Summary
The ability to define osteosarcoma (OS) patients at greatest risk for metastatic progression and non-responsiveness to conventional therapy is currently not possible. Such biomarkers are needed to predict overall prognosis, probability of metastases at diagnosis, and response to chemotherapy. The tissue microarray (TMA) serves as a powerful tool for detecting and validating protein biomarkers across a variety of patients. We constructed a novel outcome-linked TMA to add to and address shortcomings of currently available osteosarcoma tissue resources. To test the utility of our TMA we surveyed the expression of eukaryotic initiation factor 4E (eIF4E) in osteosarcoma patients using immunohistochemistry. Aberrant regulation of translation initiation is a feature of many cancers. eIF4E is central to initiation of protein synthesis. Its expression and activity have been implicated in tumor formation and potentially malignant and/or metastatic progression in some carcinomas. We found that eIF4E was uniformly expressed in osteosarcoma patient samples. No association was found between eIF4E and outcome in osteosarcoma patients. This novel osteosarcoma TMA provided a facile mechanism to assess the role of a relevant protein biomarker in osteosarcoma.
Keywords: Tissue microarray, osteosarcoma, eIF4E, immunohistochemistry
Osteosarcoma is the most common primary malignancy of bone. Approximately 400 new cases of osteosarcoma are diagnosed in pediatric patients in the United States each year (American Cancer Society, http://www.cancer.org/docroot/home/index.asp). For most pediatric osteosarcoma patients, despite successful management of the primary tumor and multi-agent adjuvant therapy, the development of metastases, commonly to the lung, is the most common cause of death. Opportunities to improve outcomes for patients who present with metastases and those at-risk for metastatic progression requires an improved understanding of tumor biology. Defining patients at the greatest risk for metastatic progression and non-responsiveness to conventional therapy is not currently possible. Such prospective identification of patients at highest risk would allow the use of novel therapeutic agents in patients at greatest need. The definition and or validation of biomarkers that predict outcome requires readily available patient samples that are linked to complete clinical follow-up. Tissue microarrays (TMAs) are well-recognized, widely used, tools that enable rapid analysis of large patient cohorts for the expression of protein biomarkers using archival paraffin embedded samples. To enable biomarker and target evaluation in osteosarcoma we report herein on the development of an outcome-linked osteosarcoma TMA available as a community resource. The tissue array is available through the National Cancer Institute (http://ttc.nci.nih.gov/forms/). As an example of the utility of the TMA we asked if the expression of eukaryotic initiation factor 4E (eIF4E) was linked to cancer progression, eIF4E is a 25kDa cytosolic protein that binds to the 7-methyl gaunosine cap at the 5’ UTR of cellular mRNAs during translation initiation. Since eIF4E is found in much lower concentrations than other translation initiation factors it is the rate-limiting component in translation initiation.1,2 eIF4E is an important modulator of cell growth and proliferation and has been shown to be overexpressed in a number of malignancies including lymphomas, cancers of the breast, lung, head and neck, bladder, prostate, colon and rectum, esophagus, skin, and cervix.2–6 Here, we asked (i) if eIF4E is expressed in osteosarcoma tissues and (ii) whether the expression levels of eIF4E were linked to clinical outcome in these patients.
We report the results of TMA immunohistochemical expression profiling of eIF4E in a variety of osteosarcoma sample types including pre-treatment excisional biopsies, post-treatment definitive resections, and lung metastases. We found eIF4E to be similarly expressed in all osteosarcoma sample subtypes.
