Abstract
In the present study, we have performed combined molecular dynamics (MD) and potential of mean force (PMF) simulations to determine the enzyme-substrate (ES) binding pathway and the corresponding free energy profiles for wild-type butyrylcholinesterase (BChE) binding with (−)/(+)-cocaine and for the A328W/Y332G mutant binding with (−)-cocaine. According to the PMF simulations, for each ES binding system, the substrate first binds with the enzyme at a peripheral anionic site around the entrance of the active site gorge to form the first ES complex (ES1-like) during the binding process. Further evolution from the ES1-like complex to the nonprereactive ES complex is nearly barrierless, with a free energy barrier being lower than 1.0 kcal/mol. So, the nonprereactive ES binding process should be very fast. The rate-determining step of the entire ES binding process is the subsequent evolution from the nonprereactive ES complex to the prereactive ES complex. Further accounting for the entire ES binding process, the PMF-based simulations qualitatively reproduced the relative order of the experimentally-derived binding free energies (ΔGbind), although the simulations systematically overestimated the magnitude of the binding affinity and systematically underestimated the differences between the ΔGbind values. The obtained structural and energetic insights into the entire ES binding process provide a valuable base for future rational design of high-activity mutants of BChE as candidates of an enzyme therapy for cocaine overdose and abuse.
Introduction
The naturally occurring and biologically active (−)-cocaine is well-known as the most-psychostimulant drug abused by millions of people worldwide.1,2,3,4 The disastrous medical and social consequences of cocaine addiction, such as increasing crimes, medical expense, and loss of lives, have made the development of an effective pharmacological treatment of cocaine abuse a high priority. 5 , 6 However, traditional pharmacodynamic approach has failed to yield a therapeutically useful small-molecule drug due to the difficulties inherent in blocking a blocker.2,3,4,6 Human butyrylcholinesterase (BChE) is the principal plasma enzyme that catalyzes the hydrolysis of cocaine into biologically inactive metabolites; thus, the BChE-catalyzed hydrolysis of (−)-cocaine is the most suitable metabolic pathway for amplification in development of a therapeutic enzyme as an anti-cocaine medication.7,8,9 However, wild-type BChE has a low catalytic efficiency against (−)-cocaine. The catalytic efficiency of wild-type BChE against (−)-cocaine is three-orders-of-magnitude lower than that against the biologically inactive (+)-cocaine.8,10 (+)-Cocaine (Scheme 1) can be cleared from plasma in seconds and prior to penetrating into the central nervous system, whereas (−)-cocaine has a plasma half-life of ~47 min or longer even for a (−)-cocaine dose as low as 0.2 mg/kg (i.v.). 11 Based on these experimental observations, it is important to understand structurally and energetically how BChE binds with cocaine enantiomers.
Scheme 1.
There are two forms of cholinesterase that coexist ubiquitously throughout our human body, one is BChE and another is acetylcholinesterase (AChE). The two enzymes share a 65% homology.12,13 AChE is responsible for the hydrolysis of neurotransmitter acetylcholine, which is released in the synaptic cleft and the neuromuscular junction.14 The catalytic efficiency of AChE against acetylcholine is close to the known diffusion-controlled rate. 15 The X-ray crystal structure of AChE12 showed that the catalytic site is located at the bottom of a 20 Å deep gorge lined mostly with aromatic residues. AChE has a peripheral anionic site (PAS) located near the entrance of the gorge.16 Ligands targeting both the active site and the PAS, or either of these two sites of AChE have been found to be potential therapeutics for Alzheimer disease.17,18 Compared with AChE, BChE is relatively abundant in plasma (~3 mg/L), and is able to hydrolyze a large number of ester-containing compounds including acetylcholine.19 The main difference between the structures of BChE and AChE exists in the residues lining the gorge. The gorge of BChE is much wider than that of AChE, as 6 of 14 aromatic residues lining the gorge of AChE are replaced by less bulky residues in BChE. Based on the structural similarity between BChE and AChE in the shape and composition of the gorge, one wants to know whether or not BChE also has a PAS around residue D70 corresponding to residue D74 at the PAS of AChE. Masson et al.20,21 proposed that BChE could form three discrete enzyme-substrate (ES) complexes for a substrate. The first one (ES1) forms when substrate binds with residue D70. The second ES complex, ES2, forms when the substrate slides down the substrate-binding gorge to bind with residue W82 and stayed vertically in the gorge between D70 and W82. The third ES complex, ES3, forms when the substrate rotates to a horizontal position at the bottom of the gorge to come within a favorable distance for nucleophilic attack and hydrolysis by residue S198. The question is whether the existence of the three discrete enzyme-substrate complexes applies to the binding pathway for BChE with (−)- and (+)-cocaine.
