, additional reactions may occur, which normally are restricted by the chaperone. Evidence for this mechanism has been presented in detail [40, 41].
CRBP1-null mice reproduce normally, show no obvious morphological abnormalities, and show no obvious signs of gross atRA deficiency [42]. Hepatic stellate cells in CRBP1-null mice, however, have fewer and smaller lipid droplets than WT after 4 weeks of age. Liver has ~50% lower liver retinol and RE concentrations, when fed a chow diet (i.e. copious vitamin A). Low liver RE levels reflect (in part?) a 6-fold faster elimination rate. atRA concentrations in serum and several target tissues do not differ significantly between CRBP1-null and WT mice, explaining lack of obvious gross abnormalities [43, 44]. These observations are consistent with CRBP1 functioning as a chaperone that conserves retinol by restricting access to retinol. In the absence of CRBP1, RDH would still convert retinol into retinal for atRA biosynthesis, but additional enzymes also would have access to retinol, prompting the drain on RE. Whether lack of CRBP1 also impairs RE biosynthesis has not been tested. Remarkably, thorough analyses of RE, retinol, retinal and atRA concentrations across the spectrum of retinoid target tissues have not been reported in the CRBP1-null mouse. Physiological effects of CRBP1 remain to be elucidated fully, especially concerning more subtle and/or long term effects on retinoid-supported processes.
5. Retinol dehydrogenases
The first of two dehydrogenations in the path of atRA biosynthesis, the conversion of retinol into retinal, is rate-limiting (lowest Vm values) in the presence of CRBP1 [45, 46]. Similar to RE formation, the rate of retinal biosynthesis from holo-CRBP1, catalyzed by microsomal members of the short-chain dehydrogenase/reductase (SDR) gene family, exceeds that allowed by the amount of free retinol in equilibrium with holo-CRBP1; and a Michaelis-Menten relationship occurs between holo-CRBP1 and microsome-catalyzed retinal biosynthesis [47]. Varying the ratio apo-CRBP1/ holo-CRBP1 (within limits), which changes the percent of unbound retinol severely, but does not affect the percent of retinol bound measurably, does not change the Km value or rate of retinal production from holo-CRBP1, catalyzed by microsomes. This also indicates involvement of holo-CRBP1 itself. Notably, the Km values of LRAT (0.2 μM) and RDH1 (~2 μM) for holo-CRBP1 are less than their Km values for unbound retinol of 0.6 and 4 μM, respectively. The points remain, each reaction rate (esterification and dehydrogenation) catalyzed by microsomes in the presence of holo-CRBP1 exceeds that supported by unbound retinol, is saturable, and shows a higher affinity than for unbound retinol. Additionally, N-ethylmaleimide, which binds covalently to sulfhydryl groups in RDH, causes 90% inhibition of holo-CRBP1 supported generation of retinal, but does not inhibit retinal synthesis from unbound retinol [48]. This indicates that N-ethylmaleimide does not affect the catalytic action of RDH1, but interferes with its approach to holo-CRBP1, interrupting retinol transfer. A substrate-enzyme relationship between holo-CRBP1 and RDH16, the human ortholog of mouse RDH1 (rat RDH2) has been confirmed [49, 50].
Retinol and other retinoids are promiscuous substrates, making it difficult to study atRA biogenesis in vitro from unbound retinol. These difficulties are exacerbated by the ability of aqueous media and/or reduced pyridine nucleotides to oxidize retinol or reduce retinal in the absence of enzymes, and contamination by commercial retinol with several percent retinal. Presenting retinol bound to CRBP1 (as holo-CRBP1) potentially avoids these problems because of its high affinity for retinol.
Studies of atRA biosynthesis focused by use of holo-CRBP1 as substrate showed that 80 to 94% of cellular retinal-generating capacity resides in microsomes, rather than cytosol, and microsomal rates in various tissues exceed cytosolic rates by 5 to 20-fold [51, 52]. Recombinant holo-CRBP1, covalently modified with a cleavable light-activated cross-linking reagent, was used as bait to identify potential RDH. This construct identified two acceptors specifically in microsomes (which obviously contain hundreds of proteins), a 25 kDa band, now known to be LRAT, and a 35 kDa band subsequently identified as an SDR family member [53]. Notably, crosslinking required the presence of a pyridine nucleotide cofactor (NAD+ or NADP+) and occurred only with holo-CRBP1, not with apo-CRBP1. This is significant because dehydrogenases, such as SDR, function via an ordered bisubstrate reaction mechanism, i.e. they must bind cofactor (aka cosubstrate) before they can bind the substrate to be oxidized. Thus, not only did the SDR bind holo-CRBP1 and not apo-CRBP1, it recognized holo-CRBP1 as expected for a substrate. This reinforced the observations noted above with respect to the Michaelis-Menten kinetics between microsomes and holo-CRBP1 for retinal generation. This information led to the first cDNA cloning of an RDH in the path of atRA biosynthesis, rat RODH1 [54]. Subsequently, the human and mouse orthologs were cloned, based on the data generated with the rat cDNA [50, 55, 56]. RDH1 (the mouse ortholog of rat RODH1) contributes to a reconstituted pathway of atRA biosynthesis, when expressed in intact cells with each of the three RALDH. These data set the stage for cloning a large sub-family of SDR associated with retinoid metabolism, reviewed in [57]. Currently, at least three RDH seem physiologically involved in converting all-trans-retinol into all-trans-retinal: RDH1, RDH10 and DHRS9 (Table 1). All are microsomal members of the SDR gene family; several retinal reductases, RRD, also have been identified. The three RDH have emerged as the primary catalysts of the first step in atRA generation from retinol, because when ablated, each demonstrates a phenotype directly associated with atRA function. It is reasonable, however, to suspect that others may emerge.
Table 1. Rdh that contribute physiologically to atRA biosynthesis. Rows depict orthologs Columns depict homologs and/or paralogs.
| Mouse | Rat | Human |
|---|---|---|
| Rdh1 | Rdh7 and Rdh2 (originally RodhI and RodhII) |
Rdh16 (originally Rodh4, RDH-E) |
|
| ||
| Rdh10 | Rdh10 | Rdh10 |
|
| ||
| Dhrs9 | Dhrs9 (originally eRolDH2) |
Dhrs9 (originally retSdr8, RDHL, Rdh-TBE, RoDH-E2, 3α-HSD |
Not all agree that RDH interact with/or recognize holo-CRBP1 [58]. Rather, another model postulates that RDH recognize only unbound retinol as substrate. This group revised their original conclusion based on reexamination of RDH16 (human ortholog of RDH1) kinetics that showed a lower rate of retinal production from holo-CRBP1 relative to unbound retinol. This argument suffers from the fact that RDH in vivo would not have access to the relatively high amounts of unbound (free) retinol substrate that were provided in vitro. In vivo, any putative retinol dehydrogenase would have to complete with apo-CRBP1 for retinol: therefore, a high catalytic rate in vitro in the absence of holo-CRBP1 cannot predict performance in CRBP1-expressing cells.
The revised conclusion also was based on RDH16 over expressed in insect cells, incorporated into proteoliposomes, and then reacted with an N-terminal fusion of CRBP1 to glutathione S-transferase or with a C-terminal fusion of CRBP1 to the chitin binding domain. A key aspect of interpreting these data is the catalytic constant (turnover number), kcat, of the RDH16 used. The kcat denotes the number of moles of product produced per sec (to keep the numbers manageably low) by one mole of enzyme. The RDH16 used had a kcat of 1.1 min−1, i.e. this formulation of RDH16 produced only one mole of product per mole of enzyme in 50 sec, indicating an enzyme preparation with severely impaired activity. As it is, endogenous RDH1 in its native mammalian environment (microsomes in vitro) generates retinal from holo-CRBP1 at a much lower rate than it does from unbound retinol, possibly because release of retinol from the very high-affinity holo-CRBP1 imposes a rate-limiting step relative to the catalytic cycle of RDH1/16 [47, 52, 59]. The appropriate issue involves whether the rate catalyzed by native RDH1 from holo-CRBP1 can sustain the atRA concentration in cells—which it can. A preparation with impaired activity, such as the Sf9 generated and solubilized enzyme, may not appear to interact with holo-CRBP1 because of its already low activity, or may not interact because the changes that impaired activity impair interactions. Use of modified CRBP1 also is problematic. Mutating a single external residue (L35) of CRBP1 reduced significantly the Vm for generating retinal from endogenous microsomal RDH1 and increased the Km value, but did not change the affinity for retinol binding (apparent kd value) [60]. This suggests that altering the CRBP1 structure, as amending it with peptides at either end, alters its ability to function.
A conclusion that RDH10 does not access holo-CRBP1 as substrate also was based on a significantly lower reaction rate with holo-CRBP1 vs. free retinol, and just as for RDH16, no test was conducted of a possible Michaelis-Menten relationship between RDH10 and holo-CRBP1 [61]. In this case, a modified RDH10 (RDH10-His6) was expressed as 10% of total insect (Sf9) cell membrane protein—recall that modifying RDH with N-ethylmaleimide inhibited holo-CRBP1-supported generation of retinal by 90%. Nor did the formulation of RDH10 duplicate the microenvironment of native RDH10 associated with mammalian membranes. Membrane-associated proteins depend on membrane composition and their stoichiometry with membrane phospholipids/proteins for native activity and function [62-64]. It is reasonable to expect that the composition and stoichiometry of smooth endoplasmic reticulum membrane phospholipids are crucial to RDH activity and any interaction with holo-CRBP1. For example, RDH1 activity during purification was maintained only with phosphotidylcholine—no other phospholipid tested produced the same reaction rate—and only when RDH1 was bound with the membrane protein, CYP2D [48, 53, 65]. In other words, the membrane context of RDH affects function.
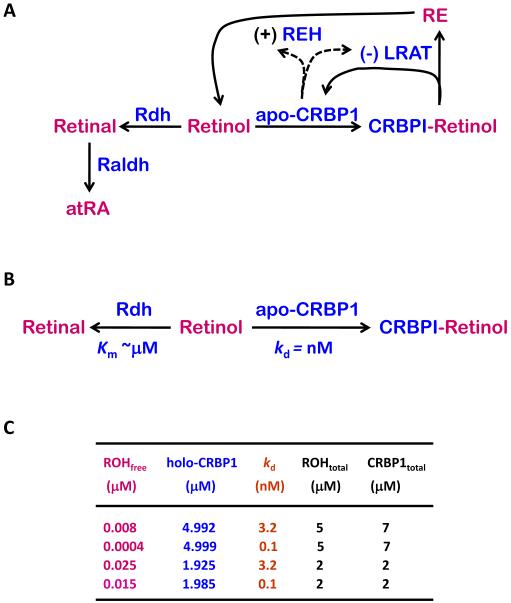
A model in which RDH would recognize only unbound retinol relies on competition between RDH and apo-CRBP1 for retinol (Figure 3A). As stated above, the kd value of apo-CRBP1 for retinol likely does not exceed 0.1 nM [38, 58]. In contrast, Km values recorded for RDH with unbound retinol vary from 1 to 6 μM—with holo-CRBP1 as substrate the Km values vary from ~0.6 to 2 μM—but in each case the values are lower than those values generated in the same report for unbound retinol [54-56, 66]. RDH10, which reportedly has a Km value of 0.035 μM for unbound retinol, may be an exception [61]. But this value was obtained with a substrate concentration range of 0.0625 to 2 μM. In other words, it is unreliable because the lowest substrate concentration tested was much higher than the inflection point of the curve; and the value was generated with RDH10-(His)6 expressed as 10% of Sf9 cell microsomal protein. In contrast, native human RDH10 assayed in membranes of COS-1 cells had an Km value of ~4 μM for all-trans-retinol [67]. To be sure, Km values do not consist only of ka and kd values of enzymes for their substrates, but Km values reflect association/dissociation constants to a large extent. This model would have two proteins competing for retinol, which differed by as much as 40,000-fold in affinity for retinol (0.1 nM kd vs. ~4 μM Km) (Figure 3B). Although this is a “back-of-the-envelope” calculation, it illustrates the issue. The lower affinity protein, in this case RDH, would have little chance of sequestering substrate (retinol). In the presence of CRBP1, the steady-state concentration of retinol would be nM, whereas the steady-state concentration of holo-CRBP1 would be μM; making holo-CRBP1 the highest concentration potential substrate (Figure 3C). In effect, RDH with μM Km values would have available low nM retinol, likely associated with membranes, or μM holo-CRBP1. Consider this with coupled retinol uptake and esterification, and it seems that generation of retinal would depend on the close relationship between holo-CRBP1 and RDH (indicated by the cross-linking that occurred only under conditions that support the catalytic cycle of RDH). Ability of CRBP1 to bind retinal, as well as retinol, and ability of RALDH1 and 2 (RALDH3 has not been tested) to generate atRA from CRBP1-retinal complete the argument for a process that does not rely on unfettered diffusion of retinol away from CRBP1.
Figure 3. An alternative model of generating atRA.
A) This model indicates a possible alternative to that shown in Figure 1, without direct involvement of holo-CRBP1 delivering retinol to RDH and RALDH, which requires diffusion of retinol from its site of release by holo-CRBP1. B) This would require competition between RDH and apo-CRBP1 for retinol (and between RALDH and apo-CRBP1 for retinal). The very high affinity of apo-CRBP1 (nM) for retinol would severely restrict RDH access to retinoids, especially with the driving force of RE formation mediating intracellular retinol uptake (see A). C) Shown are the effects of CRBP1 affinity (kd values) on concentrations of unbound (or free) retinol). With the measured kd value of ~3 nM (considered a high estimate because of technology limitations), the available free retinol concentration would vary between 8 and 25 nM, depending on the ratio of total CRBP1 to total retinol. With a kd value of ~0.1 nM, considered as likely more accurate, free retinol concentrations would vary between 0.4 and 15 nM.
6. An N-terminal leader anchors RDH and RRD in the ER facing cytoplasm
The orientation of RDH1 and similar SDR in the endoplasmic reticulum (cytoplasmic or lumenal facing) has been determined by multiple approaches [68-70]. The properties of native RDH1 were compared to a chimera with an established luminal signaling sequence, 11β-hydroxysteroid dehydrogenase(aa1-41)-RDH1(aa23-317). In addition, the green fluorescent protein fusions RDH1(1-22)-GFP and 11β-HSD1(1-41)-GFP and signaling sequence mutants were examined via confocal immunofluorescence, antibody access, proteinase K sensitivity, and deglycosylation assays [69]. The results showed clearly that an N-terminal signaling sequence of 22 residues anchors RDH1 in the endoplasmic reticulum projecting into the cytoplasm. Deleting the positive charges from the C-terminal end of the leader, and inserting two arginine residues near the N-terminus of the signaling sequence caused 95% inversion from cytoplasmic to luminal: i.e. the mutant L3R,L5R,R16Q,R19Q,R21Q faced the lumen. A retinal reductase, RDH11, with an N-terminal signaling sequence similar to that of RDH1, also orients to the cytosol, as indicated by a variety of approaches, including deglycosylation assays and mutagenesis [70]. In this case, in contrast to the native protein, the R25G,K26I mutant showed additional bands on SDS-PAGE, which were eliminated by Endo H treatment, i.e. the mutant was flipped partially from a cytosolic to a lumenal orientation by eliminating the positive charges at the C-terminal end of the signaling sequence. These results are consistent with the lack of RDH1 and RDH11 glycosylation, in contrast to lumenal facing proteins. These conclusions also are consistent with the “positive-inside rule”, which reflects the observation that positively-charged residues in leader sequences are 4-fold more abundant on the cytoplasmic side of membrane proteins, i.e. positive residues at the C-termini of leader sequences orient membrane proteins to the cytosol [71]. A refined version of the positive-inside rule takes into account the net electrical charge difference, as well as the hydrophobicity of the leader. By these criteria, the experimental data and the amino acid composition of the leaders in RDH and RRD indicate orientation to the cytosol. Alternative work concluded that the mouse RDH4 (human RDH5) orients to the lumen [72]. This conclusion was based on limited data (relative to the studies discussed above) with a mutant, and did not address effectively the extensive data discussed above, including: 1) how RDH/RRD manage to escape glycosylation, even though they harbor prototypical glycosylation signals (and lumenal proteins are glycosylated); and 2) how mutating the positive charges in the leaders of RDH/RRD allowed glycosylation, i.e. “flipped” the enzymes. Finally, a lumenal orientation for RDH4/5 would seem to preclude function as an 11-cis-retinol dehydrogenase to generate 11-cis-retinal for binding to opsin.