MATERIALS AND METHODS
Patient Selection and Pathology
Patient records from 75 osteosarcoma patients linked to 89 samples and 12 control tissues (formalin fixed paraffin embedded blocks) were provided by the Montefiore Hospital, Memorial Sloan Kettering Cancer Center, and Center for Cancer Research for the development of the osteosarcoma tissue array. Using these tissues and associated clinical annotation a schema of tissue cores, arranged according to the sample type (biopsy, definitive resection, or resection of distant metastasis) was developed. Primary biopsy specimens were those taken at the time of patient diagnosis prior to chemotherapy treatment. Definitive resection specimens were taken following neo-adjuvant chemotherapy at the time of definitive surgical treatment and metastatic specimens were collected at the time of relapse. All of the metastatic specimens included on the tissue array were taken from the lung. Diagnosis of osteosarcoma was confirmed by histologic review (TO and SH) of H&E slides. For our purposes clinical outcome was defined as overall survival of the patient. Occasionally, multiple specimens were taken from the same patient, for example, four primary biopsy patients also had definitive resection samples and two definitive resection patients had metastases at presentation. Final patient specimens included: 21 primary biopsies (19 patients), 48 definitive resections (47 patients), 20 metastases (14 patients), and 12 control tissues (Fig. 1).
Figure 1.
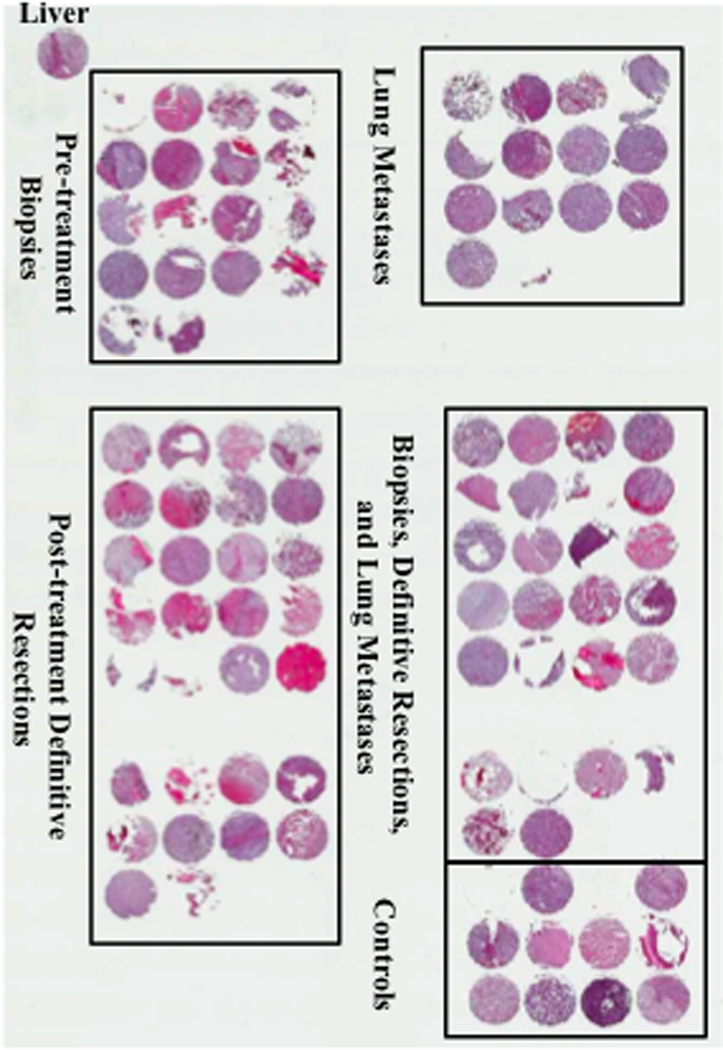
Osteosarcoma Tissue Microarray. Low magnification (0.3X) of human osteosarcoma specimens stained with H&E. Tissue microarray contains 111 tissue cores. Final patient specimens included: 21 pre-treatment biopsies (19 patients), 48 post-treatment definitive resections (47 patients), 20 lung metastases (14 patients), and 12 control tissues. A liver specimen is placed in the upper left hand corner of the TMA in order to verify proper orientation.
Construction of the Osteosarcoma Tissue Microarray
The physical construction of the TMA followed the guidelines previously used by the National Cancer Institute’s Tissue Array Project (http://ccr.cancer.gov/tech_initiatives/tarp/default.asp). Each individual case was represented by 1 tumor core of 1mm that was taken from the original paraffin block. The TMA block contained 89 osteosarcoma specimens and 12 control tissues including liver, kidney, testis, muscle, lymph node, normal bone, placenta, bone cyst, fibrosarcoma, pleomorphic liposarcoma, and malignant fibrous histiocytomas. Serial 5µm sections were cut from the TMA block and used for immunohistochemical analysis.