We performed extensive molecular dynamics (MD) simulations 22 , 23 , 24 , 25 and reaction coordinate calculations8,24 in order to uncover the fundamental reaction mechanism for BChE-catalyzed hydrolysis of (−)-cocaine. Our studies revealed that the BChE-(−)-cocaine binding involves two types of stable complexes, i.e. nonprereactive BChE-(−)-cocaine complex and prereactive BChE-(−)-cocaine complex. The nonprereactive complex is the initial enzyme-substrate (ES) binding complex which is not suitable for chemical reaction. In the nonprereactive complex, (−)-cocaine molecule stays vertically inside the substrate-binding gorge between residues D70 and W82 of the enzyme, while the cationic head of (−)-cocaine is close to W82 and other surrounding residues like E197 and Y440. After the initial, nonprereactive BChE-(−)cocaine binding, (−)-cocaine needs to rotate in the binding pocket to become the prereactive BChE-(−)-cocaine complex ready for the chemical reaction. In the prereactive complex, (−)cocaine molecule lies horizontally at the bottom of the substrate-binding gorge, with the benzoyl ester group of (−)-cocaine favorably located near the catalytic site consisting of the catalytic triad residues Ser198-His438-Glu325 of the enzyme. We recently determined the free energy profile for the transformation of the nonprereactive ES complex to the prereactive ES complex, showing two transition states during the structural transformation for the wild-type BChE-(−)-cocaine complex, and the A328W/Y332G mutation helped to reduce the free energy barrier for the rotation of (−)-cocaine in the active site of BChE.26 However, the previously reported work26 did not study the nonprereactive ES binding pathway, i.e. the pathway concerning how the nonprereactive ES complex is formed. It is necessary to examine the nonprereactive ES binding pathway and the corresponding free energy profile for understanding the entire ES binding process. It is also interesting to compare wild-type BChE with the A328W/Y332G mutant in the free energy profiles of the entire ES binding process for the binding with (−)/(+)-cocaine.
To connect our modeled BChE-cocaine complexes with the general concept of the enzyme-substrate (ES) complexes proposed by Masson et al.,20,21 our modeled nonprereactive enzyme-substrate complex may be regarded as an ES2-like complex, whereas our modeled prereactive enzyme-substrate complex may be considered to be an ES3-like complex. However, it is unknown whether an ES1-like complex (with the substrate binding at the PAS of BChE) exists in the BChE binding with (−)- or (+)-cocaine.
In the present study, we have performed MD and potential of mean force (PMF) simulations to determine the free energy profiles for the formation processes of the nonprereactive complexes for wild-type BChE binding with (−)- and (+)-cocaine and for the A328W/Y332G mutant binding with (−)-cocaine. The combined MD and PMF simulations have revealed that the three simulated nonprereactive complexes are associated with a similar binding pathway with similar free energy profiles. The relative binding free energies calculated for the prereactive complexes are qualitatively consistent with available experimental data. The simulated free energy profiles reveal that the nonprereactive binding process is nearly barrierless, because the affinity for (−)/(+)-cocaine binding at the assumed PAS of BChE (wild-type or A328W/Y332G mutant) is too weak to form a stable ES1-like complex. The novel insights obtained from our MD and PMF simulations help to better understand the detailed molecular mechanisms of BChE-cocaine binding.
Computational Methods
MD simulation
The initial structure of the nonprereactive complex for wild-type BChE-(+)-cocaine binding was prepared based on our previous molecular docking and MD simulations studies8,23,26,27 on wild-type BChE and the A328W/Y332G mutant that were derived from the X-ray crystal structure13 deposited in the Protein Data Bank28 with PDB code 1P0P. In order to further relax the wild-type BChE-(+)-cocaine structure, MD simulations were performed by using the Sander module of Amber 8 program package.29 The procedure of MD simulations was similar to that used in our previous MD simulations on the nonprereactive complexes of wild-type BChE-(−)-cocaine binding and the A328W/Y332G mutant-(−)-cocaine binding.26 In particular, the partial charges of (+)-cocaine atoms were calculated by using the restrained electrostatic potential-fitting (RESP) protocol implemented in the Antechamber module of Amber 8 program,29 following the electrostatic potential (ESP) calculation