7. Xanthine oxidase
The cytosolic enzyme xanthine oxidase (aka xanthine dehydrogenase) generates atRA from retinol (through retinal) in human mammary epithelial cells [73, 74]. The reaction depends “strictly” on CRBP1 and has a Km value for holo-CRBP1 of ~0.1 μM. This is an intriguing observation, and may represent the holo-CRBP1 recognizing activity in cytosol (recall the discussion above that 6 to 20% of cellular retinal-generating capacity occurs in cytosol), but awaits further study to confirm its contribution to atRA biosynthesis.
8. Retinal dehydrogenases
The next step in atRA biosynthesis involves a rapid (relative to retinol dehydrogenation) and irreversible dehydrogenation of retinal into atRA. At least three RALDH catalyze this step. As with identification of the first step enzymes, retinoid binding-proteins contributed to identification of RALDH candidates as physiological participants in atRA biosynthesis. Lee et al. showed that two mouse ALDH, AHD-2 and AHD-7, converted unbound retinal into atRA in vitro with Km values <1 μM [75]. Neither seemed to have widespread tissue distribution, however, prompting concern about their universal importance in atRA generation. Also, at the time, the mouse was thought to express at least 13 ALDH, but the rat was thought to express only four, not including AHD-2 [76]. Rather, the closest ALDH to AHD-2 in rat was known as phenobarbital-induced ALDH [77]. Apparent absence of an ortholog in rat liver argued against the central importance of AHD-2 to atRA biosynthesis. Fractionation of rat cytosol, however, revealed multiple RALDH activities with unbound retinal. To determine which might participate physiologically in atRA biosynthesis, retinal generated from holo-CRBP1 by microsomes was used as bait (CRBP1 binds retinal, not only retinol), and an enzyme was identified that was regulated by vitamin A status and atRA [78]. This resulted in purification of rat liver cytosolic RALDH1 as the ortholog of AHD-2 and the demonstrations that RALDH1 differs from phenobarbital-induced ALDH, is down regulated by vitamin A in liver, and is inhibited by apo-CRBP1. Subsequently, the importance of AHD-2 (RALDH1) to atRA biosynthesis was demonstrated in the dorsoventral axis of the mouse retina [79]. Further work purified RALDH1 from rat kidney [80]. Working from the amino acid sequence of RALDH1, RALDH1 was cloned and the enzyme was characterized as an RALDH with widespread tissue distribution and tissue-specific regulation by atRA [81-83]. RALDH1 recognizes CRBP1-bound retinal as substrate (K0.5 = 0.8 μM) and is inhibited by apo-CRBP1, with an IC50 of 1.4 μM.
RALDH2 was first cloned from rat testis using information from RALDH1 [84] and was cloned subsequently from P19 cells [85]. RALDH2 enjoys widespread tissue expression, often overlapping with RALDH1, but is regulated differently than RALDH1 by vitamin A status, and is not inhibited by apo-CRBP1 [86], although RALDH2 recognizes CRBP1-bound retinal as substrate with a Km value of 0.2 μM. (This differential response to apo-CRBP1 suggests RALDH1 and 2 operate during differing degrees of vitamin A status, and illustrates the complexity of atRA biosynthesis.) Like RALDH1, RALDH2 can generate atRA from retinal biosynthesized by microsomes from holo-CRBP1, and can function in a reconstituted atRA “generating system” in intact cells co-transfected with RDH1. A third RALDH, RALDH3, was later identified and originally referred to as ALDH6 [87].
RALDH1, RALDH2 and RALDH3 exhibit distinct expression patterns during mouse development, indicative of locus and stage specific individual functions for each [88].
A fourth RALDH, RALDH4 has been cloned, but RALDH4 recognizes only 9-cis-retinal as substrate and not all-trans-retinal [89]. Unlike RALDH1, 2 and 3, the contribution of RALDH4 to retinoid metabolism remains unexplored.
9. Disposition of atRA
atRA associates with binding proteins of the FABP gene family: CRABP1, CRABP2 and FABP5 [90, 91]. Holo-CRABPII interacts with and transfers atRA to RAR [92-95]. In contrast, FABP5 transfers its atRA to PPARβ/δ [96-98]. Thus, a mechanism that differentiates various atRA actions seems to be selective delivery to specific nuclear receptors mediated by binding to CRABP2 vs. FABP5 [99, 100].
The total physiological function of atRA binding to CRABP1 has not been resolved, but microsomes can access atRA bound with CRABP1 and generate catabolites with a lower Km value and higher rate than for unbound atRA, suggesting that one function of CRABP1 may include modulating atRA concentrations [101]. Consistent with this hypothesis, over expression of CRABP1 reduces atRA potency to induce F9 cell differentiation and increases its rate of catabolism [102-104].
atRA catabolites include: 5,6-epoxy-atRA, 4-oxo-atRA, 4-hydroxy-atRA, and 18-hydroxy-atRA [105-112]. Three cytochrome P-450 gene family members with widespread tissue distribution have been established as primary catalysts of atRA degradation, CYP26A1, B1 and C1[112-118]. The mouse expresses a fourth, CYP2C39, but the rat and the human do not express this P-450 [119]. Additional CYP also catabolize atRA in liver: including the rat CYP22C and the human CYP22C8 (and perhaps the human CYP22C9) [115, 120].
10. Regulation of atRA homeostasis
Biosynthesis of RA is the only established function of retinol, outside of generating retinal in the eye. Thus, it makes sense that atRA regulates retinol concentrations and autoregulates its own concentrations. CRBP1 and/or LRAT are essential for hepatic buildup of retinol and RE, and affect tissue distribution of retinol [42, 121-124]. As mentioned above, “excess” retinol is diverted into RE. Retinol cycles between its non-esterified and esterified forms, seemingly with the ratio apo-CRBP1/holo-CRBP1 contributing to the direction of flux. atRA induces RBP1 (encodes CRBP1) and LRAT gene expression [8, 125, 126]. Induction of RBP1 expression by atRA may represent a “housekeeping” function, as the amount of CRBP1 does not seem to vary extensively with variations in vitamin A status.
Challenging normal human keratinocytes in culture with increasing retinol did not substantially increase cellular retinol or atRA concentrations with a wide retinol concentration range [127]. Retinol and atRA remained low because of retinol storage via LRAT as RE. In vivo, an increase from a diet with 25 IU/g vitamin A (copious) to a very high vitamin A diet (250 IU/g), results in a 5-fold increase in liver retinol (to 0.3 μmol/g) and ~2-fold increase in RE to the very high concentration of ~7.9 μmol/g [123]. Lung, rather than adipose, stores the second highest concentration of RE in mice fed the copious diet (~1.6 μmol/g), and the greatest concentration of RE in mice fed the very high vitamin A diet (~11 μmol/g). In other words, organisms have very high capacity for storing vitamin A as RE—a substrate driven process with retinol uptake linked to RE formation. The LRAT-null mouse has very much lower tissue concentrations of retinol and RE than WT when fed both a copious and very high vitamin A diet, except for adipose retinol (with retinol variable in kidney), and maintains retinoid homeostasis by increased gastrointestinal excretion (very high diet only), redistribution of retinol to adipose (both diets), and increased retinoid catabolism via induction of liver CYP26A1 expression (both diets). These data reveal why loss of LRAT does not produce unbridled atRA biosynthesis: retinol decreases in most tissues and the increase in adipose is not sufficient to drive extraordinary atRA biosynthesis and/or compensatory mechanisms (CYP induction?) accompany loss of LRAT or CRBP1.
LRAT expression reflects dietary vitamin A, but does not show a wide range of expression in the marginal to ample dietary vitamin A range, suggesting that atRA has a “housekeeping” function to maintain LRAT expression [128]. The transition from a vitamin A-deficient diet to a diet with marginal vitamin A (1.3 IU/g) causes a 9-fold induction in LRAT, but an increase from the marginal diet to one that contained ample vitamin A (i.e. 13 IU/g) did not change LRAT expression substantially in rat liver. An increase to a very large amount of dietary vitamin A (333 IU/g) induced LRAT only 2.2-fold relative to the diet with marginal vitamin A. These data are consistent with the retinoid storage data mentioned in the previous paragraph.
The CRBP1 and LRAT knockout data show that lab mice under carefully controlled conditions do not require CRBP1 or LRAT for survival. Obviously, both are essential in the wild, because both are highly conserved, and wild mice do not enjoy the living conditions of lab mice.
atRA also induces STRA6 mRNA [129, 130]. This simultaneous regulation of STRA6, CRBP1, and LRAT reinforces the concept discussed above of coupled retinol uptake and esterification, with “excess” retinol directed into RE, rather than to large amounts of non-esterified retinol. atRA treatment in association with retinol causes maximum accumulation of RE, reflecting substrate-driven esterification during atRA-induced CRBP1 and LRAT expression.
Another aspect of autoregulating atRA concentrations involves induction of CYP, which increases the rate of atRA catabolism [131-133]. Increasing dietary vitamin A from marginal (0.6 IU/g diet) to the AIN recommended amount (4 IU/g diet) induced mouse liver CYP26A1 mRNA 8-fold, but produced no change in CYP26B1 [3]. Feeding mice a diet with copious vitamin A (30 IU/g) induced mouse liver CYP26A1 >60-fold relative to the 4 IU/g diet and 500-fold relative to the 0.6 IU/g diet. A diet with copious vitamin A also induced liver CYP26B1 ~10-fold relative to the 4 IU vitamin A/g diet. In the rat, a diet with marginal vitamin A increases CYP26A1, CYP26B1 and CYP2C22 mRNA ~5-fold, ~2-fold, and ~23-fold, respectively, relative to a vitamin A-deficient diet [128]. Progressing from the diet with marginal vitamin A to one with an ample amount of vitamin A (13 IU/g) had no further major effects on CYP26A1 and CYP2C22 mRNA, but induced CYP1B1 2-3-fold. An increase to a very large amount of dietary vitamin A (333 IU/g) induced CYP26A1 20-fold relative to the diet with ample vitamin A, but induced CYP26B1 and CYP2C22 only ~3-fold. These data show that atRA regulates its own concentrations through lowering substrate (retinol conversion into RE), and enhancing its catabolism.
Interactions between RDH and RALDH provide another level of maintaining atRA concentrations. In astrocytes, knocking down DHRS9 results in increased atRA production, despite the fact that DHRS9 functions as a retinol dehydrogenase in intact cells, not as a reductase [134]. This resulted from an increase in RALDH1 expression.
11. Possible functions for multiple RDH and RALDH
Occurrence of multiple RDH and RALDH, often in the same cells, poses the question, why so many? One might argue that the importance of atRA demands redundancy. On the other hand, the RDH10 and the RALDH2 knockouts each are embryonic lethal ~e10.5. These observations argue against only redundancy as a reason for multiple RDH and RALDH, at least under the specific experimental conditions. An alternative hypothesis has each RDH and RALDH generating atRA for a specific function, i.e. distinct intracellular pools of atRA may exist, either with respect to locality and/or in response to distinct signals. Each pool would fulfill a distinct vitamin A function. This is reminiscent of malonyl-CoA, generated by two different isozymes of acetyl-CoA carboxylase (ACC) [135]. In liver cytosol ACC1 generates malonyl-CoA as substrate for de novo fatty acid biosynthesis. In the liver outer mitochondrial membrane, ACC2 generates malonyl-CoA to inhibit carnithine-palmitoyl acyltransferase 1, which prevents acyl-CoA transport into mitochondria to support β-oxidation. The relevant point is generation of the same chemical by two isozymes at distinct subcellular sites in the same cell to serve different functions. The distinct atRA-related phenotypes that result from ablating each of RDH1, RDH10, DHRS9, RALDH1, Radlh2, and RALDH3 support such a hypothesis. These distinct phenotypes suggest that each of these enzymes fulfill a specific function that cannot be supported by the others. The ability of each of the three RDH and the three RALDH to function intracellularly has been demonstrated [134]. Primary hippocampus astrocytes express each of the six and each contributes to atRA biosynthesis, as demonstrated by sequential siRNA knockdown. In addition, immunocytochemical analysis revealed cytosolic expression of both RALDH1 and RALDH2, as expected from previous work, but substantial nuclear expression of RALDH1 and perinuclear expression of RALDH2. Even though RDH1 and RDH10 also are both expressed in the smooth endoplasmic reticulum, confocal immunofluorescence reveals their precise expression loci differ.3
12. Quantification of physiological amounts of retinoids
atRA is very difficult to detect, let alone quantify, because of its very low abundance, the small sizes of many of the loci it affects (hippocampus, limb buds, e.g.) and its liability. LC/MS/MS assays have been developed to quantify atRA in biological samples [136]. Most have been developed for use with relatively abundant and easy to manipulate matrices (serum, liver, and cells in culture), whereas other assays adapt to a wider range of samples, including samples difficult to assay such as brain sections, adipose, pancreas, and embryo sections. LC/MS quantification of atRA offers sensitive, specific and real-time atRA quantification via analytically robust methodology, if applied rigorously. Reference retinoid values are provided (Table 2). The values for RE, retinol and retinal were generated by high-performance liquid chromatography, as these retinoids are sufficiently abundant to allow quantification by UV detection.
Table 2. Representative retinoid concentrations in mice (nmol/g or ml).
| Retinoid | Liver | Serum | Exptrahepatic tissues |
|---|---|---|---|
| RE | 500a | 0.2 | 0.2-1.8 |
| Retinol | 10 | 0.8 | 0.08-0.8 |
| Retinal | 0.16 | 0.03 | 0.06-0.19 |
| atRA | 0.04 | 0.003 | 0.002-0.02 |
An RARE-lacZ reporter has been used to designate areas of atRA presence, even though it requires tissue-specific transcription cofactors obligatory for activating RARβ. Thus, the design of the reporter limits its effectiveness to those loci that express the necessary cofactors in the required stoichiometry. In addition, the RARE-lacZ reporter lacks sensitivity (false negatives), does not provide quantitative data, is not specific for atRA (false positives), and has not been validated as an assay. Such validation requires (minimally) generation of standard curves, demonstration of intra- and inter- assay reproducibility, insight into specificity, and knowledge of lower limits of detection. Moreover, the RARE-lacZ reporter requires 12 to 14 hr to produce a signal, and produces a signal whose half-life has not been established, which alone would severely limit its use it in developmental studies. For example, analysis of data generated by the RARE-lacZ reporter in the dorsal forebrain of the Foxc1 hypomorph revealed the signal mostly persisted from activation at a developmental stage earlier than the one studied [137]. These authors compared LC/MS data to RARE-lacZ reporter data, and concluded that the latter was “not useful” in assessing function of atRA in the developing cortex.