Immunohistochemistry
We have previously reported methods for immunohistochemical staining of TMAs.7 Briefly, primary rabbit polyclonal anti-eIF4E-antibody (Cell Signaling Technology, Danvers, MA) was applied to the TMA at dilution of 1:100. Substitution of non-immune serum for the primary antibody was used for negative control incubations. Liver and lymph node cores were used as positive controls for eIF4E staining. Slides were developed with avidin-biotin complex kit (DAKO, Carpintera, CA) and counter stained with Hematoxylin (DAKO).
Scoring of the TMAs
Before scoring, tissue cores containing less than 10% of the original tissue (3 tissue cores) or those containing less than 5% tumor cells (8 tissue cores) were excluded from the analysis. Scoring was based on staining intensity (Fig. 2B). The intensity of the signal was scored as 0 (no expression), 1 (mild expression), 2 (moderate expression) or 3 (marked expression). All tissue cores were co-scored independently by TO and LR who were blinded to the clinical information. If there was disagreement about scores, the tissue cores were reviewed together by TO and LR and a consensus score was reached.
Figure 2.
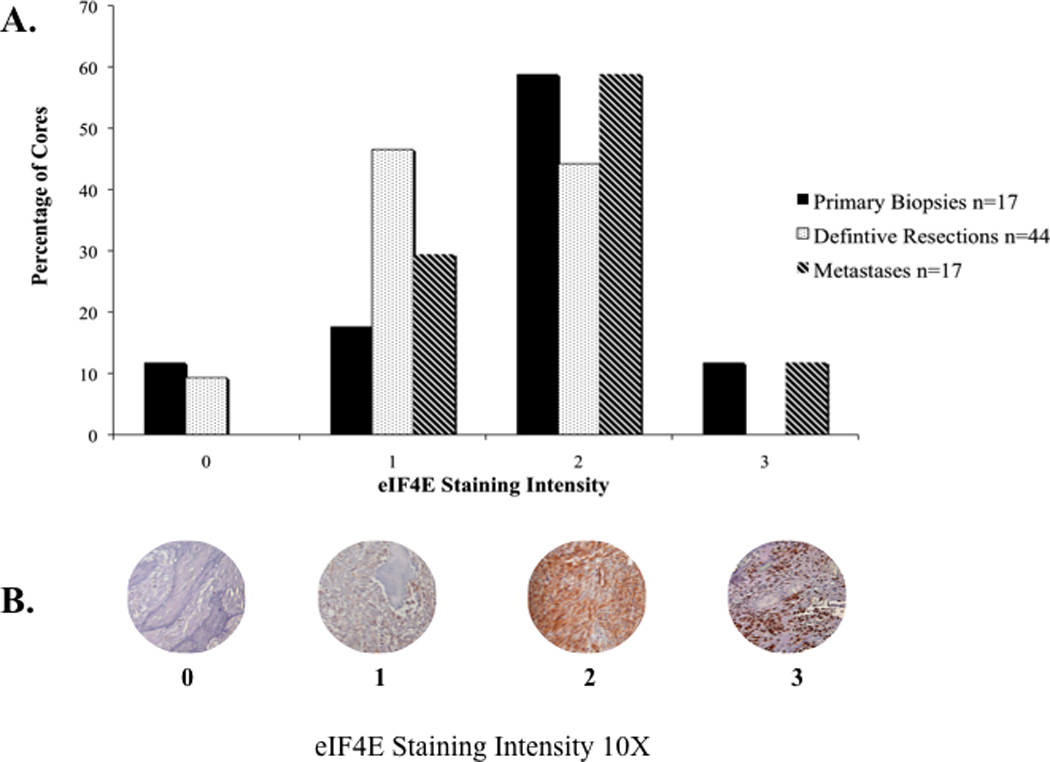
(A) Expression of eIF4E in all osteosarcoma patients. eIF4E is expressed in most osteosarcoma samples and the majority of samples have a staining intensity score of 1 or 2. (B) Representative tissue cores of osteosarcoma immunoreactivity for eIF4E (staining intensities 0, 1, 2, and 3).