at ab initio HF/6-31G* level using Gaussian 03 program.30 The geometry used in the ESP calculations was obtained from the previous ab initio reaction coordinate calculations.8 The complex structure was solvated in a rectangular box of TIP3P water molecules31 with a minimum solvent-wall distance of 10 Å. The solvated system was gradually heated to 298.15 K by weak-coupling method 32 and equilibrated for 100 ps. During the MD simulations, a 15.0 Å non-bonded interaction cutoff was used and the non-bonded list was updated every 25 steps. The motion for the mass center of the system was removed every 1,000 steps. The particle-mesh Ewald (PME) method33,34 was applied to treat long-range electrostatic interactions. The lengths of covalent bonds involving hydrogen atoms were fixed with the SHAKE algorithm,35 enabling the use of a 2-fs time step to numerically integrate the equations of motion. Finally, the production MD was kept running for 2.0 ns with a periodic boundary condition in the NTP ensemble at T = 298.15 K with Berendsen temperature coupling, and at P = 1 atm with isotropic molecule-based scaling.32,36
Potential of mean force (PMF) simulation
In order to explore the free energy profiles for the processes of (−)-cocaine binding with both wild-type and the A328W/Y332G mutant, and to explore the free energy profile for the process of (+)-cocaine binding with wild-type BChE, PMF calculations were carried out by using umbrella-sampling 37 MD simulations. The starting structures for the nonprereactive complexes of wild-type BChE-(−)-cocaine binding and the A328W/Y332G mutant-(−)-cocaine binding were adopted directly from our previous MD simulations.26 The classic PMF definition 38 can be represented by a function of reaction coordinate as
| (1) |
In Eq.(1), ρ (χ) is the probability density along the reaction coordinate χ, R is the gas constant, T is the simulation temperature, U(χ) is the biasing potential applied in the umbrella-sampling MD simulations, and F is the normalization constant. According to this approach, the reaction coordinate is usually divided into different regions, i.e. windows, and each of which is sampled separately. A biasing (umbrella) potential, i.e. U(χ), is applied for each window in order to obtain nearly uniform sampling of the potential energy surface. In the present study, the reaction coordinate was defined as the distance from the mass center of the non-hydrogen atoms of either (−)-cocaine or (+)-cocaine to the mass center of the non-hydrogen atoms at the side chains of residues E197 and I442 of the enzyme. For each of the three complex structures, the total number of windows was about 22, depending on the starting structure of each system. Each window was separated by 0.5 Å, covering the reaction coordinate from ~9.74 Å to 20.74 Å. The biasing force constant applied in all windows of umbrella-sampling was 18.0 kcal/(mol•Å2). For each umbrella-sampling window, the initial complex structure was selected from the last snapshot of the PMF simulations of the previous window. The selected structure for each window was first equilibrated for 200 ps and then kept running for 800 ps for production sampling. The frequency for data collection was set to 1 fs, which was the same as that of the time step of umbrella-sampling MD.
After all the umbrella-sampling MD simulations were finished for each system, the data collected from separate simulation windows were combined along the reaction coordinate. These data were then used to calculate the PMF for the whole binding process with the weighed histogram analysis method (WHAM) 39 , 40 using the code developed by Alan Grossfield (http://membrane.urmc.rochester.edu/Software/WHAM/WHAM.html).
Most of the MD and umbrella-sampling MD simulations were performed on an IBM Supercomputer Cluster with 1,360 processors and on a Dell Supercomputer Cluster with 4,816 processors at University of Kentucky Center for Computational Sciences. Some other modeling and computations were carried out on SGI Fuel workstations in our own lab.
Results and Discussion
Nonprereactive Structure of wild-type BChE-(+)-cocaine binding
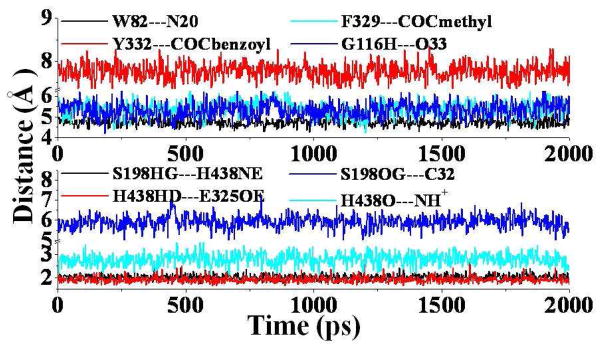
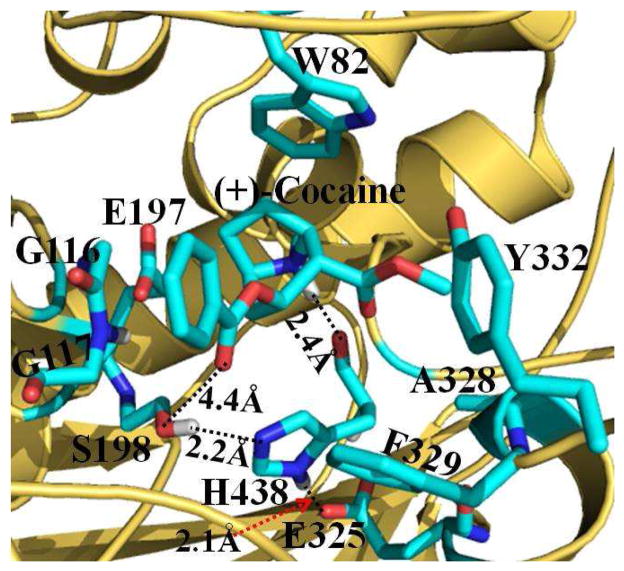