13. Retinoid-related processes served by RDH
RDH1
Although the mouse embryo expresses RDH1 as early as e7.5, and the mouse, rat and human orthologs have widespread tissue distribution, the RDH1-null mouse shows no obvious developmental phenotype [3, 54-56, 66]. Decreases in expression of CYP that degrade atRA and increases in other RDH and RALDH may account for survival of the RDH1-null mouse. Some developmental biologists have concluded that this shows RDH1 is not “essential” for development. This is understandable, concerning a morphological view of development. On the other hand, the concept that a well-conserved gene expressed early and widely (species and tissue) is not “essential” for development seems contradictory to the concept of natural selection, especially when its expression increases 40-fold between e7.5 and e18.5 and drops precipitously at P2. The ablated enzyme might cause (partial) cell function failure that could contribute to serious diseases that develop over a lifetime, such as type II diabetes, even though they do not produce a morphologically defective embryo. Another issue is the artificial environment of laboratory mice. Laboratory mice are products of unnatural selection and are larger, fatter and quicker to mature than mice in the wild [138-141]. Laboratory mice have larger litters, and are slower, weaker, and less active than wild mice. In addition, mouse chow contains copious amounts of nutrients that exceed even the generous amounts recommended by the AIN for rodent chow, which does affect the outcome of gene deletion experiments [15].
A chow diet contains copious vitamin A (15 to 30 IU/g, depending on the formula), which presumably overwhelms retinoid binding-proteins and allows generation of atRA by enzymes that normally would not have access to retinol. In contrast to a lack of phenotype when fed a chow diet, when fed a vitamin A-deficient diet (note: feeding a vitamin A-deficient diet to a mouse bred from a dam fed a chow diet does not induce vitamin A-deficiency up to 40 weeks), or the AIN recommended amount of vitamin A , the RDH1-null mouse gains weight faster than WT starting after weaning, and becomes up to 30% fatter than WT controls. This is remarkable, as most other models of obesity require feeding a high-fat diet for many months, even to genetically altered mice. Prevention of the phenotype by a chow diet emphasizes the ability of excess vitamin A to rescue the phenotype and the pitfalls of using chow diets to investigate knockouts related to vitamin A action. The precise mechanism of the phenotype remains unresolved, but the highly specific phenotype reinforces the hypothesis that each of the multiple RDH generate atRA to serve specific functions.
atRA protects against nephropathy in HIV-1 transgenic mice (Tg26 mice). Conversely, atRA was reduced 50% in the kidney cortex of Tg26 mice and 70% in their glomeruli [142]. This was attributed to a 70% reduction in RDH1 mRNA and ~2-fold increase in CYP26A1 mRNA. These data indicate that a defect in atRA synthesis contributes to the pathology of HIV in kidney and illustrates a physiological function for RDH1 in generating atRA.
RDH10
RDH10 was first cloned and evaluated kinetically as a retinol dehydrogenase in the retinal pigment epithelium, and later was found in Muller cells [143, 144]. RDH10 was subsequently correlated with several sites of atRA biosynthesis in the mouse embryo, suggesting functions outside of the eye [145, 146]. Like, RDH1, RDH10 is active with both all-trans-retinol and 9-cis-retinol [61].
RDH10 in conjunction with RALDH2 generates atRA in human dendritic cells [147]. The PPARγ agonist, rosiglitazone, induced the mRNA of RDH10 and RALDH2, followed by induction of atRA biosynthesis. The atRA generated, functioning via RARα induced CD1d, a receptor for xenobiotic lipids. NKT cell development requires CD1d and Cd1d regulates T-lymphocyte function [148]. These data demonstrate contributions for RDH10 and RALDH2 in generating atRA for immune system performance.
ENU-mutagenesis generated a point mutation that impaired embryogenesis, which has been traced to loss of RDH10 activity [149]. A single amino acid substitution destabilizes RDH10 and eliminates its activity. This impact on retinol dehydrogenase activity is consistent with the demonstration that a single mutation in RDH1 causes a 1250-fold decrease in activity [150]. The mutations in both cases were not within the cofactor binding domain or in active site residues. The RDH10 mutant produced defects at e10.5 in limbs, optic vesicles and caudal pharyngeal arches, and morphological defects suggestive of neural crest cell defects. Mice died by e13.5. Significantly, an admittedly “imperfect” reporter, i.e. an RARE-lacZ transgene, detected residual atRA biosynthetic activity in these mutants. If the reporter data are taken at face value, these data suggest contributions of other RDH to embryogenesis, as does the fact that RDH10 mutants did not suffer defects in all atRA-directed processes during embryogenesis.
A more comprehensive study tied developmental abnormalities of Foxc1 hypomorphs to decreased atRA signaling resulting from decreased RDH10 and RALDH2 expression [137]. Foxc1 hypomorphs lack dorsal forebrain meninges, which relate to decreases in intermediate progenitor cells and neurons by e14.5, resulting in cortical abnormalities. This suggested a diffusible factor that directs progression of progenitor cells, which was identified as atRA. atRA generation was related to RDH10 and RALDH2, which had substantial, but not complete expression loci overlap, again suggesting contributions from other RDH and/or RALDH to atRA biogenesis in the embryo. This study then examined an RDH10 hypomorph viable until e16.5 and noted that the dorsal forebrain defects were similar to those of the Foxc1 hypomorph, as was a reduction in cortical neurons. Reductions in atRA were not total, but rather approached 50% in the cortices (quantified by LC/MS), again indicating alternative sources of atRA. These data support the conclusions that atRA functions as a meningeal-derived signal for corticogenesis, and RDH10 contributes significantly to generating the atRA, but isn’t the only RDH that contributes to the path of atRA biosynthesis during embryogenesis.
DHRS9
DHRS9 has been cloned multiple times resulting in several designations. DHRS9 recognizes both free- and CRBP1-bound retinol as substrate [50, 59]. Additional data that DHRS9 functions in the physiological generation of atRA were developed in the zebrafish, which require DHRS9 for normal gut development and differentiation [151]. Zebrafish express DHRS9 ubiquitously in early embryogenesis, but occurs only in the gut 3 days after fertilization. DHRS9 knockdown at the one-cell stage produced a phenotype revealing a lack of atRA-directed embryogenesis, which was rescued by dosing with atRA. In addition, adenomatous polyposis coli mutant zebrafish turn out as hypomorphs in DHRS9 expression: these mutants also are rescued by dosing with atRA, or by replacing DHRS9.
Human colon and epidermis in the atRA–dependent strata epidermis express DHRS9 most intensely. Interestingly, relative to normal tissue, human colon adenoma and carcinoma samples showed reduced expression of DHRS9 and RDH5—but the latter SDR is better known for its activity in the retinal pigment epithelium with 11-cis-retinol [152]. Colon carcinoma cell lines, which had low rates of atRA biosynthesis from retinol, albeit relative to normal human mammary epithelia cells, did not express either DHRS9 or RDH5. Introduction of the tumor suppressor adenomatous polyposis coli into the colon carcinoma cell line HT29 induced DHRS9 5-fold (qPCR) via the transcription factor Cdx2, but did not induce RDH5, and increased atRA biosynthesis about 2-fold. Consistent with a function of DHRS9 in generating atRA to suppress tumorigenesis, screening of 19 tumor cell lines found that the Burkett lymphoma-derived cell line DAUDI does not express DHRS9. Similar observations were made with the MOLT-14 cell line (derived from Burkett lymphoma) and the T-cell acute lymphoblastic leukemia cell line Raji [153].
DHRS9 also seems important to atRA function during the estrus cycle. The rat uterine lining epithelium expresses DHRS9 only during estrus and co-expresses the SDR with CRBP1 and CRABP2 [154] and estrogen induces DHRS9 expression [155].
DHRS9, however, has not been knocked out in mice to understand its precise contribution to atRA biogenesis in the intact animal. Yet, its clear association with atRA supported processes render it an “RDH of interest”. Recalling the evidence that RDH1 and RDH10 are not capable of fulfilling all needs for atRA in the embryo, a function for DHRS9 in atRA biogenesis seems likely.
14. Visual cycle RDH
Multiple RDH/SDR function in the visual cycle to convert 11-cis-retinol into 11-cis-retinal in the retinal pigment epithelium, and to reduce all-trans-retinal into all-trans-retinol in photoreceptor outer segments [156]. Although, the precise RDH involved in each step remain a matter of discussion, RDH5, RDH10 and RDH11 seem to contribute to 11-cis-retinal production, whereas RDH8, RDH12 and retSDR1 reduce all-trans-retinal [157-161].
15. Retinoid-related processes served by RALDH (Aldh1A)
RALDH1 (Aldh1A1)
RALDH1-null mice are fertile and indistinguishable from WT littermates [162]. In the adult, RALDH1 accounts for ~90% of the all-trans-retinal dehydrogenase activity in kidney and liver of rat and mouse, and contributes substantially to atRA biosynthesis in many other tissues [75, 163-165]. RALDH1 has been associated with obesity in the adult. RALDH1 expression is up-regulated 3-fold in the kidney of db/db mice, whereas atRA concentrations are lower, consistent with regulation feedback regulation of RALDH1 by atRA [82, 166]. RALDH1-null mice resist glucose intolerance and obesity when fed a high-fat diet [167]. This observation triggered a hypothesis that retinal itself functions as a “distinct transcriptional regulator”, based (in part) on an increase in retinal in fat in knockout mice. But atRA in fat was not measured, and therefore changes in atRA were not excluded as underlying the phenomena. Although dosing with retinal reduced subcutaneous fat as a percentage of total fat in ob/ob mice more efficiently than dosing with atRA (15.5% vs. 19.1%), the conclusion that retinal must be functioning per se did not consider the distinctive pharmacokinetic characteristics of retinoids, and the metabolic vulnerability of retinal, and also was not supported with atRA measurements. The mechanism proposed, that retinal suppresses PPARγ-mediated processes, seems unlikely because the kd value of all-trans-retinal for PPARγ was reported as ~12 μM, far above the usual kd values for ligands of nuclear receptors, and the physiological concentration of retinal in white adipose (~64 pmol/g) [34]. A subsequent study of RALDH1-null mice addressed the atRA issue, and concluded that RALDH1 generates the major amount of atRA used to promote adipogenesis (relative to RALDH2 and 3), and RALDH1 deficiency impairs atRA production, which decreases atRA signaling, ZFP423 expression, PPARγ expression, and adipogenesis, specifically in visceral fat [168]. These later data provide a viable mechanism for RALDH1-null mice resisting weight gain when fed a high-fat diet.
A 56 kDa low-affinity androgen-binding protein, which occurs only in genital skin fibroblasts and not in non-genital fibroblasts, has been identified as RALDH1 [169, 170]. Curiously, RALDH1 is not expressed in most humans with androgen receptor mutations that cause androgen insensitivity. These mutations lead to testicular feminization and/or reduced fertility. The hypothesis has been suggested that lack of RALDH1 is causal in these physiological impairments, given the function of RALDH1 in atRA biosynthesis [171, 172].
The RALDH1-null mouse develop cataracts by 6-9 months old, indicating a function in protecting the eye other than by generating atRA [173].
Molecular insight into regulation of RALDH1 expression was generated from a RALDH1-CAT construct consisting of a proximal promoter upstream of the transcription start site, evaluated in the mouse cell line Hepa-1. The CCAAT/enhancer-binding protein, C/EBPβ, induced RALDH1 expression via a CCAAT box, whereas atRA decreased RALDH1 expression via RARα by decreasing the C/EBPβ concentration [174]. A subsequent study verified the role of the CCAAT-binding protein and added a role for the Oct transcription factor in the kidney, both within −80 and +43 of the 5′-region [175]. This was latter confirmed and extended by the observation that atRA increases GADD153, which sequesters C/EBPβ [176].
Notably, RALDH1 is the only RALDH expressed in the adult rat pituitary gland [177]. Estradiol decreases RALDH1 expression via ERα in the pituitary [178, 179].
Mice fed an ethanol-containing diet for one month had increased hippocampus atRA and RALDH1 activity, despite a decrease in RALDH1 protein and no notable RALDH2 activity or high RALDH3 activity, suggesting post-translational modification of RALDH1 [180]. atRA stimulates protein tyrosine kinase [181], and RALDH1 has been identified as a tyrosine phosphorylated protein in nasopharyngeal tissues [182]. This indicates that RALDH1 may undergo post-translational regulation by atRA, in addition to transcriptional regulation.
Even though there is no observed phenotype for in the embryo of the RALDH1-null mouse, RALDH1 denotes cancer stem cells and seems to designate a poor prognosis [183]. In addition, as well as other members of the ALDH gene family, also serve as markers of normal stem cells [184]. For example, the developing mouse and human express RALDH1 in pancreas at various stages during β-cell generation; atRA induces Ngn3+ endocrine progenitor cell formation and stimulates their further differentiation into β-cells [185].
RALDH2 (Aldh1A2)
RALDH2-null mice do not live beyond e10.5. The embryos are malformed and do not undergo the axial rotation that normally occurs on or about e8.5 [186]. Malformations include shortening of the anterioposterior axis, lack of limb buds, a deformed heart, and a truncated frontonasal region. Defects in embryonic rats that model congenital diaphragmatic hernia in humans have been related to inhibiting RALDH2 [187]. Nitrofen, 4-biphenylcarboxylic acid, bisdiamine, and SB-210661 inhibit RALDH2 and produce defects in the rat diaphragm. Interestingly, the atRA-mediated processes that go awry in the RALDH2-null mouse differ from those that go awry in the RDH10 knockout, again suggesting that each enzyme contributes to paths that generate atRA for specific aspects of development, and each may be coupled (partially) with a different RDH or RALDH. The different developmental expression patterns of RALDH1 and 2 also suggest distinctive biological functions during embryogenesis [188]. These observations have stimulated many studies of RALDH2 expression during embryogenesis, which are beyond the scope of this review. One conclusion, however, should be emphasized: even though RALDH2 ablation causes embryonic lethality, RA generating activities occur in embryo loci that are RA dependent and devoid of RALDH2 expression, including but not limited to RALDH1 and RALDH3 [189]. These data show clearly that routes of atRA biosynthesis alternative to RALDH2 occur during all phases of embryonic development.
In contrast to the plethora of studies during embryogenesis, relatively little has been done with RALDH2 postnatally. In the adult rodent, RALDH2 has widespread expression, but is most highly expressed in the testis. Interstitial cells of the testis, spermatogonia, and spermatocytes express RALDH2 [86, 190]. Restricting dietary vitamin A causes a 70-fold decrease in RALDH2 mRNA, which reflects massive loss of germ cells, and possibly decreased expression in the remaining cells. RALDH2 expression often overlaps with that of RALDH1, including in hepatocytes and airway epithelia. A so-called “testis-specific aldehyde dehydrogenase” has been confirmed as RALDH2 [191].
atRA modulates nephrogenesis [192]. El Kares et al. have identified a common variant (20% of the human population) in RALDH2 that was associated with increased cord blood atRA and a 22% increase in kidney volume [193].