Clinical Outcome Measures
The clinical endpoint of this study was overall survival (Fig. 3A). Overall survival was defined as the time from diagnosis until death or last patient contact. Staining results were compared to overall survival (Fig. 3B).
Figure 3.
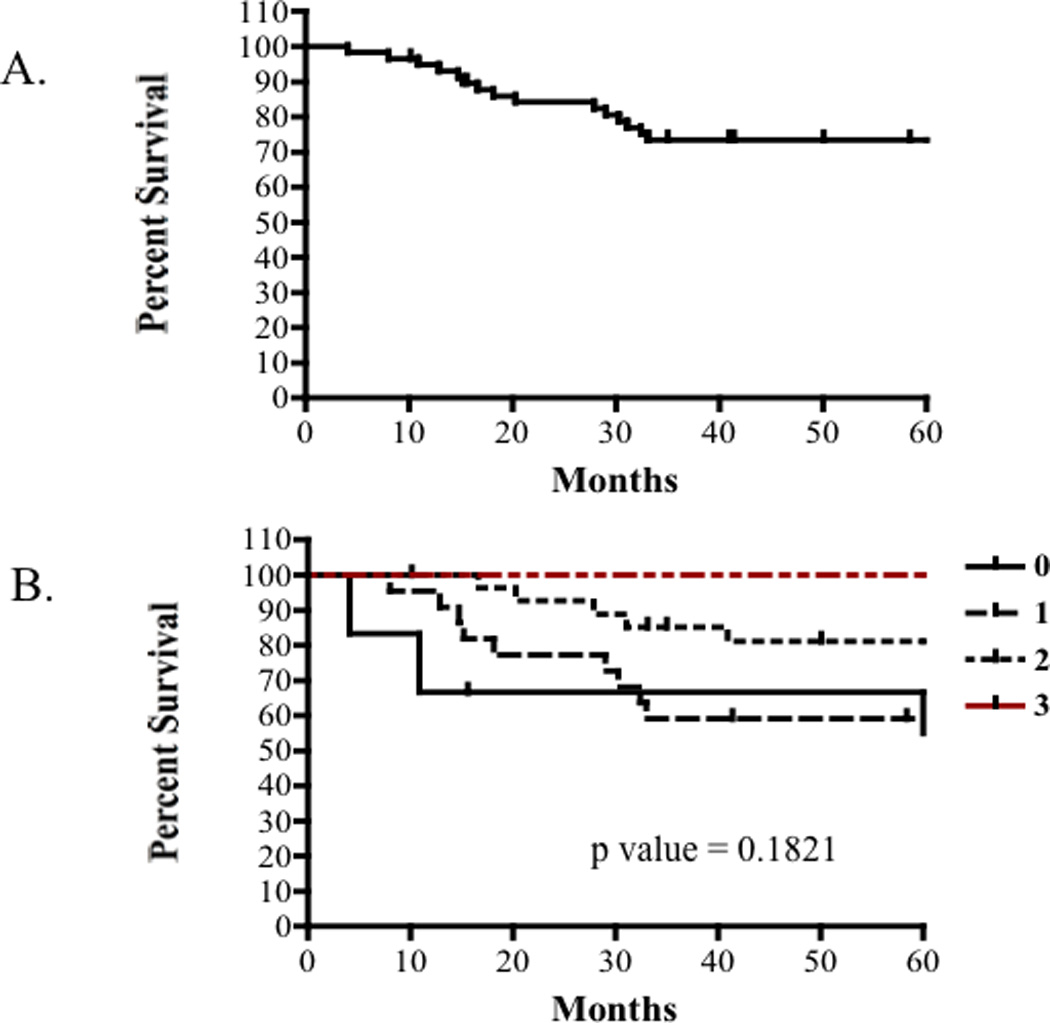
(A) Five year survival rate for patients with localized osteosarcoma without clinically detectable metastases is approximately 75%. (B) Kaplan-Meier survival analysis showed no significant differences in survival among distinct eIF4E staining intensities (P= 0.1821). Therefore, there is no correlation between eIF4E expression and overall survival in osteosarcoma patients with localized disease.