As suggested in our previous studies,8,25,26,27 (+)-cocaine can also form both the nonprereactive and prereactive complexes with wild-type BChE. Once the initial nonprereactive structure of wild-type BChE-(+)-cocaine complex was constructed, it was subjected to structural relaxation through MD simulations (Figure 1). The performed MD simulations were also served to obtain stable complex structure used as the starting structure in subsequent PMF simulations (discussed below). The time-evolved curves (Figure 1) of important distances indicate that the nonprereactive complex structure for wild-type BChE-(+)-cocaine binding has been well stabilized by the intermolecular interactions between wild-type BChE and (+)-cocaine. In the nonprereactive complex (Figure 2), the cationic head of (+)-cocaine is located at a sub-binding site around residue W82. It interacts with residue E197 through tight electrostatic attractions; and with aromatic side chain of W82 through a strong cation-π interaction. The cationic head of (+)cocaine is also weakly hydrogen-bonded with the backbone of residue H438 at an average hydrogen bonding distance of ~2.6 Å (see the cyan curve in the lower panel of Figure 1 and the structure in Figure 2). Several other residues, including Y128, Y440, and I442, are also around the bottom of the sub-binding site for the cationic head of (+)-cocaine. The methyl ester group of (+)-cocaine stays above residues H438 and Y440 of wild-type BChE, and contacts closely with residues A328, F329, M437, and Y332 (Figure 2). However, the benzoyl ester group of (+)cocaine is far away from the side chains of residues F329 and Y332. The distance from the center of aromatic side chain of Y332 to the center of the benzoyl group of (+)-cocaine fluctuates around 7.7 Å (red curve in the upper panel of Figure 1), indicating no direct interactions. The benzoyl group of (+)-cocaine is close to the loop containing residue S287 and the loop containing residues G115 and G116 (Figure 2). The distance from the backbone hydrogen of G116 to the carbonyl oxygen atom of the benzoyl group of (+)-cocaine is around 5.3 Å (blue curve at the upper panel of Figure 1). The binding mode of the benzoyl group of (+)-cocaine in wild-type BChE-(+)-cocaine complex simulated in the present study is quite different from the binding mode of the benzoyl group of (−)-cocaine in the wild-type BChE-(−)-cocaine complex simulated in our previous study.26 According to the results of our previous MD simulations,26 the benzoyl group of (−)-cocaine was found in close packing with the side chains of residues F329 and Y332 of wild-type BChE, and such intermolecular interactions contributed to the first free energy barrier associated with the first rotational transition state (TS1rot) during the structural transformation from the nonprereactive complex to the prereactive complex of wild-type BChE-(−)-cocaine binding. The loose packing of the benzoyl group of (+)-cocaine with the surrounding loops in wild-type BChE-(+)-cocaine complex (Figure 2) makes it much easier for the rotation of (+)-cocaine in the binding site to form the prereactive complex. It can be expected that the free energy barrier for the transformation from the nonprereactive complex to the prereactive complex of wild-type BChE-(+)-cocaine binding should be much lower than that of the (−)cocaine rotation in the binding site of wild-type BChE. Such BChE-(+)-cocaine binding mode also indicates that the rate-determining step for the BChE-catalyzed hydrolysis of (+)-cocaine will not be the (+)-cocaine rotation in the binding site of wild-type BChE from the nonprereactive complex to the prereactive complex. This is in consistent with the well-established mechanistic understanding that the rate-determining step for wild-type BChE-catalyzed hydrolysis of (+)-cocaine is the first step of the chemical reaction.8
Figure 1.
Plots of important distances tracked through MD simulation on the nonprereactive complex for wild-type BChE-(+)-cocaine binding. W82---N20 represents the distance from the center of aromatic side chain of W82 to the nitrogen atom at the cationic head of (+)-cocaine; Y332---COCbenzoyl means the distance from the center of aromatic side chain of Y332 to the center of benzoyl group of (+)-cocaine; F329---COCmethyl represents the distance from the center of aromatic side chain of F329 to the center of methyl ester group of (+)-cocaine; G116H---O33 represents the distance between the backbone hydrogen of residue G116 to the carbonyl oxygen at the benzoyl ester of (+)-cocaine; S198HG-H438NE and H438HD-E325OE represent the hydrogen bonding distances within the catalytic triad residues S198-H438-E325 of the enzyme; S198OG---C32 represents the distance from the OG atom of the hydroxyl group at the side chain of residue S198 to the carbonyl carbon atom at the benzoyl ester of (+)-cocaine, and the H438O---NH+ for the distance from the backbone oxygen atom of H438 to the proton at the cationic head of (+)-cocaine.
Figure 2.