RALDH2 mRNA decreases during rat neonate suckling, whereas RALDH1 mRNA increases during weaning of the rat pup [194]. The mechanisms of these effects have not been established. Estradiol induces RALDH2, with most intense induction during metestrus [195]. Promoter analysis indicated an estrogen receptor response element [190]. Subsequent work revealed that estradiol induces RALDH2 in uterine stromal cells, contrasting with estradiol suppression of RALDH1 in the glandular epithelium [196]. Gonadotropin also induces RALDH2 expression intensely. Spinal cord injury induces RALDH2 in what may be a unique atRA-synthesizing subpopulation of activated oligodendrocyte precursors or “polydendrocytes” [197]. In epicardial cells, RALDH2 is a transcriptional target of Wt1 [198]. Both TAL1 and LIM-only protein genes, but neither alone, induce RALDH2 expression when co-transfected into T-cell acute lymphoblastic leukemia cell lines [199]. A GATA site in the second intron binds GATA3 to initiate transcription.
Further analysis of the RALDH2 promoter indicated the presence of a sterol regulatory element binding protein (SREBP) [190]. In fact, cholesterol induces RALDH1 and RALDH2 expression in liver, brain, kidney and heart [200]. Oxysterols mediate the cholesterol effect via SREBP-1c and LXRα/β, establishing that “cross-talk” occurs between atRA biosynthesis and cholesterol metabolism.
RALDH3 (Aldh1A3)
The RALDH3-null mouse suffers from choanal atresia, because of persistence of nasal fins at e18.5 [201]. Rupture of nasal fins is a normal development process, which allows communication between oral and nasal cavities. Persistence of the fins causes death from respiratory distress in newborn mice. All malformations of RALDH3-null mice are rescued by supplementing dams with atRA during pregnancy—providing a direct link to its physiological importance in generating atRA.
RALDH3 is the only one of RALDH1, 2 and 3 that does not recognize 9-cis-retinal as substrate [202].
Whereas the developing mouse and human pancreas express RALDH1 during β-cell generation, abnormally high pancreas expression of RALDH3 has been associated with decreased glucose-stimulated insulin secretion in diabetic mouse models [203]. A 9-fold increase in pancreatic islet RALDH3 mRNA was observed in mice fed a high-fat diet or in db/db mice. A high glucose concentration (25 mM) caused a 4-fold induction of RALDH3, relative to 5 mM in the rat insulinoma cell line Min6 and a 2-fold increase in the glucagon-producing cell line αTC1. Transfecting an RALDH3 expression vector into Min6 cells caused a 3-fold decrease in insulin secretion, but transfecting αTC1 cells with RALDH3 produced a 2-fold increase in glucagon secretion. The precise retinoid responsible for these results was not determined, as only 13-cis-RA was investigated, an isomer without known activity. Regardless of the mechanism, these data are consistent with retinoids contributing to pancreas function [204]. These observations, along with different expression patterns of RALDH2 and RALDH3 in the cycling hair follicle of the adult mouse again indicate that each RALDH provides distinct pools of atRA for specialized functions, and that each contributes essentially to adult atRA-mediated functions [205].
RALDH3 seems to generate atRA to maintain differentiation in mammary epithelia, which suppresses tumorigenesis. Primary cultures of normal human mammary epithelial cells convert retinol into atRA, as do the non-tumorigenic mammary epithelial cell lines tested [206]. In contrast, five of six mammary cancer cell lines had either no or vastly diminished ability to biosynthesize atRA. Retinol caused growth inhibition in cells capable of converting retinol into atRA, but not in cells incompetent in atRA biosynthesis. The “missing” enzyme was identified as RALDH3 and re-introduction of RALDH3 into MCF-7 cells, a tumorigenic breast epithelial cell line, reinstituted ability to convert retinal into atRA [207]. In support of this conclusion, Mtsm1 a dominant modifier of mammary tumor susceptibility, has been mapped to a locus with five candidate genes for mammary tumorigenesis, one of which is RALDH3 [208]. In contrast to these data, RALDH3 has been identified as a cancer stem cell marker in human mammary tumors that correlates very well with tumor grade, metastasis, and cancer stage [209]. Moreover, treating normal human mammary epithelial cells with benzo(a)pyrene caused 3-fold up regulation of only two genes, one of which was RALDH3 [210]. In HL1-1 human liver cells, 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure induced RALDH3, as well as a number of other genes [211]. Although these data seem contrary to the lack of RALDH3 in mammary epithelial tumors, they may reflect a compensatory mechanism to carcinogenic insult, or may reflect differences caused by comparing samples from a small patient population to cells maintained in culture for long durations.
Much less work has been done on regulation of RALDH3 expression than with RALDH1 and 2 expression. Phenobarbital induces RALDH3 2 to 10-fold in rat liver, depending on the strain [212]. In glioblastoma cells, a fusion protein of IL-13 with a mutated form of Pseudomonas exotoxin induces RALDH3 via the IL-13 receptor, suggesting a mechanism of regulating atRA biosynthesis [213]. In primary keratinocytes, atRA (1 μM) up regulates RALDH3 ~10-fold, but does not affect RALDH1 and RALDH2 expression [214]. atRA also increases RALDH3 expression in organotypic human skin cultures and in an epidermal explant, but has no effect in dermal fibroblasts or HeLa cells. The authors proposed that these data suggest a “feed forward” loop, although this really requires much more study to understand, especially because of the high atRA concentration evaluated. Normally, atRA exerts negative feedback on its concentrations.
Dihydrotestosterone induces RALDH3 mRNA 4-fold in the prostate cell line LNCaP via activating the androgen receptor [215]. An ~8-fold increase in retinal conversion into atRA accompanied RALDH3 induction, and induction of CYP26A1 followed the atRA increase, thus indicating androgen regulation of RALDH3 expression and atRA biosynthesis.
16. CYP functions in atRA homeostasis
Several reviews discuss CYP contributions to atRA homeostasis in detail; therefore, this section will refer only to key points, to emphasize participation of CYP in autoregulation of atRA homeostasis [8, 117, 216, 217]. The first observation that atRA degradation contributes essentially to maintaining atRA function involved the demonstration that inhibiting atRA metabolism enhances atRA potency in vivo and in retinoid responsive cells [218-222]. This realization motivated synthesis of a variety of atRA catabolism inhibitors known as retinoic acid metabolism blocking agents, or RAMBA, for treating cancer and dermatological diseases, as an alternative to dosing with atRA or synthetic retinoids, to avoid the toxicity of the later two [223-226].
The importance of atRA degradation to controlling atRA function has been illustrated by knockout of CYP26A1, the only atRA-dedicated CYP knocked out to date [227, 228]. CYP26A1-null mice die during mid to late gestation with morphological defects that involve the hindbrain, vertebrae, and tailbud. The mutants have spina bifida, tail truncation, and kidney abnormalities, among other deformities, which mimic those produced by treating WT mice with excess atRA. These outcomes contribute to the insight that concentrations of atRA must be tightly controlled spatially and temporarily during development. Whereas developing embryos require atRA, they also require it in measured amounts and/or not at all in some loci, consistent with the well-known teratogenicity of atRA, or of retinol given in sufficiently high doses to defeat control mechanisms and produce atRA pharmacologically. Interestingly, crossing CYP26A1-null mice with RALDH2-null mice (heterozygotes) results in 12 of the 17 offspring living until weaning [229]. These data indicate that oxidized derivatives of atRA, such as 4-oxo-RA, 4-OH-RA, and 18-OH-RA, represent catabolites of atRA, and by preventing atRA catabolism, a reduction in atRA generation can be made tolerable. This is the same phenomenon observed in the RDH1-null mice, which have diminished CYP26A1 expression, and illustrates the principle underlying the efficacy of RAMBA [3, 221].
17. ADH1 catalyzes metabolism of high retinol doses
The medium-chain alcohol dehydrogenases, ADH1 and ADH4, convert retinol into retinal in vitro, but Han et al. detected no activity of ADH3 with 100 μM retinol and 930 nM enzyme subunit, which confirmed earlier findings of no ADH3 activity with all-trans-retinol [58, 230-232]. Using a more sensitive hplc assay, a recombinant ADH3 purified by affinity chromatography had extremely low activity with 30 μM retinol (105 pmol/mg/min)—a concentration that exceeds unbound retinol concentrations in vivo by orders of magnitude (see Figure 3)—and no kinetic values were reported [233]. This rate is unfavorable even compared to non-purified RDH. For example, 1 μM recombinant RDH1, using an 800 x g supernatant of CHO cells, generated 2 nmol retinal/mg protein/min. Thus, these data do not establish ability of the soluble ADH3, which does not recognize holo-CRBP1, to access sufficient unbound retinol in vivo (not likely present in the soluble matrix) to contribute to atRA biosynthesis. Moreover, ethanol inhibits atRA production catalyzed by rat liver cytosol with an IC50 ~20 μM, but up to 10 mM ethanol does not inhibit microsomal retinol metabolism [51]. This reflects inhibition of the cytosolic ADH by ethanol in vitro. In vivo, however, ethanol dosing does not decrease tissue or serum atRA concentrations; rather, either no changes occur, or increases in atRA occur in select locations [180, 234].
Data have not been published demonstrating ADH catalysis of physiological retinol concentrations in vivo, or metabolism of retinol by ADH expressed in cultured cells, or of impaired ADH expression reducing metabolism of physiological retinol concentrations in vivo or in cultured cells. Conclusions that ADH knockouts reveal function in atRA biogenesis have relied in part on correlations between atRA biosynthesis sites and ADH expression patterns [235, 236]. Correlations have limitations, but these correlations are especially vulnerable, as atRA biosynthesis is widespread among many structures/tissues in the developing embryo and in the adult, and in multiple cell types of both. In fact, ADH1 expression does not correlate well with atRA biosynthesis sites, and ADH4 has restricted expression [237]. ADH3, however, is expressed ubiquitously. Thus, overlap between atRA biosynthesis and ADH3 expression, and ADH4 in some sites, seems assured, which imparts limited insight about causal relationships.
ADH-null mice have not been informative with respect to a function for ADH in atRA generation under physiological conditions. Studies of ADH knockout mice have not reported essential experiments, such as quantifying atRA during normal vitamin A nutriture, documenting phenotypic alterations consistent with reduced atRA, and assaying for gene expression changes indicating lack of atRA signaling or compensation for disrupted atRA function. Thus, genetic, metabolic, and function-related data are not available to support the notion that ADH metabolize physiological levels of retinol in vivo.
When stressed by feeding a vitamin A-deficient diet over a longer term, ADH3 and ADH4-null mice do not thrive, whereas the ADH1-null mouse remains unaffected [233]. Although this has been interpreted to indicate that ADH3 and ADH4 generate atRA during low vitamin A intake, crucial data are lacking to support this hypothesis. atRA was not quantified in serum or tissues, and the cause(s) of death was/were not reported. The notion was not discussed that knocking out ADH3 and ADH4 causes stress, and vitamin A deprivation itself stresses animals and would compound stress afforded by gene ablation. These observations did not distinguish the possibility that atRA compensates for stress caused by absence of ADH3 and 4, or that additional stress in the absence of atRA causes death by mechanisms unrelated to ADH3 and 4 function, from the proposal that ADH participate in atRA biosynthesis.
More recently, a counterintuitive hypothesis proposes that ADH1 and perhaps ADH2 function to eliminate excess retinol to avoid retinol toxicity [238]. Deliberately generating atRA to avoid retinol toxicity seems analogous to generating prostanoids to avoid polyunsaturated fatty acid toxicity. The issue throughout evolution has been to sequester limited amounts of retinol. Evolutionary pressure has not been exerted in humans, mice or rats to eliminate “excess” retinol; rather the experimental data show clearly that increased dietary retinol leads to increased RE, reaching mM values in liver of rodents fed a chow diet [123]. In addition, CYP occur that recognize retinol as substrate and convert it into more polar products (other than atRA). RA is more toxic than retinol. Retinol/RE may contribute further to retinoid toxicity when organisms are overwhelmed by exposure to vast amounts of vitamin A (a rarity), such that catabolism and generation of RE can no longer relieve the stress and the accumulated retinol and RE begin to disrupt membranes. Unless such rare and extreme conditions, multiple xenobiotic clearing enzymes would catabolize retinoids. The ADH1-null mouse has vastly decreased ability to convert high doses of retinol (50 mg/kg) into RA. But this bolus provides ~300-fold more than the recommended daily intake for a mouse, and drives serum atRA ~1600-fold over the steady-state value of ~2 pmol/ml in WT mice. A much lower single retinol dose (3 mg/kg) induces 71% incidence of cleft palate in mice, and a somewhat lower dose (39 mg/kg) induces 76% incidence of neural tube defects [239]. Emphasis has been directed toward a >80% decrease in serum atRA in the ADH1-null mouse after the 50 mg/kg dose, but the ADH1-null mouse still allows an atRA concentration ~100 to 200-fold greater than normal with this toxic dose. These data, therefore, do not indicate that ADH detoxifies retinol, because prolonged exposure to even modestly increased concentrations of atRA cause teratology [240, 241]. Rather, these data show that high (toxic) vitamin A doses overwhelm homeostatic mechanisms that evolved to concentrate scarce retinol, chaperone its disposition in vivo, and store it as RE and produce toxic amounts of RA.
Liver RE in the double ADH1/CRBP1 knockout exceed those in the single CRBP1 knockout, and decline more slowly when the animals are switched from a chow to a vitamin A-deficient diet, indicating that the absence of CRBP1 releases retinol from “supervision” and allows ADH1 access to retinol [242]. Liver RE values in the single ADH1 knockout exhibited too much variation to compare to WT. One and 5 weeks after switching from a chow to a vitamin A-deficient diet, RE in liver of the single ADH1-null differed significantly from WT, but RE values at 9, 13 and 18 weeks did not differ significantly from WT. No simple explanation is apparent for such a phenomenon, and the large SE and few mice/group (n = 3) contribute to confounding interpretation. It is not clear which of these points may be valid, but if the last three points are, then the data do not support the contention that ADH1 serves a physiological function to deplete RE. Also, recall that the amount of dietary vitamin A and the STRA6/LRAT/CRBP1 mediated uptake and esterification of retinol determines the liver RE concentration. In addition, ethanol, a competitive inhibitor of ADH1, increases liver atRA within 1 hr after dosing [180]. In summary, retinol metabolism catalyzed by ADH likely contributes to retinoid toxicity by generating high concentrations of atRA during exposure to vitamin A doses that saturate CRBP1 and defeat the mechanism of innocuous retinoid storage as RE.
18. Summary
A plethora of biochemical and genetic data consistently identify precise functions for the retinol dehydrogenases, RDH1, RDH10, and perhaps DHRS9 as physiological catalysts of the first step in atRA biosynthesis. Much data also conclusively identify RALDH1, RALDH2, and RALDH3 as participating in the second and irreversible step. Each of these enzymes has been conclusively related to a biological process mediated by atRA. Most of these enzymes interact with retinoid binding-proteins and, in several cases, influence the expression of one another. The cumulative data support the model of Figure 1 that proposes interactions among these enzymes and retinoid binding-proteins, with feedback regulation and/or control by atRA via (tissue specific) modulation of RALDH, CYP, LRAT, STRA6, and CRBP1. In addition to transcriptional regulation, the ratio apo-CRBP1/holo-CRBP1 likely influences flux of retinol into and out of RE. Finally, preliminary data suggest post-translation regulation of at least RALDH1. Understanding where and when atRA biosynthesis occurs increasingly depends on placing the topic into the larger context of the associated enzymes and binding proteins, rather than focusing on a single contributor to this intricate process.