Statistical Analysis
Descriptors of eIF4E staining intensities, including mean, median, confidence intervals, and log-rank tests were determined using Microsoft Excel and GraphPad Prism version 4c for Macintosh software. Log-rank and log-rank trend statistics were used for assessment of survival curves. A P value less than 0.05 was considered significant.
RESULTS
TMA Patient Characteristics
Eighty-nine tissue specimens collected between 1984 and 2001 were obtained from 75 patients (35 males and 40 females) undergoing resection of osteosarcoma of the extremities, pelvis, and craniofacial bones. Written informed consent in accordance with the Ethics Committee and Institutional Review Board of the Memorial Sloan Kettering Cancer Center and/or Montefiore Hospital was obtained prior to tissue procurement. Sixty-seven percent of samples were taken from patients 21 years of age or less (mean-14.5, median-15). Thirty-three percent of samples included patients that were older than 21 years (mean-43.3, median-37). Specimens included tissues collected at biopsy (n=21), during definitive resection of the tumor (n=48), and during resection of distant metastasis to the lung (n=20) for a total of 89 specimens. A variety of anatomic sites and histologic subtypes (Table 1) were included. The majority of tumors were high-grade (47 of 48 patients; 98%) chondroblastic osteosarcomas (14 of 43 patients; 33%) that were located in the femur (20 of 52 patients; 38%). Nine patients had lung metastases at the time of diagnosis. One patient had a low-grade osteosarcoma of the metatarsus. Following procurement, samples of all specimens were decalcified, formalin-fixed, and paraffin embedded.
TABLE 1.
Patient Characteristics
| Number of Patients | |
|---|---|
| Gender | |
| Females | 40 |
| Males | 35 |
| Age (years) | |
| ≤21 | 50 |
| ≥21 | 25 |
| Sample Types | |
| Pre-treatment Biopsies | 19a |
| Post-treatment Definitive Resections | 47b |
| Lung Metastases | 14 |
| Anatomic Location | |
| Femur | 20 |
| Proximal tibia | 11 |
| Humerus | 6 |
| Pelvis | 5 |
| Craniofacial bones | 4 |
| Metatarsal | 2 |
| Extraskeletal | 2 |
| Ulna | 1 |
| Knee | 1 |
| Not Specified | 22 |
| Histologic Subtype | |
| Osteoblastic | 11 |
| Chondroblastic | 14 |
| Fibroblastic | 4 |
| Giant cell rich | 4 |
| Mixed | 10 |
| Not Specified | 31 |
Four pre-treatment biopsy patients also had post-treatment definitive resection samples
Two post-treatment definitive resection patients had lung metastases at presentation
eIF4E expression and relationship with overall survival in osteosarcoma patients
To assess the utility of our TMA and test a hypothesis of interest in our studies of osteosarcoma biology, the expression of eIF4E in osteosarcoma tissues was surveyed using immunohistochemistry. Tissue controls (positive controls; liver and lymph node) exhibited diffuse cytoplasmic expression of eIF4E with staining intensities ranging from mild to marked. An assessment for eIF4E was possible in the TMA for 78 of 89 cores. Positive osteosarcoma immunoreactivity for eIF4E was detected in 71 of 78 (91%) tumors analyzed. Diffuse, mild (intensity score 1) to moderate (intensity score 2) cytoplasmic expression was observed in 67 of 78 (86%) specimens. Specifically, eIF4E was expressed in 15 of 17 (88%) primary biopsy samples, in 40 of 44 (91%) definitive resections, and in 17 of 17 (100%) metastatic lesions (Fig. 2A). Kaplan-Meier survival curves were used to examine differences in overall survival between each defined staining intensity (quartiles) and between high and low eIF4E expressers (Fig. 3B). Based on the uniform expression of eIF4E in most patient samples, it was not surprising to find that the expression intensity of eIF4E protein alone was not an independent predictor of overall survival (P = 0.1821) (Fig. 3B).