MD-simulated nonprereactive complex structure for (+)-cocaine binding with wild-type BChE. BChE is shown as gold ribbon. (+)-cocaine molecule and residues of BChE within 5 Å around (+)-cocaine are shown in the stick style and colored by the atom type. The distances from the proton at the cationic head of (+)-cocaine to the backbone oxygen atom of residue H438 and from the carbonyl oxygen at the benzoyl group of (+)-cocaine to the OG atom at the hydroxyl group of residue S198 are represented as dashed lines and labeled. Labeled also are the hydrogen bonds formed within the catalytic triad S198-H438-E325 of BChE.
Nonprereactive binding pathway and free energy profile
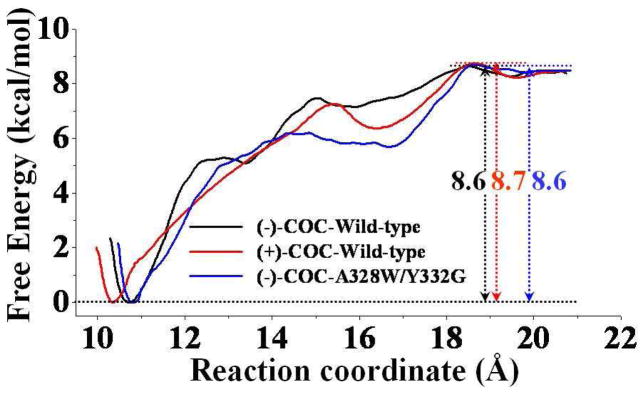
Based on the data collected from the umbrella-sampling MD simulations, we have determined the PMF for each of the three simulated enzyme-substrate binding complexes, including the wild-type BChE-(+)-cocaine binding, wild-type BChE-(−)-cocaine binding, and A328W/Y332G mutant-(−)-cocaine binding. Figure 3 depicts the calculated PMF results for the formation of these three nonprereactive complexes. The PMF results depicted in Figure 3 were based on the MD simulations for 1 ns for each window. In order to examine the convergence of the PMF results, we also performed longer MD simulations on the wild-type BChE-(+)-cocaine complex in each PMF window. The additional PMF results (provided as Figure S1 in Supporting Information) revealed that when extending the MD simulations in each window from the original 1.0 ns to 2.0 ns did not noticeably change the PMF curve. In fact, the three curves (associated with the MD simulations during 0.2 to 1.0 ns, 0.2 to 1.4 ns, and 0.2 to 2.0 ns) depicted in Figure S1 are indistinguishable or identical, which suggests 1.0 ns is sufficient for each window of the PMF simulations.
Figure 3.
Free energy profiles determined for (−)-cocaine binding with wild-type BChE (black curve) and the A328W/Y332G mutant (blue curve) and for (+)-cocaine binding with wild-type BChE (red curve). The reaction coordinate was defined as the distance between the mass center of all non-hydrogen atoms of either (−)-cocaine or (+)-cocaine and the mass center of all non-hydrogen atoms belonging to the E197 and I442 side chains of BChE.
During the PMF simulations, the distance between the mass center of the non-hydrogen atoms of cocaine and the mass center of the non-hydrogen atoms at the side chains of residues E197 and I442 was used as the reaction coordinate for each of the PMF simulations. Such selection of reaction coordinate was based on the structural features of our modeled BChE-cocaine binding structures in the present study and those obtained in our previous study,26 as we found that the direction of reaction coordinate roughly went along the axis of active site gorge of BChE to reach the entrance of the gorge.
A detailed check on the umbrella-sampling MD simulations on wild-type BChE-(−)-cocaine binding revealed that the (−)-cocaine molecule stays around the entrance of the gorge of wild-type BChE and interacts mainly with residues D70 and S287 when the reaction coordinate has the value of ~17 Å. The (−)-cocaine molecule diffuses into the active-site gorge of wild-type BChE when the value of the reaction coordinate becomes smaller than 17 Å. After then, the (−)cocaine molecule slides down smoothly along the active-site gorge, and its cationic head reaches the sub-binding site around residue W82 of BChE in the end of the binding process. According to our umbrella-sampling MD simulations, the binding processes for the wild-type BChE-(+)cocaine binding and the A328W/Y332G mutant-(−)-cocaine binding are all very similar to that of the nonprereactive binding process of wild-type BChE-(−)-cocaine binding.