Acknowledgment
Thanks to Charles R. Krois for reading and critiquing this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- ADH
- medium-chain alcohol dehydrogenase(s)
- ALDH
- aldehyde dehydrogenase(s)
- ARAT
- acyl-CoA:retinol acyltransferase
- atRA
- all-trans-retinoic acid
- CRABP
- cellular retinoic acid binding-protein(s)
- CRBP
- cellular retinol binding-protein(s)
- FABP
- fatty acid binding protein
- LRAT
- lecithin:retinol acyltransferase
- RAR
- retinoic acid receptor(s)
- RBP
- serum retinol binding-protein
- RDH
- retinol dehydrogenase
- RE
- retinyl ester(s)
- REH
- retinyl ester hydrolase
- RRD
- retinal reductase
- SDR
- short-chain dehydrogenase/reductase
- STRA
- (gene) stimulated by RA
- WT
- wild-type
Hui Sun, personal communication.
Weiya Jiang and J. Napoli, unpublished data.
References
- [1].Olson JA. Serum levels of vitamin A and carotenoids as reflectors of nutritional status. J Natl Cancer Inst. 1984;73:1439–1444. [PubMed] [Google Scholar]
- [2].E X, Zhang L, Lu J, Tso P, Blaner WS, Levin MS, Li E. Increased neonatal mortality in mice lacking cellular retinol-binding protein II. J Biol Chem. 2002;277:36617–36623. doi: 10.1074/jbc.M205519200. [DOI] [PubMed] [Google Scholar]
- [3].Zhang M, Hu P, Krois CR, Kane MA, Napoli JL. Altered vitamin A homeostasis and increased size and adiposity in the rdh1-null mouse. FASEB J. 2007;21:2886–2896. doi: 10.1096/fj.06-7964com. [DOI] [PubMed] [Google Scholar]
- [4].Raghu P, Sivakumar B. Interactions amongst plasma retinol-binding protein, transthyretin and their ligands: implications in vitamin A homeostasis and transthyretin amyloidosis. Biochim Biophys Acta. 2004;1703:1–9. doi: 10.1016/j.bbapap.2004.09.023. [DOI] [PubMed] [Google Scholar]
- [5].Senoo H, Kojima N, Sato M. Vitamin A-storing cells (stellate cells) Vitam Horm. 2007;75:131–159. doi: 10.1016/S0083-6729(06)75006-3. [DOI] [PubMed] [Google Scholar]
- [6].Wolf G. Serum retinol-binding protein: a link between obesity, insulin resistance, and type 2 diabetes. Nutr Rev. 2007;65:251–256. doi: 10.1111/j.1753-4887.2007.tb00302.x. [DOI] [PubMed] [Google Scholar]
- [7].Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ross AC, Zolfaghari R. Regulation of hepatic retinol metabolism: perspectives from studies on vitamin A status. J Nutr. 2004;134:269S–275S. doi: 10.1093/jn/134.1.269S. [DOI] [PubMed] [Google Scholar]
- [9].Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- [10].Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ottonello S, Petrucco S, Maraini G. Vitamin A uptake from retinol-binding protein in a cell-free system from pigment epithelial cells of bovine retina. Retinol transfer from plasma retinol-binding protein to cytoplasmic retinol-binding protein with retinyl-ester formation as the intermediate step. J Biol Chem. 1987;262:3975–3981. [PubMed] [Google Scholar]
- [12].Ong DE, MacDonald PN, Gubitosi AM. Esterification of retinol in rat liver. Possible participation by cellular retinol-binding protein and cellular retinol-binding protein II. J Biol Chem. 1988;263:5789–5796. [PubMed] [Google Scholar]
- [13].Yost RW, Harrison EH, Ross AC. Esterification by rat liver microsomes of retinol bound to cellular retinol-binding protein. J Biol Chem. 1988;263:18693–18701. [PubMed] [Google Scholar]
- [14].Harrison EH, Blaner WS, Goodman DS, Ross AC. Subcellular localization of retinoids, retinoid-binding proteins, and acyl-CoA:retinol acyltransferase in rat liver. J Lipid Res. 1987;28:973–981. [PubMed] [Google Scholar]
- [15].Reeves PG. Components of the AIN-93 diets as improvements in the AIN-76A diet. J Nutr. 1997;127:838S–841S. doi: 10.1093/jn/127.5.838S. [DOI] [PubMed] [Google Scholar]
- [16].Reeves PG, Nielsen FH, Fahey GC., Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- [17].Batres RO, Olson JA. Relative amount and ester composition of vitamin A in rat hepatocytes as a function of the method of cell preparation and of total liver stores. J Nutr. 1987;117:77–82. doi: 10.1093/jn/117.1.77. [DOI] [PubMed] [Google Scholar]
- [18].MacDonald PN, Ong DE. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem Biophys Res Commun. 1988;156:157–163. doi: 10.1016/s0006-291x(88)80818-0. [DOI] [PubMed] [Google Scholar]
- [19].Szuts EZ, Harosi FI. Solubility of retinoids in water. Arch Biochem Biophys. 1991;287:297–304. doi: 10.1016/0003-9861(91)90482-x. [DOI] [PubMed] [Google Scholar]
- [20].Orland MD, Anwar K, Cromley D, Chu CH, Chen L, Billheimer JT, Hussain MM, Cheng D. Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: diacylglycerol acyltransferase 1. Biochim Biophys Acta. 2005;1737:76–82. doi: 10.1016/j.bbalip.2005.09.003. [DOI] [PubMed] [Google Scholar]
- [21].Yen CL, Monetti M, Burri BJ, Farese RV., Jr. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J Lipid Res. 2005;46:1502–1511. doi: 10.1194/jlr.M500036-JLR200. [DOI] [PubMed] [Google Scholar]
- [22].Randolph RK, Winkler KE, Ross AC. Fatty acyl CoA-dependent and - independent retinol esterification by rat liver and lactating mammary gland microsomes. Arch Biochem Biophys. 1991;288:500–508. doi: 10.1016/0003-9861(91)90227-a. [DOI] [PubMed] [Google Scholar]
- [23].Ross AC. Retinol esterification by rat liver microsomes. Evidence for a fatty acyl coenzyme A: retinol acyltransferase. J Biol Chem. 1982;257:2453–2459. [PubMed] [Google Scholar]
- [24].Shih MY, Kane MA, Zhou P, Yen CL, Streeper RS, Napoli JL, Farese RV., Jr. Retinol Esterification by DGAT1 Is Essential for Retinoid Homeostasis in Murine Skin. J Biol Chem. 2009;284:4292–4299. doi: 10.1074/jbc.M807503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Harrison EH. Lipases and carboxylesterases: possible roles in the hepatic metabolism of retinol. Annu Rev Nutr. 1998;18:259–276. doi: 10.1146/annurev.nutr.18.1.259. [DOI] [PubMed] [Google Scholar]
- [26].Linke T, Dawson H, Harrison EH. Isolation and characterization of a microsomal acid retinyl ester hydrolase. J Biol Chem. 2005;280:23287–23294. doi: 10.1074/jbc.M413585200. [DOI] [PubMed] [Google Scholar]
- [27].Sun G, Alexson SE, Harrison EH. Purification and characterization of a neutral, bile salt-independent retinyl ester hydrolase from rat liver microsomes. Relationship To rat carboxylesterase ES-2. J Biol Chem. 1997;272:24488–24493. doi: 10.1074/jbc.272.39.24488. [DOI] [PubMed] [Google Scholar]
- [28].Gad MZ, Harrison EH. Neutral and acid retinyl ester hydrolases associated with rat liver microsomes: relationships to microsomal cholesteryl ester hydrolases. J Lipid Res. 1991;32:685–693. [PubMed] [Google Scholar]
- [29].Harrison EH, Gad MZ. Hydrolysis of retinyl palmitate by enzymes of rat pancreas and liver. Differentiation of bile salt-dependent and bile salt-independent, neutral retinyl ester hydrolases in rat liver. J Biol Chem. 1989;264:17142–17147. [PubMed] [Google Scholar]
- [30].Napoli JL, Pacia EB, Salerno GJ. Cholate effects on all-trans-retinyl palmitate hydrolysis in tissue homogenates: solubilization of multiple kidney membrane hydrolases. Arch Biochem Biophys. 1989;274:192–199. doi: 10.1016/0003-9861(89)90430-x. [DOI] [PubMed] [Google Scholar]
- [31].Harrison EH, Napoli JL. Bile salt-independent retinyl ester hydrolase activities associated with membranes of rat tissues. Methods Enzymol. 1990;189:459–469. doi: 10.1016/0076-6879(90)89323-a. [DOI] [PubMed] [Google Scholar]
- [32].Boerman MH, Napoli JL. Cholate-independent retinyl ester hydrolysis. Stimulation by Apo-cellular retinol-binding protein. J Biol Chem. 1991;266:22273–22278. [PubMed] [Google Scholar]
- [33].Herr FM, Ong DE. Differential interaction of lecithin-retinol acyltransferase with cellular retinol binding proteins. Biochemistry. 1992;31:6748–6755. doi: 10.1021/bi00144a014. [DOI] [PubMed] [Google Scholar]
- [34].Kane MA, Folias AE, Napoli JL. HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal Biochem. 2008;378:71–79. doi: 10.1016/j.ab.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ong DE, Chytil F. Cellular retinol-binding protein from rat liver. Purification and characterization. J Biol Chem. 1978;253:828–832. [PubMed] [Google Scholar]
- [36].Newcomer ME, Jamison RS, Ong DE. Structure and function of retinoid-binding proteins. Subcell Biochem. 1998;30:53–80. doi: 10.1007/978-1-4899-1789-8_3. [DOI] [PubMed] [Google Scholar]
- [37].Kane MA, Bright FV, Napoli JL. Binding affinities of CRBPI and CRBPII for 9-cis-retinoids. Biochem Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.02.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Malpeli G, Stoppini M, Zapponi MC, Folli C, Berni R. Interactions with retinol and retinoids of bovine cellular retinol-binding protein. Eur J Biochem. 1995;229:486–493. [PubMed] [Google Scholar]
- [39].Cowan SW, Newcomer ME, Jones TA. Crystallographic studies on a family of cellular lipophilic transport proteins. Refinement of P2 myelin protein and the structure determination and refinement of cellular retinol-binding protein in complex with all-trans-retinol. J Mol Biol. 1993;230:1225–1246. doi: 10.1006/jmbi.1993.1238. [DOI] [PubMed] [Google Scholar]
- [40].Napoli JL. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim Biophys Acta. 1999;1440:139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- [41].Napoli JL. A gene knockout corroborates the integral function of cellular retinol-binding protein in retinoid metabolism. Nutr Rev. 2000;58:230–236. doi: 10.1111/j.1753-4887.2000.tb01870.x. [DOI] [PubMed] [Google Scholar]
- [42].Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Hakansson H, Sauvant P, Azais-Braesco V, Frasson M, Picaud S, Chambon P. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Matt N, Schmidt CK, Dupe V, Dennefeld C, Nau H, Chambon P, Mark M, Ghyselinck NB. Contribution of cellular retinol-binding protein type 1 to retinol metabolism during mouse development. Dev Dyn. 2005;233:167–176. doi: 10.1002/dvdy.20313. [DOI] [PubMed] [Google Scholar]
- [44].Kane MA, Chen N, Sparks S, Napoli JL. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem J. 2005;388:363–369. doi: 10.1042/BJ20041867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Napoli JL. Retinol metabolism in LLC-PK1 Cells. Characterization of retinoic acid synthesis by an established mammalian cell line. J Biol Chem. 1986;261:13592–13597. [PubMed] [Google Scholar]
- [46].Wang C, Kane MA, Napoli JL. Multiple Retinol and Retinal Dehydrogenases Catalyze All-trans-retinoic Acid Biosynthesis in Astrocytes. J Biol Chem. 2011;286:6542–6553. doi: 10.1074/jbc.M110.198382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Posch KC, Boerman MH, Burns RD, Napoli JL. Holocellular retinol binding protein as a substrate for microsomal retinal synthesis. Biochemistry. 1991;30:6224–6230. doi: 10.1021/bi00239a021. [DOI] [PubMed] [Google Scholar]
- [48].Boerman MH, Napoli JL. Effects of sulfhydryl reagents, retinoids, and solubilization on the activity of microsomal retinol dehydrogenase. Arch Biochem Biophys. 1995;321:434–441. doi: 10.1006/abbi.1995.1415. [DOI] [PubMed] [Google Scholar]
- [49].Lapshina EA, Belyaeva OV, Chumakova OV, Kedishvili NY. Differential recognition of the free versus bound retinol by human microsomal retinol/sterol dehydrogenases: characterization of the holo-CRBP dehydrogenase activity of RoDH-4. Biochemistry. 2003;42:776–784. doi: 10.1021/bi026836r. [DOI] [PubMed] [Google Scholar]
- [50].Jurukovski V, Markova NG, Karaman-Jurukovska N, Randolph RK, Su J, Napoli JL, Simon M. Cloning and characterization of retinol dehydrogenase transcripts expressed in human epidermal keratinocytes. Mol Genet Metab. 1999;67:62–73. doi: 10.1006/mgme.1999.2840. [DOI] [PubMed] [Google Scholar]
- [51].Boerman MH, Napoli JL. Cellular retinol-binding protein-supported retinoic acid synthesis. Relative roles of microsomes and cytosol. J Biol Chem. 1996;271:5610–5616. doi: 10.1074/jbc.271.10.5610. [DOI] [PubMed] [Google Scholar]
- [52].Ottonello S, Scita G, Mantovani G, Cavazzini D, Rossi GL. Retinol bound to cellular retinol-binding protein is a substrate for cytosolic retinoic acid synthesis. J Biol Chem. 1993;268:27133–27142. [PubMed] [Google Scholar]
- [53].Boerman MH, Napoli JL. Characterization of a microsomal retinol dehydrogenase: a short-chain alcohol dehydrogenase with integral and peripheral membrane forms that interacts with holo-CRBP (type I) Biochemistry. 1995;34:7027–7037. doi: 10.1021/bi00021a014. [DOI] [PubMed] [Google Scholar]
- [54].Chai X, Boerman MH, Zhai Y, Napoli JL. Cloning of a cDNA for liver microsomal retinol dehydrogenase. A tissue-specific, short-chain alcohol dehydrogenase. J Biol Chem. 1995;270:3900–3904. doi: 10.1074/jbc.270.8.3900. [DOI] [PubMed] [Google Scholar]
- [55].Gough WH, VanOoteghem S, Sint T, Kedishvili NY. cDNA cloning and characterization of a new human microsomal NAD+-dependent dehydrogenase that oxidizes all-trans-retinol and 3alpha-hydroxysteroids. J Biol Chem. 1998;273:19778–19785. doi: 10.1074/jbc.273.31.19778. [DOI] [PubMed] [Google Scholar]
- [56].Zhang M, Chen W, Smith SM, Napoli JL. Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. J Biol Chem. 2001;276:44083–44090. doi: 10.1074/jbc.M105748200. [DOI] [PubMed] [Google Scholar]
- [57].Belyaeva OV, Kedishvili NY. Comparative genomic and phylogenetic analysis of short-chain dehydrogenases/reductases with dual retinol/sterol substrate specificity. Genomics. 2006;88:820–830. doi: 10.1016/j.ygeno.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [58].Gallego O, Belyaeva OV, Porte S, Ruiz FX, Stetsenko AV, Shabrova EV, Kostereva NV, Farres J, Pares X, Kedishvili NY. Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids. Biochem J. 2006;399:101–109. doi: 10.1042/BJ20051988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Markova NG, Pinkas-Sarafova A, Karaman-Jurukovska N, Jurukovski V, Simon M. Expression pattern and biochemical characteristics of a major epidermal retinol dehydrogenase. Mol Genet Metab. 2003;78:119–135. doi: 10.1016/s1096-7192(02)00226-3. [DOI] [PubMed] [Google Scholar]