DISCUSSION
Successful management of primary tumors and advances in multimodal chemotherapy regimens have improved the overall 5-year relapse-free survival rate from 20% to approximately 60–70% for osteosarcoma patients who present with localized disease.8 However, long-term outcomes for patients have not substantively improved in over 20 years despite intensification of therapy before, during, and after the management of the primary tumor. Furthermore, the 5-year survival for patients who present with metastatic disease is 10–30% and has remained static over this time period.8 The importance of defining patients with the poorest outcomes at the time of diagnosis is critical for improvements to be seen in this disease. Unfortunately, the biological heterogeneity of this disease coupled with the relative rarity of the cancer and limited availability of outcome-linked patient data, has made such advances difficult. TMAs are well-recognized tools that play an important role in the evaluation of biological targets in tumors. The value of TMAs in the study of rare cancers is further amplified. Currently, there are six osteosarcoma TMAs reported in the literature that are consistently used within the research community (Table 2).9–14 Each of these TMAs contains relatively small patient numbers, varied and limited clinical patient data, and are primarily derived from pre-treatment excisional biopsies. These small sample/patient numbers have often dampened the opportunities to assess proteins of interest in osteosarcoma and have limited the opportunity to make important associations between expressed proteins and patient outcomes. The lack of TMAs with relatively large sample sizes that contain well-documented clinical data and are derived from wide variety of sample types poses a serious challenge to physicians and researchers attempting to improve their understanding of osteosarcoma biology. The described characteristics of our array contribute begin to address this unmet need. Although used recently, we provide herein a complete description of the patient population as well as the patient numbers and sample types.15
TABLE 2.
Osteosarcoma tissue microarrays reported
| TMAa (Reference) |
Samples (Patient #)b |
Age Range (Mean)c |
Sample Type |
Grade | Huvos Score |
||
|---|---|---|---|---|---|---|---|
| BXd | DRe | Mf | |||||
| Di Cristofano | 58 (50) | Unknown | 22 | 31 | 5 | High-40 | Unknown |
| (9) | Low-8 | ||||||
| Folio | 46 (18) | 6–29 | 11 | 0 | 7 | High | Unknown |
| (13) | 1986–2001 | (14.6) | |||||
| Kim | 64 (64) | 4–58 | |||||
| (12) | 1995–2000 | (19.4) | 64 | 0 | 0 | High | Unknown |
| Males-45 | |||||||
| Females-19 | |||||||
| Do | 47 (47) | I-20 | |||||
| (14) | 1983–2005 | 7–66 | 39 | 0 | 8 | Unknown | II-15 |
| Males-25 | (25) | III-7 | |||||
| Females-22 | IV-1 | ||||||
| Salas | 73 (73) | Unknown | 73 | 0 | 0 | Unknown | Unknown |
| (11) | 1993–2007 | ||||||
| Somers | 34 (18) | I-3 | |||||
| (10) | Males-12 | 7–17 | 13 | 11 | 10 | High | II-5 |
| Females-6 | (12) | III-3 | |||||
| IV-0 | |||||||
TMA Tissue microarray
Number of patients the samples are derived from
Age range and mean are measured in years
BX pre-treatment biopsy
DR post-treatment definitive resection
M lung metastases
The majority of samples included in this TMA were taken from patients who had received neoadjuvant chemotherapy but still had a large percentage of viable neoplastic cells at the primary tumor site. This could represent the source of a potential bias in the TMA design towards a patient population of patients with poor response to chemotherapy. However, the 5-year overall survival rates for patients included in our TMA with localized osteosarcoma (approximately 75%) is consistent with survival rates reported in the literature.16,17
As an example of our TMA’s utility we evaluated the expression of eIF4E in osteosarcoma tissues formatted on this array and assessed correlations between eIF4E expression, sample type, and overall survival. Our findings demonstrated that relatively uniform expression of eIF4E in both primary tumors and metastatic lesions of these osteosarcoma patients (Fig. 2A) and that eIF4E expression intensity was not an independent predictor of overall survival (Fig. 3B). It is reasonable that the uniform expression of eIF4E in osteosarcoma tissues precluded its association with outcome.
