Abstract
Accurate assessment of brain-implantable microdevice bio-integration remains a formidable challenge. Prevailing histological methods require device extraction prior to tissue processing, often disrupting and removing the tissue of interest which had been surrounding the device. The Device-Capture Histology method, presented here, overcomes many limitations of the conventional Device-Explant Histology method, by collecting the device and surrounding tissue intact for subsequent labeling. With the implant remaining in situ, accurate and precise imaging of the morphologically preserved tissue at the brain/microdevice interface can then be collected and quantified. First, this article presents the Device-Capture Histology method for obtaining and processing the intact, undisturbed microdevice-tissue interface, and images using fluorescent labeling and confocal microscopy. Second, this article gives examples of how to quantify features found in the captured peridevice tissue. We also share histological data capturing 1) the impact of microdevice implantation on tissue, 2) the effects of an experimental anti-inflammatory coating, 3) a dense grouping of cell nuclei encapsulating a long-term implant, and 4) atypical oligodendrocyte organization neighboring a longterm implant. Data sets collected using the Device-Capture Histology method are presented to demonstrate the significant advantages of processing the intact microdevice-tissue interface, and to underscore the utility of the method in understanding the effects of the brain-implantable microdevices on nearby tissue.
1. Introduction
Brain-implantable microdevices, such as microelectrode arrays (MEAs) (Drake et al., 1988), optogenetic devices (Zhang et al., 2009), and chemical sensors (Johnson et al., 2008), have the potential to provide reliable, direct interfaces with small populations of neurons in the central nervous system (CNS) over time (Berger et al., 2008). Future applications of these microdevices in the human CNS include micro-stimulation to replace lost sensory function (Otto et al., 2005) and recording of neural activity to replace lost motor function (Velliste et al., 2008). Fundamental insights in brain research, such as the investigation of Parkinsonian neural circuitry, are being made thanks to these implantable devices (Gradinaru et al., 2009). However, both clinical and research applications employing these devices are currently limited by the poorly understood effects of the devices themselves on the nearby tissue, including local neurodegeneration (Biran et al., 2005; McConnell et al., 2009), and accumulation of microglial and astrocyte cells (Turner et al., 1999; Hascup et al., 2009).
Current histological methods require removing the implanted devices prior to tissue sectioning and labeling due to a significant elastic modulus mismatch between typical microdevice materials (Weppelmann et al., 1993) and brain tissue (Hirakawa et al., 1981). This Device-Explant Method severely limits quantification of data collected proximally to the device/tissue interface and complicates interpretations of this data (Holecko et al., 2005). Furthermore, misleading fluorescent labeling intensities with proximity to labeling solution in the explant hole can further confound histological analysis once the interfacing device is removed.
In this article we describe a novel method of overcoming the limitations of the extraction-based histological method by collecting, labeling and imaging the CNS implanted microdevices within a slice of fixed tissue. We have developed and validated this Device-Capture Histology (DCHist) method to collect the device in situ, and have tested multiple antibody and chemical markers relevant to analyzing the tissue response to implanted microdevices. This paper describes each component of the DCHist method while demonstrating its superior performance relative to conventional, extraction-based histological methods. Examples of quantification strategies, allowing objective analyses of the image-based data sets, are also presented. Specifically, we share data sets analyzed by cell-count assessment, label intensity measurement, and directionality assessment, each with respect to a nearby implanted microdevice. Applications of these methods will aid in unraveling the biological impact that different microdevice materials (Rousche et al., 2001; Moxon et al., 2004), design features (Williams et al., 2005; Seymour and Kipke, 2007; Jackson, 2010), coating technologies (Kim and Martin, 2006; Winter et al., 2007; Azemi et al., 2011), and implantation methods (Rousche and Normann, 1992; Bjornsson et al., 2006; Jaroch et al., 2009) have on surrounding brain tissue.
2. Materials and Methods
To evaluate the utility of the DCHist method for collecting, processing, and imaging the intact device/tissue interface, we collected histological data from rats implanted with single shank MEAs for 6, 12 and 24 hrs, as well as from rats implanted for 1, 2, and 4 weeks. All experimental animal procedures were performed in conformity with the guidelines of the US National Institutes of Health. The laboratory animal protocol for this work (#06–042) was approved by the Institutional Animal Care and Use Committee of Purdue University (West Lafayette, IN, USA).