- [60].Penzes P, Napoli JL. Holo-cellular retinol-binding protein: distinction of ligand-binding affinity from efficiency as substrate in retinal biosynthesis. Biochemistry. 1999;38:2088–2093. doi: 10.1021/bi982228t. [DOI] [PubMed] [Google Scholar]
- [61].Belyaeva OV, Johnson MP, Kedishvili NY. Kinetic analysis of human enzyme RDH10 defines the characteristics of a physiologically relevant retinol dehydrogenase. J Biol Chem. 2008;283:20299–20308. doi: 10.1074/jbc.M800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Heinemann FS, Ozols J. Isolation and structural analysis of microsomal membrane proteins. Front Biosci. 1998;3:d483–493. doi: 10.2741/a295. [DOI] [PubMed] [Google Scholar]
- [63].Brenner RR. Effect of unsaturated acids on membrane structure and enzyme kinetics. Prog Lipid Res. 1984;23:69–96. doi: 10.1016/0163-7827(84)90008-0. [DOI] [PubMed] [Google Scholar]
- [64].Ahn T, Kim M, Yun CH, Chae HJ. Functional regulation of hepatic cytochrome p450 enzymes by physicochemical properties of phospholipids in biological membranes. Curr Protein Pept Sci. 2007;8:496–505. doi: 10.2174/138920307782411392. [DOI] [PubMed] [Google Scholar]
- [65].Imaoka S, Wan J, Chow T, Hiroi T, Eyanagi R, Shigematsu H, Funae Y. Cloning and characterization of the CYP2D1-binding protein, retinol dehydrogenase. Arch Biochem Biophys. 1998;353:331–336. doi: 10.1006/abbi.1998.0644. [DOI] [PubMed] [Google Scholar]
- [66].Chai X, Zhai Y, Popescu G, Napoli JL. Cloning of a cDNA for a second retinol dehydrogenase type II. Expression of its mRNA relative to type I. J Biol Chem. 1995;270:28408–28412. doi: 10.1074/jbc.270.47.28408. [DOI] [PubMed] [Google Scholar]
- [67].Takahashi Y, Moiseyev G, Farjo K, Ma JX. Characterization of key residues and membrane association domains in retinol dehydrogenase 10. Biochem J. 2009;419:113–122. doi: 10.1042/BJ20080812. 111 p following 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang J, Bongianni JK, Napoli JL. The N-terminus of retinol dehydrogenase type 1 signals cytosolic orientation in the microsomal membrane. Biochemistry. 2001;40:12533–12540. doi: 10.1021/bi011396+. [DOI] [PubMed] [Google Scholar]
- [69].Zhang M, Hu P, Napoli JL. Elements in the N-terminal signaling sequence that determine cytosolic topology of short-chain dehydrogenases/reductases. Studies with retinol dehydrogenase type 1 and cis-retinol/androgen dehydrogenase type 1. J Biol Chem. 2004;279:51482–51489. doi: 10.1074/jbc.M409051200. [DOI] [PubMed] [Google Scholar]
- [70].Belyaeva OV, Stetsenko AV, Nelson P, Kedishvili NY. Properties of short-chain dehydrogenase/reductase RalR1: characterization of purified enzyme, its orientation in the microsomal membrane, and distribution in human tissues and cell lines. Biochemistry. 2003;42:14838–14845. doi: 10.1021/bi035288u. [DOI] [PubMed] [Google Scholar]
- [71].Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Romert A, Tuvendal P, Tryggvason K, Dencker L, Eriksson U. Gene structure, expression analysis, and membrane topology of RDH4. Exp Cell Res. 2000;256:338–345. doi: 10.1006/excr.2000.4817. [DOI] [PubMed] [Google Scholar]
- [73].Taibi G, Di Gaudio F, Nicotra CM. Xanthine dehydrogenase processes retinol to retinoic acid in human mammary epithelial cells. J Enzyme Inhib Med Chem. 2008;23:317–327. doi: 10.1080/14756360701584539. [DOI] [PubMed] [Google Scholar]
- [74].Taibi G, Nicotra CM. Xanthine oxidase catalyzes the oxidation of retinol. J Enzyme Inhib Med Chem. 2007;22:471–476. doi: 10.1080/14756360701408739. [DOI] [PubMed] [Google Scholar]
- [75].Lee MO, Manthey CL, Sladek NE. Identification of mouse liver aldehyde dehydrogenases that catalyze the oxidation of retinaldehyde to retinoic acid. Biochem Pharmacol. 1991;42:1279–1285. doi: 10.1016/0006-2952(91)90266-8. [DOI] [PubMed] [Google Scholar]
- [76].Sladek NE, Manthey CL, Maki PA, Zhang Z, Landkamer GJ. Xenobiotic oxidation catalyzed by aldehyde dehydrogenases. Drug Metab Rev. 1989;20:697–720. doi: 10.3109/03602538909103572. [DOI] [PubMed] [Google Scholar]
- [77].Dunn TJ, Koleske AJ, Lindahl R, Pitot HC. Phenobarbital-inducible aldehyde dehydrogenase in the rat. cDNA sequence and regulation of the mRNA by phenobarbital in responsive rats. J Biol Chem. 1989;264:13057–13065. [PubMed] [Google Scholar]
- [78].Posch KC, Burns RD, Napoli JL. Biosynthesis of all-trans-retinoic acid from retinal. Recognition of retinal bound to cellular retinol binding protein (type I) as substrate by a purified cytosolic dehydrogenase. J Biol Chem. 1992;267:19676–19682. [PubMed] [Google Scholar]
- [79].McCaffery P, Lee MO, Wagner MA, Sladek NE, Drager UC. Asymmetrical retinoic acid synthesis in the dorsoventral axis of the retina. Development. 1992;115:371–382. doi: 10.1242/dev.115.2.371. [DOI] [PubMed] [Google Scholar]
- [80].Labrecque J, Bhat PV, Lacroix A. Purification and partial characterization of a rat kidney aldehyde dehydrogenase that oxidizes retinal to retinoic acid. Biochem Cell Biol. 1993;71:85–89. doi: 10.1139/o93-013. [DOI] [PubMed] [Google Scholar]
- [81].Penzes P, Wang X, Napoli JL. Enzymatic characteristics of retinal dehydrogenase type I expressed in Escherichia coli. Biochim Biophys Acta. 1997;1342:175–181. doi: 10.1016/s0167-4838(97)00102-7. [DOI] [PubMed] [Google Scholar]
- [82].Penzes P, Wang X, Sperkova Z, Napoli JL. Cloning of a rat cDNA encoding retinal dehydrogenase isozyme type I and its expression in E. coli. Gene. 1997;191:167–172. doi: 10.1016/s0378-1119(97)00054-1. [DOI] [PubMed] [Google Scholar]
- [83].Bhat PV, Labrecque J, Boutin JM, Lacroix A, Yoshida A. Cloning of a cDNA encoding rat aldehyde dehydrogenase with high activity for retinal oxidation. Gene. 1995;166:303–306. doi: 10.1016/0378-1119(96)81752-5. [DOI] [PubMed] [Google Scholar]
- [84].Wang X, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J Biol Chem. 1996;271:16288–16293. doi: 10.1074/jbc.271.27.16288. [DOI] [PubMed] [Google Scholar]
- [85].Zhao D, McCaffery P, Ivins KJ, Neve RL, Hogan P, Chin WW, Drager UC. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- [86].Zhai Y, Sperkova Z, Napoli JL. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin A status on testis mRNA. J Cell Physiol. 2001;186:220–232. doi: 10.1002/1097-4652(200102)186:2<220::AID-JCP1018>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [87].Grun F, Hirose Y, Kawauchi S, Ogura T, Umesono K. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J Biol Chem. 2000;275:41210–41218. doi: 10.1074/jbc.M007376200. [DOI] [PubMed] [Google Scholar]
- [88].Niederreither K, Fraulob V, Garnier JM, Chambon P, Dolle P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- [89].Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- [90].Ong DE. Cellular retinoid-binding proteins. Arch Dermatol. 1987;123:1693–1695a. [PubMed] [Google Scholar]
- [91].Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348(Pt 3):481–495. [PMC free article] [PubMed] [Google Scholar]
- [92].Budhu A, Gillilan R, Noy N. Localization of the RAR interaction domain of cellular retinoic acid binding protein-II. J Mol Biol. 2001;305:939–949. doi: 10.1006/jmbi.2000.4340. [DOI] [PubMed] [Google Scholar]
- [93].Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- [95].Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18:343–353. doi: 10.1016/j.molcel.2005.03.026. [DOI] [PubMed] [Google Scholar]
- [96].Lin Q, Ruuska SE, Shaw NS, Dong D, Noy N. Ligand selectivity of the peroxisome proliferator-activated receptor alpha. Biochemistry. 1999;38:185–190. doi: 10.1021/bi9816094. [DOI] [PubMed] [Google Scholar]
- [97].Shaw N, Elholm M, Noy N. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J Biol Chem. 2003;278:41589–41592. doi: 10.1074/jbc.C300368200. [DOI] [PubMed] [Google Scholar]
- [98].Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46:13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- [100].Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fiorella PD, Napoli JL. Expression of cellular retinoic acid binding protein (CRABP) in Escherichia coli. Characterization and evidence that holo-CRABP is a substrate in retinoic acid metabolism. J Biol Chem. 1991;266:16572–16579. [PubMed] [Google Scholar]
- [102].Boylan JF, Gudas LJ. Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells. J Cell Biol. 1991;112:965–979. doi: 10.1083/jcb.112.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Boylan JF, Gudas LJ. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem. 1992;267:21486–21491. [PubMed] [Google Scholar]
- [104].Won JY, Nam EC, Yoo SJ, Kwon HJ, Um SJ, Han HS, Kim SH, Byun Y, Kim SY. The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma. Metabolism. 2004;53:1007–1012. doi: 10.1016/j.metabol.2003.12.015. [DOI] [PubMed] [Google Scholar]
- [105].Fiorella PD, Napoli JL. Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J Biol Chem. 1994;269:10538–10544. [PubMed] [Google Scholar]
- [106].Frolik CA, Roberts AB, Tavela TE, Roller PP, Newton DL, Sporn MB. Isolation and identification of 4-hydroxy- and 4-oxoretinoic acid. In vitro metabolites of all-trans-retinoic acid in hamster trachea and liver. Biochemistry. 1979;18:2092–2097. doi: 10.1021/bi00577a039. [DOI] [PubMed] [Google Scholar]
- [107].Frolik CA, Roller PP, Roberts AB, Sporn MB. In vitro and in vivo metabolism of all-trans- and 13-cis-retinoic acid in hamsters. Identification of 13-cis-4-oxoretinoic acid. J Biol Chem. 1980;255:8057–8062. [PubMed] [Google Scholar]
- [108].Roberts AB, Nichols MD, Newton DL, Sporn MB. In vitro metabolism of retinoic acid in hamster intestine and liver. J Biol Chem. 1979;254:6296–6302. [PubMed] [Google Scholar]
- [109].McCormick AM, Kroll KD, Napoli JL. 13-cis-retinoic acid metabolism in vivo. The major tissue metabolites in the rat have the all-trans configuration. Biochemistry. 1983;22:3933–3940. doi: 10.1021/bi00285a032. [DOI] [PubMed] [Google Scholar]
- [110].Napoli JL, Khalil H, McCormick AM. Metabolism of 5,6-epoxyretinoic acid in vivo: isolation of a major intestinal metabolite. Biochemistry. 1982;21:1942–1949. doi: 10.1021/bi00537a038. [DOI] [PubMed] [Google Scholar]
- [111].McCormick AM, Napoli JL. Identification of 5,6-epoxyretinoic acid as an endogenous retinol metabolite. J Biol Chem. 1982;257:1730–1735. [PubMed] [Google Scholar]
- [112].Chithalen JV, Luu L, Petkovich M, Jones G. HPLC-MS/MS analysis of the products generated from all-trans-retinoic acid using recombinant human CYP26A. J Lipid Res. 2002;43:1133–1142. doi: 10.1194/jlr.m100343-jlr200. [DOI] [PubMed] [Google Scholar]
- [113].Ray WJ, Bain G, Yao M, Gottlieb DI. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J Biol Chem. 1997;272:18702–18708. doi: 10.1074/jbc.272.30.18702. [DOI] [PubMed] [Google Scholar]
- [114].White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem. 1997;272:18538–18541. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- [115].McSorley LC, Daly AK. Identification of human cytochrome P450 isoforms that contribute to all-trans-retinoic acid 4-hydroxylation. Biochem Pharmacol. 2000;60:517–526. doi: 10.1016/s0006-2952(00)00356-7. [DOI] [PubMed] [Google Scholar]
- [116].Nelson DR. A second CYP26 P450 in humans and zebrafish: CYP26B1. Arch Biochem Biophys. 1999;371:345–347. doi: 10.1006/abbi.1999.1438. [DOI] [PubMed] [Google Scholar]
- [117].Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol. 2009;5:875–886. doi: 10.1517/17425250903032681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, Korczak B, Petkovich M. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and all-trans isomers of retinoic acid. J Biol Chem. 2004;279:77–85. doi: 10.1074/jbc.M308337200. [DOI] [PubMed] [Google Scholar]
- [119].Andreola F, Hayhurst GP, Luo G, Ferguson SS, Gonzalez FJ, Goldstein JA, De Luca LM. Mouse liver CYP2C39 is a novel retinoic acid 4-hydroxylase. Its down-regulation offers a molecular basis for liver retinoid accumulation and fibrosis in aryl hydrocarbon receptor-null mice. J Biol Chem. 2004;279:3434–3438. doi: 10.1074/jbc.M305832200. [DOI] [PubMed] [Google Scholar]
- [120].Qian L, Zolfaghari R, Ross AC. Liver-specific cytochrome P450 CYP2C22 is a direct target of retinoic acid and a retinoic acid-metabolizing enzyme in rat liver. J Lipid Res. 2010;51:1781–1792. doi: 10.1194/jlr.M002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Liu L, Gudas LJ. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J Biol Chem. 2005;280:40226–40234. doi: 10.1074/jbc.M509643200. [DOI] [PubMed] [Google Scholar]
- [123].Liu L, Tang XH, Gudas LJ. Homeostasis of retinol in lecithin: retinol acyltransferase gene knockout mice fed a high retinol diet. Biochem Pharmacol. 2008;75:2316–2324. doi: 10.1016/j.bcp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kim YK, Wassef L, Hamberger L, Piantedosi R, Palczewski K, Blaner WS, Quadro L. Retinyl ester formation by lecithin: retinol acyltransferase is a key regulator of retinoid homeostasis in mouse embryogenesis. J Biol Chem. 2008;283:5611–5621. doi: 10.1074/jbc.M708885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zolfaghari R, Ross AC. Lecithin:retinol acyltransferase from mouse and rat liver. CDNA cloning and liver-specific regulation by dietary vitamin a and retinoic acid. J Lipid Res. 2000;41:2024–2034. [PubMed] [Google Scholar]