There has been a wealth of evidence in both experimental cancer models and in human cancer tissues implicating eIF4E with tumor development and progression. However, the majority of this work has been conducted in primary epithelial tumors. There is very limited information about eIF4E expression and activity in cancers of mesenchymal origin, including osteosarcoma. Because eIF4E is overexpressed in many cancers and its role in oncogenic transformation and tumor progression,6,18–20 many groups are working to develop strategies to effectively target eIF4E. Novel agents in various stages of development include antisense oligonucleotides to eIF4E, RNAi or antisense RNAs that suppress eIF4E, a physical mimic of the natural ligand, suicide gene therapy, and peptide-based inhibition of eIF4E.21 The widespread expression of eIF4E in osteosarcoma may suggest the potential utility of these approaches in osteosarcoma.
We report herein the development of a relatively large, outcome-linked TMA for use by the community. Our TMA was built to compliment other TMAs currently available and to address the limitation of small sample size and to provide a wider variety of sample types that characterize the disease including pre-treatment excisional biopsies, post-treatment definitive resections, and lung metastases. We hope that our TMA will facilitate rapid evaluation of potential protein biomarkers and ultimately result in an improved understanding of osteosarcoma biology and therapy.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tanasa S. Osborne, Tumor and Metastasis Biology Section, Pediatric Oncology Branch, National Institutes of Health, Center for Cancer Research, National Cancer Institute, 37 Convent Drive, Rm 2144, Bethesda, Maryland 20892, Tel: 301-496-9464, Fax: 301-402-4422, osbornet@mail.nih.gov.
Ling Ren, Tumor and Metastasis Biology Section, Pediatric Oncology Branch, National Institutes of Health, Center for Cancer Research, National Cancer Institute, 37 Convent Drive, Rm 2144, Bethesda, Maryland 20892, Tel: 301-402-0011, Fax: 301-402-4422, renl@mail.nih.gov.
John H. Healey, Department of Surgery, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, Tel: 212-639-7610, Fax: 212-794-4015, healeyj@mskcc.org.
Lauren Q. Shapiro, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, Tel: 212-639-6800, shapirol@mskcc.org.
Alexander J. Chou, Department of Pediatrics, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10065, Tel: 212-639-6057, Fax: 212-717-3239, choua@mskcc.org.
Richard G. Gorlick, Montefiore Medical Center, 111 East 210th Street, Rosenthal Pavilion, Room 3, Bronx, NY 10467, Tel: 718-741-2342, Fax: 718-920-6506, richard.gorlick@einstein.yu.edu.
Stephen M. Hewitt, Tarp Lab/Applied Molecular Pathology Lab, NCI Advanced Technology Center, MSC 4605, Bethesda, Maryland 20892-4605, Tel: 301-496-0040, Fax: 301-402-3134, hewitts@mail.nih.gov.
Chand Khanna, Head, Tumor and Metastasis Biology Section, Pediatric Oncology Branch, Center for Cancer Research, National Cancer Institute Director, Comparative Oncology Program, Center for Cancer Research, National Cancer Institute, 37 Convent Drive, Rm 2144, Bethesda, Maryland 20892, Tel: 301-594-3406, Fax: 301-402-4422, khannac@mail.nih.gov.