According to reported X-ray crystal structure analysis,13 there was no significant difference between the conformation of the Ω-loop (residue #65 to #92) of BChE and that of AChE when the X-ray crystal structure of BChE was superimposed with that of AChE. Previous studies9,21 indicated that residue D70 from the Ω-loop of BChE played an important role in the process of substrate binding, and that the D70G mutant had a ~10-fold decreased binding affinity (corresponding to a 10-fold increased KM value) for cocaine or butyrlthiocholine (BTC). In order to explore the possible conformational change of the Ω-loop of BChE during the binding process, we tracked the positional root-mean square deviation (RMSD) of this loop along the reaction coordinate of the PMF simulations using wild-type BChE-(+)-cocaine complex as an example. Based on the MD trajectories from all the PMF simulation windows, each snapshot of the MD-simulated structure was superimposed with the X-ray crystal structure of BChE (PDB entry code 1P0P),13 the positional RMSD for the backbone atoms of the Ω-loop was calculated for each PMF window. The numerical data are provided in Supporting Information (as Tables S1 and S2). As listed in Table S1, the RMSD of the Ω-loop of BChE had the largest value when (+)-cocaine was entering into the active site gorge, and gradually decreased as (+)-cocaine slid down along the active site gorge. The RMSD of this Ω-loop was also calculated based on the starting structure, i.e. the nonprereactive ES structure as shown in Figure 2, and the results are listed in Table S2 (Supporting Information). The general trend of the change in the RMSD for the Ω-loop of BChE was the same, no matter whether the RMSD was relative to the X-ray crystal structure or the MD-simulated nonprereactive ES complex structure (see tables in Supporting Information). These RMSD data suggest that the Ω-loop of BChE changed from the opening conformation (before the substrate reached the entrance of the active site gorge) to the closed conformation after the nonprereactive ES complex was formed. The distance from the cationic head of (−)/(+)cocaine to the negatively charged atom (OD1 or OD2) of the D70 side chain of BChE was also tracked along the reaction coordinate for each of the three ES complexes, and the numerical results are provided as Supporting Information (Tables S3 to S5). For the wild-type BChE-(−)- cocaine binding, the tracked minimum distance (Dmin) was as short as ~3 Å when (−)-cocaine reached the entrance of the active site gorge, indicating that (−)-cocaine interacted directly with residue D70 of BChE. Such distance increased gradually as (−)-cocaine slid down along the active site gorge of BChE.
Based on the calculated PMF profiles (Figure 3), we identified a local free-energy minimum (i.e. a local minimum on the free energy surface) at a reaction coordinate value of ~16 to 17 Å for each of the enzyme-substrate binding complex systems. Such a value of the reaction coordinate corresponds to the local binding event on the entrance of gorge for wild-type BChE binding with (−)-cocaine and (+)-cocaine and for the A328W/Y332G mutant-(−)-cocaine binding. The binding structure corresponding to such a local free-energy minimum may be regarded as the ES1-like complex.20,21 Each of the ES1-like structures was identified based on the criterion that the distance from the cationic head of (−)/(+)-cocaine to the negatively charged atom (OD1 or OD2) on the side chain of residue D70 reached the minimum (average) value while the reaction coordinate is associated with the identified local free-energy minimum (i.e. in the range of ~16–17 Å). As shown in Figure S2 (Supporting Information) for the ES1-like structure for each of the enzyme-substrate complexes, (−)/(+)-cocaine is located nearby the entrance of the active site gorge, and contacts closely with residue D70 from the Ω-loop and/or residue Y332 of BChE. According to the results of PMF simulations (Figure 3), further binding process starting from this local minimum is nearly barrierless, with a free energy barrier of < 1.0 kcal/mol: 0.2 kcal/mol for the ES1-like complex of wild-type BChE-(−)-cocaine binding (black curve in Figure 3); 0.8 kcal/mol for the ES1-like complex of wild-type BChE-(+)-cocaine binding (red curve in Figure 3); and 0.3 kcal/mol for the ES1-like complex of A328W/Y332G mutant-(−)-cocaine binding (blue curve in Figure 3).
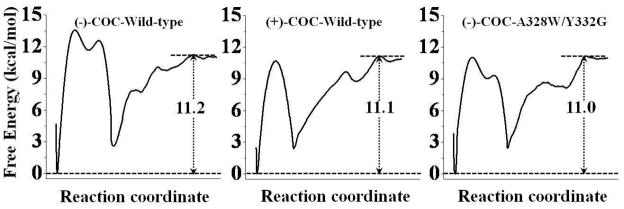
Figure 3 also suggests that the binding free energies calculated for the three nonprereactive complexes are close to each other. Based on the results obtained from the PMF simulations, the binding free energy (ΔGbind) for the nonprereactive complex of wild-type BChE-(−)-cocaine binding has a value of −8.6 kcal/mol. The calculated binding free energy of the nonprereactive complex for A328W/Y332G/mutant-(−)-cocaine binding is −8.6 kcal/mol, and that for wild-type BChE-(+)-cocaine binding is −8.7 kcal/mol. According to our previous study,26 the free energy difference between the prereactive and nonprereactive complexes for wild-type BChE-(−)cocaine structure was determined to be −2.6 kcal/mol, and such difference for the A328W/Y332G mutant-(−)-cocaine structure was −2.4 kcal/mol. We also simulated the transformation from the nonprereactive complex to the prereactive complex for wild-type BChE-(+)-cocaine structure (Figure S3 in Supporting Information), and the calculated free energy difference between the prereactive and nonprereactive complexes was −2.4 kcal/mol. Combining our previous results26 with the calculated free energies in the present study (Figure 3), we have obtained the free energy profile (shown in Figure 4) for the entire binding process from the free enzyme + free substrate to the prereactive complex. Based on the results depicted in Figure 4, the binding free energy (ΔGbind) of the prereactive binding complex is predicted to be −11.2 kcal/mol for wild-type BChE binding with (−)-cocaine, −11.1 kcal/mol for wild-type BChE binding with (+)-cocaine, and −11.0 kcal/mol for the A328W/Y332G mutant binding with (−)-cocaine.