- [126].Matsuura T, Gad MZ, Harrison EH, Ross AC. Lecithin:retinol acyltransferase and retinyl ester hydrolase activities are differentially regulated by retinoids and have distinct distributions between hepatocyte and nonparenchymal cell fractions of rat liver. J Nutr. 1997;127:218–224. doi: 10.1093/jn/127.2.218. [DOI] [PubMed] [Google Scholar]
- [127].Jurukovski V, Simon M. Reduced lecithin:retinol acyl transferase activity in cultured squamous cell carcinoma lines results in increased substrate-driven retinoic acid synthesis. Biochim Biophys Acta. 1999;1436:479–490. doi: 10.1016/s0005-2760(98)00154-4. [DOI] [PubMed] [Google Scholar]
- [128].Ross AC, Cifelli CJ, Zolfaghari R, Li NQ. Multiple cytochrome P-450 genes are concomitantly regulated by vitamin A under steady-state conditions and by retinoic acid during hepatic first-pass metabolism. Physiol Genomics. 2011;43:57–67. doi: 10.1152/physiolgenomics.00182.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wu L, Ross AC. Acidic retinoids synergize with vitamin A to enhance retinol uptake and STRA6, LRAT, and CYP26B1 expression in neonatal lung. J Lipid Res. 2010;51:378–387. doi: 10.1194/jlr.M001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63:173–186. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- [131].Roberts AB, Frolik CA, Nichols MD, Sporn MB. Retinoid-dependent induction of the in vivo and in vitro metabolism of retinoic acid in tissues of the vitamin A-deficient hamster. J Biol Chem. 1979;254:6303–6309. [PubMed] [Google Scholar]
- [132].Yamamoto Y, Zolfaghari R, Ross AC. Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. FASEB J. 2000;14:2119–2127. doi: 10.1096/fj.00-0061com. [DOI] [PubMed] [Google Scholar]
- [133].Ross AC. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic Acid oxidation. J Nutr. 2003;133:291S–296S. doi: 10.1093/jn/133.1.291S. [DOI] [PubMed] [Google Scholar]
- [134].Wang C, Napoli JL. Multiple retinol and retinal dehydrogenases catalyze all-trans-retinoic acid biosynthesis in astrocytes. J Biol Chem. 2010 doi: 10.1074/jbc.M110.198382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 2009;50(Suppl):S138–143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kane MA, Napoli JL. Quantification of endogenous retinoids. Methods Mol Biol. 2010;652:1–54. doi: 10.1007/978-1-60327-325-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, Napoli JL, Peterson AS, Pleasure SJ. Retinoic acid from the meninges regulates cortical neuron generation. Cell. 2009;139:597–609. doi: 10.1016/j.cell.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Yoshiki A, Moriwaki K. Mouse phenome research: implications of genetic background. ILAR J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]
- [139].Guenet JL, Bonhomme F. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- [140].Harper JM, Durkee SJ, Dysko RC, Austad SN, Miller RA. Genetic modulation of hormone levels and life span in hybrids between laboratory and wild-derived mice. J Gerontol A Biol Sci Med Sci. 2006;61:1019–1029. doi: 10.1093/gerona/61.10.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Miller RA, Harper JM, Dysko RC, Durkee SJ, Austad SN. Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med (Maywood) 2002;227:500–508. doi: 10.1177/153537020222700715. [DOI] [PubMed] [Google Scholar]
- [142].Ratnam KK, Feng X, Chuang PY, Verma V, Lu TC, Wang J, Jin Y, Farias EF, Napoli JL, Chen N, Kaufman L, Takano T, D’Agati VD, Klotman PE, He JC. Role of the retinoic acid receptor-alpha in HIV-associated nephropathy. Kidney Int. 2011;79:624–634. doi: 10.1038/ki.2010.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Wu BX, Chen Y, Fan J, Rohrer B, Crouch RK, Ma JX. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest Ophthalmol Vis Sci. 2002;43:3365–3372. [PubMed] [Google Scholar]
- [144].Wu BX, Moiseyev G, Chen Y, Rohrer B, Crouch RK, Ma JX. Identification of RDH10, an All-trans Retinol Dehydrogenase, in Retinal Muller Cells. Invest Ophthalmol Vis Sci. 2004;45:3857–3862. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]
- [145].Cammas L, Romand R, Fraulob V, Mura C, Dolle P. Expression of the murine retinol dehydrogenase 10 (Rdh10) gene correlates with many sites of retinoid signalling during embryogenesis and organ differentiation. Dev Dyn. 2007;236:2899–2908. doi: 10.1002/dvdy.21312. [DOI] [PubMed] [Google Scholar]
- [146].Romand R, Kondo T, Cammas L, Hashino E, Dolle P. Dynamic expression of the retinoic acid-synthesizing enzyme retinol dehydrogenase 10 (rdh10) in the developing mouse brain and sensory organs. J Comp Neurol. 2008;508:879–892. doi: 10.1002/cne.21707. [DOI] [PubMed] [Google Scholar]
- [147].Szatmari I, Pap A, Ruhl R, Ma JX, Illarionov PA, Besra GS, Rajnavolgyi E, Dezso B, Nagy L. PPARgamma controls CD1d expression by turning on retinoic acid synthesis in developing human dendritic cells. J Exp Med. 2006;203:2351–2362. doi: 10.1084/jem.20060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- [149].Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Song MS, Chen W, Zhang M, Napoli JL. Identification of a mouse short-chain dehydrogenase/reductase gene, retinol dehydrogenase-similar. Function of non-catalytic amino acid residues in enzyme activity. J Biol Chem. 2003;278:40079–40087. doi: 10.1074/jbc.M304910200. [DOI] [PubMed] [Google Scholar]
- [151].Nadauld LD, Shelton DN, Chidester S, Yost HJ, Jones DA. The zebrafish retinol dehydrogenase, rdh1l, is essential for intestinal development and is regulated by the tumor suppressor adenomatous polyposis coli. J Biol Chem. 2005;280:30490–30495. doi: 10.1074/jbc.M504973200. [DOI] [PubMed] [Google Scholar]
- [152].Jette C, Peterson PW, Sandoval IT, Manos EJ, Hadley E, Ireland CM, Jones DA. The tumor suppressor adenomatous polyposis coli and caudal related homeodomain protein regulate expression of retinol dehydrogenase L. J Biol Chem. 2004;279:34397–34405. doi: 10.1074/jbc.M314021200. [DOI] [PubMed] [Google Scholar]
- [153].Limongi ZM, Curatolo A, Pelliccia F, Rocchi A. Biallelic deletion and loss of expression analysis of genes at FRA2G common fragile site in tumor-derived cell lines. Cancer Genet Cytogenet. 2005;161:181–186. doi: 10.1016/j.cancergencyto.2005.01.018. [DOI] [PubMed] [Google Scholar]
- [154].Rexer BN, Ong DE. A novel short-chain alcohol dehydrogenase from rats with retinol dehydrogenase activity, cyclically expressed in uterine epithelium. Biol Reprod. 2002;67:1555–1564. doi: 10.1095/biolreprod.102.007021. [DOI] [PubMed] [Google Scholar]
- [155].Li XH, Kakkad B, Ong DE. Estrogen directly induces expression of retinoic acid biosynthetic enzymes, compartmentalized between the epithelium and underlying stromal cells in rat uterus. Endocrinology. 2004;145:4756–4762. doi: 10.1210/en.2004-0514. [DOI] [PubMed] [Google Scholar]
- [156].Parker RO, Crouch RK. Retinol dehydrogenases (RDHs) in the visual cycle. Exp Eye Res. 2010;91:788–792. doi: 10.1016/j.exer.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Chrispell JD, Feathers KL, Kane MA, Kim CY, Brooks M, Khanna R, Kurth I, Hubner CA, Gal A, Mears AJ, Swaroop A, Napoli JL, Sparrow JR, Thompson DA. Rdh12 activity and effects on retinoid processing in the murine retina. J Biol Chem. 2009;284:21468–21477. doi: 10.1074/jbc.M109.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, Kedishvili NY. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry. 2005;44:7035–7047. doi: 10.1021/bi050226k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J Biol Chem. 1998;273:21790–21799. doi: 10.1074/jbc.273.34.21790. [DOI] [PubMed] [Google Scholar]
- [160].Rattner A, Smallwood PM, Nathans J. Identification and characterization of all-trans-retinol dehydrogenase from photoreceptor outer segments, the visual cycle enzyme that reduces all-trans-retinal to all-trans-retinol. J Biol Chem. 2000;275:11034–11043. doi: 10.1074/jbc.275.15.11034. [DOI] [PubMed] [Google Scholar]
- [161].Farjo KM, Moiseyev G, Takahashi Y, Crouch RK, Ma JX. The 11-cis-retinol dehydrogenase activity of RDH10 and its interaction with visual cycle proteins. Invest Ophthalmol Vis Sci. 2009;50:5089–5097. doi: 10.1167/iovs.09-3797. [DOI] [PubMed] [Google Scholar]
- [162].Matt N, Dupe V, Garnier JM, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- [163].el Akawi Z, Napoli JL. Rat liver cytosolic retinal dehydrogenase: comparison of 13-cis-, 9-cis-, and all-trans-retinal as substrates and effects of cellular retinoid-binding proteins and retinoic acid on activity. Biochemistry. 1994;33:1938–1943. doi: 10.1021/bi00173a042. [DOI] [PubMed] [Google Scholar]
- [164].Yoshida A, Hsu LC, Yanagawa Y. Biological role of human cytosolic aldehyde dehydrogenase 1: hormonal response, retinal oxidation and implication in testicular feminization. Adv Exp Med Biol. 1993;328:37–44. doi: 10.1007/978-1-4615-2904-0_5. [DOI] [PubMed] [Google Scholar]
- [165].Lopez-Fernandez LA, del Mazo J. The cytosolic aldehyde dehydrogenase gene (Aldh1) is developmentally expressed in Leydig cells. FEBS Lett. 1997;407:225–229. doi: 10.1016/s0014-5793(97)00352-9. [DOI] [PubMed] [Google Scholar]
- [166].Starkey JM, Zhao Y, Sadygov RG, Haidacher SJ, Lejeune WS, Dey N, Luxon BA, Kane MA, Napoli JL, Denner L, Tilton RG. Altered retinoic acid metabolism in diabetic mouse kidney identified by O isotopic labeling and 2D mass spectrometry. PLoS One. 2010;5:e11095. doi: 10.1371/journal.pone.0011095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, Tang G, Dolnikowski GG, Vogel S, Duester G, Plutzky J. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Reichert B, Yasmeen R, Jeyakumar SM, Yang F, Thomou T, Alder H, Duester G, Maiseyeu A, Mihai G, Harrison EH, Rajagopalan S, Kirkland JL, Ziouzenkova O. Concerted Action of Aldehyde Dehydrogenases Influences Depot-Specific Fat Formation. Mol Endocrinol. 2011 doi: 10.1210/me.2010-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Pereira F, Rosenmann E, Nylen E, Kaufman M, Pinsky L, Wrogemann K. The 56 kDa androgen binding protein is an aldehyde dehydrogenase. Biochem Biophys Res Commun. 1991;175:831–838. doi: 10.1016/0006-291x(91)91640-x. [DOI] [PubMed] [Google Scholar]
- [170].Pereira F, Belsham D, Duerksen K, Rosenmann E, Kaufman M, Pinsky L, Wrogemann K. The 56 kDa androgen-binding protein in human genital skin fibroblasts: its relation to the human androgen receptor. Mol Cell Endocrinol. 1990;68:195–204. doi: 10.1016/0303-7207(90)90193-c. [DOI] [PubMed] [Google Scholar]
- [171].Yoshida A, Dave V, Han H, Scanlon KJ. Enhanced transcription of the cytosolic ALDH gene in cyclophosphamide resistant human carcinoma cells. Adv Exp Med Biol. 1993;328:63–72. doi: 10.1007/978-1-4615-2904-0_8. [DOI] [PubMed] [Google Scholar]
- [172].Pereira FA, Rosenmann E, Nylen EG, Wrogemann K. Human cytosolic aldehyde dehydrogenase in androgen insensitivity syndrome. Adv Exp Med Biol. 1993;328:45–50. doi: 10.1007/978-1-4615-2904-0_6. [DOI] [PubMed] [Google Scholar]
- [173].Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 2007;282:25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Elizondo G, Corchero J, Sterneck E, Gonzalez FJ. Feedback inhibition of the retinaldehyde dehydrogenase gene ALDH1 by retinoic acid through retinoic acid receptor alpha and CCAAT/enhancer-binding protein beta. J Biol Chem. 2000;275:39747–39753. doi: 10.1074/jbc.M004987200. [DOI] [PubMed] [Google Scholar]
- [175].Guimond J, Devost D, Brodeur H, Mader S, Bhat PV. Characterization of the rat RALDH1 promoter. A functional CCAAT and octamer motif are critical for basal promoter activity. Biochim Biophys Acta. 2002;1579:81–91. doi: 10.1016/s0167-4781(02)00510-9. [DOI] [PubMed] [Google Scholar]
- [176].Elizondo G, Medina-Diaz IM, Cruz R, Gonzalez FJ, Vega L. Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPbeta interaction. Biochem Pharmacol. 2009;77:248–257. doi: 10.1016/j.bcp.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Fujiwara K, Kikuchi M, Takigami S, Kouki T, Yashiro T. Expression of retinaldehyde dehydrogenase 1 in the anterior pituitary glands of adult rats. Cell Tissue Res. 2007;329:321–327. doi: 10.1007/s00441-007-0423-5. [DOI] [PubMed] [Google Scholar]
- [178].Fujiwara K, Kikuchi M, Horiguchi K, Kusumoto K, Kouki T, Kawanishi K, Yashiro T. Estrogen receptor alpha regulates retinaldehyde dehydrogenase 1 expression in rat anterior pituitary cells. Endocr J. 2009;56:963–973. doi: 10.1507/endocrj.k09e-115. [DOI] [PubMed] [Google Scholar]
- [179].Fujiwara K, Davaadash B, Kikuchi M, Takigami S, Yashiro T. Estrogen suppresses retinaldehyde dehydrogenase 1 expression in the anterior pituitary glands of female rats. Endocr J. 2008;55:91–96. doi: 10.1507/endocrj.k07-101. [DOI] [PubMed] [Google Scholar]
- [180].Kane MA, Folias AE, Wang C, Napoli JL. Ethanol elevates physiological all-trans-retinoic acid levels in select loci through altering retinoid metabolism in multiple loci: a potential mechanism of ethanol toxicity. FASEB J. 2010;24:823–832. doi: 10.1096/fj.09-141572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Calvert-Evers JL, Hammond KD. Temporal variations in protein tyrosine phosphatase activity during cell proliferation and differentiation. Cell Biol Int. 2000;24:559–568. doi: 10.1006/cbir.2000.0570. [DOI] [PubMed] [Google Scholar]
- [182].Chen Y, Tang CE, Ouyang GL, Ruan L, Li MY, Zhang PF, Li C, Yi H, Peng F, Li JL, Chen ZC, Xiao ZQ. Identification of RKIP as a differentially tyrosine-phosphorylated protein in nasopharyngeal carcinoma and normal nasopharyngeal epithelial tissues by phosphoproteomic approach. Med Oncol. 2009;26:463–470. doi: 10.1007/s12032-008-9147-y. [DOI] [PubMed] [Google Scholar]