REFERENCES
- 1.Rhoads RE, Joshi-Barve S, Rinker-Schaeffer C. Mechanism of action and regulation of protein synthesis initiation factor 4E: effects on mRNA discrimination, cellular growth rate, and oncogenesis. Prog Nucleic Acid Res Mol Biol. 1993;46:183–219. doi: 10.1016/s0079-6603(08)61022-3. [DOI] [PubMed] [Google Scholar]
- 2.Sonenburg N, Gingras A. The mRNA 5' cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 3.Matthews-Greer J CG, de Benedetti A, Herrera GA, Dominguez-Malagon H, Chanona-Vilchis J, Turbat-Herrera EA. eIF4E as a marker for cervical neoplasia. Appl Immunohistochem Mol Morphol. 2005;13:367–370. doi: 10.1097/01.pai.0000170625.98446.3e. [DOI] [PubMed] [Google Scholar]
- 4.Salehi Z MF, Shahosseini F. Significance of eIF4E expression in skin squamous cell carcinoma. Cell Biol Int. 2007;31:1400–1404. doi: 10.1016/j.cellbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Salehi Z MF. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clin Biochem. 2006;39:404–409. doi: 10.1016/j.clinbiochem.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 6.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 7.Hewitt SM. Design, construction, and use of tissue microarrays. Methods Mol Biol. 2004;264:61–72. doi: 10.1385/1-59259-759-9:061. [DOI] [PubMed] [Google Scholar]
- 8.Meyers PA. Muramyl tripeptide (mifamurtide) for the treatment of osteosarcoma. Expert Rev Anticancer Ther. 2009;9:1035–1049. doi: 10.1586/era.09.69. [DOI] [PubMed] [Google Scholar]
- 9.Di Cristofano C, Leopizzi M, Miraglia A, et al. Phosphorylated ezrin is located in the nucleus of the osteosarcoma cell. Modern Pathology. 2010:1–9. doi: 10.1038/modpathol.2010.77. [DOI] [PubMed] [Google Scholar]
- 10.Somers GR, Ho M, Zielenska M, et al. HER2 amplification and overexpression is not present in pediatric osteosarcoma: a tissue microarray study. Pediatr Dev Pathol. 2005;8:525–532. doi: 10.1007/s10024-005-0044-5. [DOI] [PubMed] [Google Scholar]
- 11.Salas S, Jezequel P, Campion L, et al. Molecular characterization of the response to chemotherapy in conventional osteosarcomas: predictive value of HSD17B10 and IFITM2. Int J Cancer. 2009;125:851–860. doi: 10.1002/ijc.24457. [DOI] [PubMed] [Google Scholar]
- 12.Kim MS, Song WS, Cho WH, et al. Ezrin expression predicts survival in stage IIB osteosarcomas. Clin Orthop Relat Res. 2007;459:229–236. doi: 10.1097/BLO.0b013e3180413dbf. [DOI] [PubMed] [Google Scholar]
- 13.Folio C, Mora MI, Zalacain M, et al. Proteomic analysis of chemonaive pediatric osteosarcomas and corresponding normal bone reveals multiple altered molecular targets. J Proteome Res. 2009;8:3882–3888. doi: 10.1021/pr900113w. [DOI] [PubMed] [Google Scholar]
- 14.Do SI, Kim YW, Park HR, et al. Expression of insulin-like growth factor-II mRNA binding protein 3 (IMP3) in osteosarcoma. Oncol Res. 2008;17:269–272. doi: 10.3727/096504008786991639. [DOI] [PubMed] [Google Scholar]
- 15.Abdeen A, Chou AJ, Healey JH, et al. Correlation between clinical outcome and growth factor pathway expression in osteogenic sarcoma. Cancer. 2009;115:5243–5250. doi: 10.1002/cncr.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurney JG SA, Bulterys M. Malignant Bone Tumors. NIH Pub. No. 99-4649. 1999:99–110. [Google Scholar]
- 17.Meyers PA SC, Krailo MD, Healey JH, Bernstein ML, Betcher D, Ferguson WS, Gebhardt MC, Goorin AM, Harris M, Kleinerman E, Link MP, Nadel H, Nieder M, Siegal GP, Weiner MA, Wells RJ, Womer RB, Grier HE. Osteosarcoma: The Addition of Muramyl Tripeptide to Chemotherapy Improves Overall Survival—A Report From the Children’s Oncology Group. J Clin Oncol. 2008;26:633–638. doi: 10.1200/JCO.2008.14.0095. [DOI] [PubMed] [Google Scholar]
- 18.Mamane Y, Petroulakis E, Rong L, et al. eIF4E--from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 19.CG P. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 20.Ruggero D ML, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 21.Ko SY GH, Barengo N, Naora H. Inhibition of ovarian cancer growth by a tumor-targeting peptide that binds eukaryotic translation initiation factor 4E. Clin Cancer Res. 2009;15:4336–4347. doi: 10.1158/1078-0432.CCR-08-2924. [DOI] [PubMed] [Google Scholar]