Figure 4.
Free energy profiles for the formation of prereactive complex for wild-type BChE binding with (−)-cocaine (left), wild-type BChE binding with (+)-cocaine (middle), and the A328W/Y332G mutant binding with (−)-cocaine (right).
In order to examine how good the calculated binding free energies are, we also estimated the corresponding experimental binding free energies from available experimental data (i.e. the experimental values of Michaelist-Menten constant KM) under the well-known rapid equilibration assumption, i.e. KM ≈ Kd (dissociation constant). Under the assumption, we may have
| (2) |
It has been reported that KM = 4.5 μM for wild-type BChE against (−)-cocaine,6 KM = 17 μM for the A328W/332G mutant against (−)-cocaine,6 and KM = 8.5 μM for wild-type BChE against (+)-cocaine.9 Thus, when T = 298.15 K, we may have ΔGbind(expt) ≈ −7.3 kcal/mol for wild-type BChE binding with (−)-cocaine, −6.9 kcal/mol for wild-type BChE binding with (+)-cocaine, and −6.5 kcal/mol for the A328W/Y332G mutant binding with (−)-cocaine. Compared to the experimentally-derived binding free energies, the PMF-based computational simulations systematically overestimated the enzyme-substrate binding affinity by ~4 kcal/mol. The systematical overestimation is likely due to the inherent limits of the classical MD simulations based on the empirical force field. Nevertheless, despite of the overestimation of the binding affinity, the PMF-based simulations qualitatively reproduced the relative order of the experimental data, i.e. ΔGbind(wild-type BChE-(−)-cocaine) < ΔGbind(wild-type BChE-(+)-cocaine) < ΔGbind(A328W/Y332G BChE-(−)-cocaine), although the simulations also systematically underestimated the differences between the ΔGbind values.
Conclusion
The combined molecular dynamics (MD) and potential of mean force (PMF) simulations have allowed us to determine the nonprereactive enzyme-substrate (ES) binding pathway and the corresponding free energy profiles for wild-type BChE binding with (−)/(+)-cocaine and for the A328W/Y332G mutant binding with (−)-cocaine. It has been shown that the nonprereactive binding pathway for (+)-cocaine binding with wild-type BChE is similar to that for (−)-cocaine binding with wild-type BChE and the A328W/Y332G mutant. For each ES binding system, the simulated PMF free energy profile reveals a local minimum on the free energy surface corresponding to the formation of ES1-like complex (in which the substrate binds with the enzyme at the presumed peripheral anionic site around the entrance of the active site gorge) prior to the formation of the prereactive ES complex during the enzyme-substrate binding process. However, further evolution from the ES1-like complex to the nonprereactive ES complex is nearly barrierless, with a free energy barrier being lower than 1.0 kcal/mol. So, the nonprereactive ES binding process should be very fast. The rate-determining step of the entire ES binding process is the further evolution from the nonprereactive ES complex to the prereactive ES complex. The combined MD and PMF simulations also demonstrate that all of the three simulated nonprereactive complexes have similar free energy profiles for the binding processes, and the calculated binding free energies are close to each other. Further accounting for the PMF-simulated free energy change from the nonprereactive ES complex to the prereactive ES complex have allowed us to estimate the binding free energy for the entire ES binding process. The PMF simulations qualitatively reproduced the relative order of the experimentally-derived binding free energies, i.e. ΔGbind(wild-type BChE-(−)-cocaine) < ΔGbind(wild-type BChE-(+)-cocaine) < ΔGbind(A328W/Y332G BChE-(−)-cocaine), although the simulations systematically overestimated the magnitude of the binding affinity and systematically underestimated the differences between the ΔGbind values. The obtained structural and energetic insights into the entire ES binding process provide a valuable base for future rational design of high-activity mutants of BChE as candidates of an enzyme therapy for cocaine overdose and abuse.
Supplementary Material
Acknowledgments
This work was supported by NIH (Grants R01 DA013930, R01 DA025100, and R01 DA021416 to CGZ). The authors also acknowledge the Center for Computational Sciences (CCS) at the University of Kentucky for supercomputing time on IBM X-series Cluster consisting of 340 nodes or 1,360 processors and on a Dell Supercomputer Cluster consisting of 388 nodes or 4,816 processors.