- [183].Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, Connolly DC, Zhang Y, Montone K, Butzow R, Coukos G, Zhang L. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Ma I, Allan AL. The Role of Human Aldehyde Dehydrogenase in Normal and Cancer Stem Cells. Stem Cell Rev. 2010 doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- [185].Ostrom M, Loffler KA, Edfalk S, Selander L, Dahl U, Ricordi C, Jeon J, Correa-Medina M, Diez J, Edlund H. Retinoic acid promotes the generation of pancreatic endocrine progenitor cells and their further differentiation into beta-cells. PLoS One. 2008;3:e2841. doi: 10.1371/journal.pone.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- [187].Mey J, Babiuk RP, Clugston R, Zhang W, Greer JJ. Retinal dehydrogenase-2 is inhibited by compounds that induce congenital diaphragmatic hernias in rodents. Am J Pathol. 2003;162:673–679. doi: 10.1016/s0002-9440(10)63861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Hsu LC, Chang WC, Yoshida A. Mouse type-2 retinaldehyde dehydrogenase (RALDH2): genomic organization, tissue-dependent expression, chromosome assignment and comparison to other types. Biochim Biophys Acta. 2000;1492:289–293. doi: 10.1016/s0167-4781(00)00108-1. [DOI] [PubMed] [Google Scholar]
- [189].Niederreither K, Vermot J, Fraulob V, Chambon P, Dolle P. Retinaldehyde dehydrogenase 2 (RALDH2)- independent patterns of retinoic acid synthesis in the mouse embryo. Proc Natl Acad Sci U S A. 2002;99:16111–16116. doi: 10.1073/pnas.252626599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Wang X, Sperkova Z, Napoli JL. Analysis of mouse retinal dehydrogenase type 2 promoter and expression. Genomics. 2001;74:245–250. doi: 10.1006/geno.2001.6546. [DOI] [PubMed] [Google Scholar]
- [191].Maly IP, Crotet V, Toranelli M. The so-called “testis-specific aldehyde dehydrogenase” corresponds to type 2 retinaldehyde dehydrogenase in the mouse. Histochem Cell Biol. 2003;119:169–174. doi: 10.1007/s00418-002-0488-x. [DOI] [PubMed] [Google Scholar]
- [192].Mendelsohn C, Batourina E, Fung S, Gilbert T, Dodd J. Stromal cells mediate retinoid-dependent functions essential for renal development. Development. 1999;126:1139–1148. doi: 10.1242/dev.126.6.1139. [DOI] [PubMed] [Google Scholar]
- [193].El Kares R, Manolescu DC, Lakhal-Chaieb L, Montpetit A, Zhang Z, Bhat PV, Goodyer P. A human ALDH1A2 gene variant is associated with increased newborn kidney size and serum retinoic acid. Kidney Int. 2010;78:96–102. doi: 10.1038/ki.2010.101. [DOI] [PubMed] [Google Scholar]
- [194].Ogura Y, Suruga K, Mochizuki H, Yamamoto T, Takase S, Goda T. Postnatal changes in gene expression of retinal dehydrogenase and retinoid receptors in liver of rats. Life Sci. 2004;74:1519–1528. doi: 10.1016/j.lfs.2003.08.020. [DOI] [PubMed] [Google Scholar]
- [195].Vermot J, Fraulob V, Dolle P, Niederreither K. Expression of enzymes synthesizing (aldehyde dehydrogenase 1 and reinaldehyde dehydrogenase 2) and metabolizaing (Cyp26) retinoic acid in the mouse female reproductive system. Endocrinology. 2000;141:3638–3645. doi: 10.1210/endo.141.10.7696. [DOI] [PubMed] [Google Scholar]
- [196].Ruhl R, Fritzsche B, Vermot J, Niederreither K, Neumann U, Schmidt A, Schweigert FJ, Dolle P. Regulation of expression of the retinoic acid-synthesising enzymes retinaldehyde dehydrogenases in the uteri of ovariectomised mice after treatment with oestrogen, gestagen and their combination. Reprod Fertil Dev. 2006;18:339–345. doi: 10.1071/rd05056. [DOI] [PubMed] [Google Scholar]
- [197].Kern J, Schrage K, Koopmans GC, Joosten EA, McCaffery P, Mey J. Characterization of retinaldehyde dehydrogenase-2 induction in NG2-positive glia after spinal cord contusion injury. Int J Dev Neurosci. 2007;25:7–16. doi: 10.1016/j.ijdevneu.2006.11.006. [DOI] [PubMed] [Google Scholar]
- [198].Guadix JA, Ruiz-Villalba A, Lettice L, Velecela V, Munoz-Chapuli R, Hastie ND, Perez-Pomares JM, Martinez-Estrada OM. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Ono Y, Fukuhara N, Yoshie O. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol Cell Biol. 1998;18:6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Huq MD, Tsai NP, Gupta P, Wei LN. Regulation of retinal dehydrogenases and retinoic acid synthesis by cholesterol metabolites. EMBO J. 2006;25:3203–3213. doi: 10.1038/sj.emboj.7601181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Sima A, Parisotto M, Mader S, Bhat PV. Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim Biophys Acta. 2009;1790:1660–1664. doi: 10.1016/j.bbagen.2009.09.004. [DOI] [PubMed] [Google Scholar]
- [203].Shimamura M, Karasawa H, Sakakibara S, Shinagawa A. Raldh3 expression in diabetic islets reciprocally regulates secretion of insulin and glucagon from pancreatic islets. Biochem Biophys Res Commun. 2010;401:79–84. doi: 10.1016/j.bbrc.2010.09.013. [DOI] [PubMed] [Google Scholar]
- [204].Kane MA, Folias AE, Pingitore A, Perri M, Obrochta KM, Krois CR, Cione E, Ryu JY, Napoli JL. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1008859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Everts HB, King LE, Jr., Sundberg JP, Ong DE. Hair cycle-specific immunolocalization of retinoic acid synthesizing enzymes Aldh1a2 and Aldh1a3 indicate complex regulation. J Invest Dermatol. 2004;123:258–263. doi: 10.1111/j.0022-202X.2004.23223.x. [DOI] [PubMed] [Google Scholar]
- [206].Mira YLR, Zheng WL, Kuppumbatti YS, Rexer B, Jing Y, Ong DE. Retinol conversion to retinoic acid is impaired in breast cancer cell lines relative to normal cells. J Cell Physiol. 2000;185:302–309. doi: 10.1002/1097-4652(200011)185:2<302::AID-JCP15>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- [207].Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001;61:7065–7070. [PubMed] [Google Scholar]
- [208].Koch JG, Gu X, Han Y, El-Naggar AK, Olson MV, Medina D, Jerry DJ, Blackburn AC, Peltz G, Amos CI, Lozano G. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm Genome. 2007;18:300–309. doi: 10.1007/s00335-007-9028-2. [DOI] [PubMed] [Google Scholar]
- [209].Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- [210].John K, Keshava C, Richardson DL, Weston A, Nath J. Transcriptional profiles of benzo(a)pyrene exposure in normal human mammary epithelial cells in the absence or presence of chlorophyllin. Mutat Res. 2008;640:145–152. doi: 10.1016/j.mrfmmm.2008.01.003. [DOI] [PubMed] [Google Scholar]
- [211].Kim S, Dere E, Burgoon LD, Chang CC, Zacharewski TR. Comparative analysis of AhR-mediated TCDD-elicited gene expression in human liver adult stem cells. Toxicol Sci. 2009;112:229–244. doi: 10.1093/toxsci/kfp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Pappas P, Stephanou P, Karamanakos P, Vasiliou V, Marselos M. Phenobarbital inducibility and differences in protein expression of an animal model. Chem Biol Interact. 2001;130-132:275–283. doi: 10.1016/s0009-2797(00)00271-4. [DOI] [PubMed] [Google Scholar]
- [213].Han J, Yang L, Puri RK. Analysis of target genes induced by IL-13 cytotoxin in human glioblastoma cells. J Neurooncol. 2005;72:35–46. doi: 10.1007/s11060-004-3119-7. [DOI] [PubMed] [Google Scholar]
- [214].Koenig U, Amatschek S, Mildner M, Eckhart L, Tschachler E. Aldehyde dehydrogenase 1A3 is transcriptionally activated by all-trans-retinoic acid in human epidermal keratinocytes. Biochem Biophys Res Commun. 2010;400:207–211. doi: 10.1016/j.bbrc.2010.08.035. [DOI] [PubMed] [Google Scholar]
- [215].Trasino SE, Harrison EH, Wang TT. Androgen regulation of aldehyde dehydrogenase 1A3 (ALDH1A3) in the androgen-responsive human prostate cancer cell line LNCaP. Exp Biol Med (Maywood) 2007;232:762–771. [PubMed] [Google Scholar]
- [216].Njar VC. Cytochrome p450 retinoic acid 4-hydroxylase inhibitors: potential agents for cancer therapy. Mini Rev Med Chem. 2002;2:261–269. doi: 10.2174/1389557023406223. [DOI] [PubMed] [Google Scholar]
- [217].Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res A Clin Mol Teratol. 2010;88:883–894. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- [218].Williams JB, Napoli JL. Metabolism of retinoic acid and retinol during differentiation of F9 embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1985;82:4658–4662. doi: 10.1073/pnas.82.14.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Van Wauwe JP, Coene MC, Goossens J, Van Nijen G, Cools W, Lauwers W. Ketoconazole inhibits the in vitro and in vivo metabolism of all-trans-retinoic acid. J Pharmacol Exp Ther. 1988;245:718–722. [PubMed] [Google Scholar]
- [220].Vanden Bossche H, Willemsens G, Janssen PA. Cytochrome-P-450-dependent metabolism of retinoic acid in rat skin microsomes: inhibition by ketoconazole. Skin Pharmacol. 1988;1:176–185. doi: 10.1159/000210771. [DOI] [PubMed] [Google Scholar]
- [221].Williams JB, Napoli JL. Inhibition of retinoic acid metabolism by imidazole antimycotics in F9 embryonal carcinoma cells. Biochem Pharmacol. 1987;36:1386–1388. doi: 10.1016/0006-2952(87)90102-x. [DOI] [PubMed] [Google Scholar]
- [222].Van Wauwe JP, Coene MC, Goossens J, Cools W, Monbaliu J. Effects of cytochrome P-450 inhibitors on the in vivo metabolism of all-trans-retinoic acid in rats. J Pharmacol Exp Ther. 1990;252:365–369. [PubMed] [Google Scholar]
- [223].Miller WH., Jr. The emerging role of retinoids and retinoic acid metabolism blocking agents in the treatment of cancer. Cancer. 1998;83:1471–1482. doi: 10.1002/(sici)1097-0142(19981015)83:8<1471::aid-cncr1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- [224].Patel JB, Huynh CK, Handratta VD, Gediya LK, Brodie AM, Goloubeva OG, Clement OO, Nanne IP, Soprano DR, Njar VC. Novel retinoic acid metabolism blocking agents endowed with multiple biological activities are efficient growth inhibitors of human breast and prostate cancer cells in vitro and a human breast tumor xenograft in nude mice. J Med Chem. 2004;47:6716–6729. doi: 10.1021/jm0401457. [DOI] [PubMed] [Google Scholar]
- [225].Njar VC, Gediya L, Purushottamachar P, Chopra P, Vasaitis TS, Khandelwal A, Mehta J, Huynh C, Belosay A, Patel J. Retinoic acid metabolism blocking agents (RAMBAs) for treatment of cancer and dermatological diseases. Bioorg Med Chem. 2006;14:4323–4340. doi: 10.1016/j.bmc.2006.02.041. [DOI] [PubMed] [Google Scholar]
- [226].Lotan Y, Lotan R. Prevention of bladder cancer recurrence by retinoic acid-ketoconazole: a promising strategy? Cancer Biol Ther. 2008;7:101–102. doi: 10.4161/cbt.7.1.5860. [DOI] [PubMed] [Google Scholar]
- [227].Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- [230].Han CL, Liao CS, Wu CW, Hwong CL, Lee AR, Yin SJ. Contribution to first-pass metabolism of ethanol and inhibition by ethanol for retinol oxidation in human alcohol dehydrogenase family--implications for etiology of fetal alcohol syndrome and alcohol-related diseases. Eur J Biochem. 1998;254:25–31. doi: 10.1046/j.1432-1327.1998.2540025.x. [DOI] [PubMed] [Google Scholar]
- [231].Yang ZN, Davis GJ, Hurley TD, Stone CL, Li TK, Bosron WF. Catalytic efficiency of human alcohol dehydrogenases for retinol oxidation and retinal reduction. Alcohol Clin Exp Res. 1994;18:587–591. doi: 10.1111/j.1530-0277.1994.tb00914.x. [DOI] [PubMed] [Google Scholar]
- [232].Boleda MD, Saubi N, Farres J, Pares X. Physiological substrates for rat alcohol dehydrogenase classes: aldehydes of lipid peroxidation, omega-hydroxyfatty acids, and retinoids. Arch Biochem Biophys. 1993;307:85–90. doi: 10.1006/abbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- [233].Molotkov A, Fan X, Deltour L, Foglio MH, Martras S, Farres J, Pares X, Duester G. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proc Natl Acad Sci U S A. 2002;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].McCaffery P, Koul O, Smith D, Napoli JL, Chen N, Ullman MD. Ethanol increases retinoic acid production in cerebellar astrocytes and in cerebellum. Brain Res Dev Brain Res. 2004;153:233–241. doi: 10.1016/j.devbrainres.2004.09.003. [DOI] [PubMed] [Google Scholar]
- [235].Duester G, Mic FA, Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem Biol Interact. 2003;143-144:201–210. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- [236].Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Vonesch JL, Nakshatri H, Philippe M, Chambon P, Dolle P. Stage and tissue-specific expression of the alcohol dehydrogenase 1 (Adh-1) gene during mouse development. Dev Dyn. 1994;199:199–213. doi: 10.1002/aja.1001990305. [DOI] [PubMed] [Google Scholar]
- [238].Pares X, Farres J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families : Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol Life Sci. 2008;65:3936–3949. doi: 10.1007/s00018-008-8591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Biesalski HK. Comparative assessment of the toxicology of vitamin A and retinoids in man. Toxicology. 1989;57:117–161. doi: 10.1016/0300-483x(89)90161-3. [DOI] [PubMed] [Google Scholar]
- [240].Tzimas G, Collins MD, Burgin H, Hummler H, Nau H. Embryotoxic doses of vitamin A to rabbits result in low plasma but high embryonic concentrations of all-trans-retinoic acid: risk of vitamin A exposure in humans. J Nutr. 1996;126:2159–2171. doi: 10.1093/jn/126.9.2159. [DOI] [PubMed] [Google Scholar]
- [241].Tzimas G, Thiel R, Chahoud I, Nau H. The area under the concentration-time curve of all-trans-retinoic acid is the most suitable pharmacokinetic correlate to the embryotoxicity of this retinoid in the rat. Toxicol Appl Pharmacol. 1997;143:436–444. doi: 10.1006/taap.1997.8105. [DOI] [PubMed] [Google Scholar]
- [242].Molotkov A, Ghyselinck NB, Chambon P, Duester G. Opposing actions of cellular retinol-binding protein and alcohol dehydrogenase control the balance between retinol storage and degradation. Biochem J. 2004;383:295–302. doi: 10.1042/BJ20040621. [DOI] [PMC free article] [PubMed] [Google Scholar]