Footnotes
Supporting Information Available. Additional three figures (Figures S1 to S3) for the ES1-like binding structures and the free energy profiles of the ES binding processes and five tables (Tables S1 to S5) for the structural changes during the ES binding process. This material is available free of charge via the Internet http://pubs.acs.org.
References
- 1.Mendelson JH, Mello NK. New Eng J Med. 1996;334:965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Sparenborg S, Vocci F, Zukin S. Drug Alcohol Depend. 1997;48:149–151. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 3.Singh S. Chem Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 4.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr J Med Chem. 2004;47:133–142. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 5.Gorelick DA. Drug Alcohol Depend. 1997;48:159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Pan Y, Zheng F, Cho H, Tai HH, Zhan CG. Biophys J. 2009;96:1931–1938. doi: 10.1016/j.bpj.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena A, Redman AM, Jiang X, Lockridge O, Doctor BP. Biochem. 1997;36:14642–14651. doi: 10.1021/bi971425+. [DOI] [PubMed] [Google Scholar]
- 8.Zhan CG, Zheng F, Landry DW. J Am Chem Soc. 2003;125:2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie W, Altamirano CV, Bartels CF, Speirs RJ, Cashman JR, Lockridge O. Mol Pharmacol. 1999;55:83–91. doi: 10.1124/mol.55.1.83. [DOI] [PubMed] [Google Scholar]
- 10.Gatley SJ. Biochem Pharmcol. 1991;41:1249–1254. doi: 10.1016/0006-2952(91)90665-r. [DOI] [PubMed] [Google Scholar]
- 11.Landry DW, Zhao K, Yang GXQ, Glickman M, Georgiadis TM. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 12.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 13.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. J Biol Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 14.Massoulie J, Sussman J, Bon S, Silman I. Prog Brain Res. 1993;98:139–146. doi: 10.1016/s0079-6123(08)62391-2. [DOI] [PubMed] [Google Scholar]
- 15.Quinn QM. Chem Rev. 1987;87:955–979. [Google Scholar]
- 16.Szegletes T, Mallender WD, Thomas PJ, Rosenberry TL. Biochem. 1999;38:122–133. doi: 10.1021/bi9813577. [DOI] [PubMed] [Google Scholar]
- 17.Saxena A, Fedorko JM, Vinayaka CR, Medhekar R, Radiæ Z, Taylor P, Lockridge O, Doctor BP. Eur J Biochem. 2003;270:4447–4458. doi: 10.1046/j.1432-1033.2003.03837.x. [DOI] [PubMed] [Google Scholar]
- 18.Viayna E, Gómez T, Galdeano C, Ramírez L, Ratia M, Badia A, Clos MV, Verdaguer E, Junyent F, Camins A, et al. ChemMedChem. 2010;5:1855–1870. doi: 10.1002/cmdc.201000322. [DOI] [PubMed] [Google Scholar]
- 19.Lockridge O, Masson P. Neurotox. 2000;21:113–126. [PubMed] [Google Scholar]
- 20.Masson P, Legrand P, Bartels CF, Froment MT, Schopfer LM, Lockridge O. Biochem. 1997;36:2266–2277. doi: 10.1021/bi962484a. [DOI] [PubMed] [Google Scholar]
- 21.Masson P, Xie W, Froment MT, Levitsky V, Fortier PL, Albaret C, Lockridge O. Biochim Biophys Acta. 1999;1433:281–293. doi: 10.1016/s0167-4838(99)00115-6. [DOI] [PubMed] [Google Scholar]
- 22.Gao D, Zhan CG. Proteins. 2006;62:99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao D, Zhan CG. J Phys Chem B. 2005;109:23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 24.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai HH, Zhan CG. Angew Chem Int Ed. 2006;45:653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan CG, Gao D. Biophys J. 2005;89:3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Pan Y, Zheng F, Zhan CG. J Phys Chem B. 2010;114:13545–13554. doi: 10.1021/jp106539w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamza A, Cho H, Tai HH, Zhan CG. J Phys Chem B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. AMBER. Vol. 8. University of California; San Francisco: 2004. [Google Scholar]
- 30.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, et al. Gaussian 03, Revision C.02. Gaussian, Inc; Wallingford CT: 2004. [Google Scholar]
- 31.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 32.Morishita T. J Chem Phys. 2000;113:2976–2982. [Google Scholar]
- 33.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 34.Toukmaji A, Sagui C, Board J, Darden T. J Chem Phys. 2000;113:10913–10927. [Google Scholar]
- 35.Ryckaert J, Ciccotti PG, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 36.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 37.Torrie GM, Valleau JP. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 38.Kirkwood JG. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 39.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg J. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 40.Roux B. Comput Phys Commu. 1995;91:275–282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.