2.1. Surgery
Surgical tools were sterilized in an autoclave while heat-sensitive surgical items were plasma peroxide sterilized. The surgeon prepped with single-use surgical hand scrubs (Biomedical Systems, Maryland Heights, MO), donned bouffant caps (Boundary Medical, Dubuque, IA), molded surgical masks (3M Health Care, St. Paul, MN), sterile surgical gowns (Medline, Mundelein, IL), and sterile gloves (Micro-Touch Technologies, Ansell Healthcare Inc. Dothan, AL). A surgical assistant, masked and capped, aided during each procedure; a surgical microscope with an in-line camera (Model MZ6 equipped with IC-D camera, Leica Microsystems GmbH, Wetzlar, Germany) aided the surgeon and enabled image and video acquisition by the surgical assistant during surgeries.
Male Sprague-Dawley and Long-Evans rats (10 total, 250 to 450 g, single-housed, Harlan Laboratories, Indianapolis, IN) were used to evaluate the method. A cocktail of Ketamine (75–95 mg/kg) and Xylazine (5 mg/kg) was delivered via intraperitoneal injection to induce long lasting anesthesia. Adequate anesthesia was measured by foot pressure reflex test, with administration of additional Ketamine dosages between 0.05–0.15 ml delivered by the surgical assistant when appropriate. Heads were shaved with clippers (Model BTF, Andis Company Inc., Sturtevant, WI) and cleaned with alternating swabs of Betadine solution and ethanol, ending with ethanol. Eye lubricating ointment (Akwa Tears, Akorn Pharmaceutical Products Inc., Buffalo Grove, IL) was applied, and subjects were positioned on a circulating-water heating pad (T/Pump, Gaymar Industries Inc., Orchard Park, NY) resting on a steel plate. A pulse-oximeter (8600V, Nonin Medical Inc., Minneapolis, MN) was attached to the subject’s foot, and Oxygen was given between 0.5 and 2.0 L/min during surgery to maintain a high SpO2 (Saturation of peripheral Oxygen) level. The surgeon covered the surgical area with sterilized surgical drape, and cut a small hole directly over the site of surgery. Lidocaine was administered (0.2 ml total/rat) in multiple positions under the skin at the surgical area above the skull. A midline incision was made using a scalpel, starting at the base (posterior portion) of the skull and proceeding 2 cm anterior. Muscle and connective tissue overlaying the skull was retracted to expose the skull surface.
Commercially available MEAs (NeuroNexus Technologies, Inc, Ann Arbor, MI) were used to develop the presented method (cross section ~15 × 150 μm). Silicon MEAs were first cleaned by three alternating washes of ethanol and Milli-Q water. MEAs underwent Sterrad System (Johnson & Johnson Medical Ltd) plasma peroxide sterilization with the device shanks suspended in the air to improve uniformity of sterilization.
The subject’s heads were temporarily held with a stainless steel bar attached anterior to bregma during implantation of unmounted, inactive MEA shanks (Figs. 1b, 1e). The bar was secured to the cleaned, dry skull with Loctite 454 (Henkel Corp., USA) and dental cement (Lang Dental Mfg. Co. Inc., Wheeling, IL). A magnetically mounted base (World Precision Instruments Inc., Sarasota, FL) held the steel bar to the surgical table. The skull over the cortical implant site was removed using a dental drill (Z-35, Henry Schein Inc., USA), with intermittent applications of sterile saline (0.9% NaCl, wt/vol) to reduce local heating. Craniotomies of approximately 2.5 mm in diameter centered approximately −1.5 mm posterior to bregma and 2.1 mm lateral to midline were created by drilling a small circle in the skull and carefully removing the center bone fragment. To prevent drying, Gelfoam (Pfizer Inc., New York, NY) was wetted with sterile saline and applied to the exposed dura. A small incision was made in the dura using micro surgical scissors.
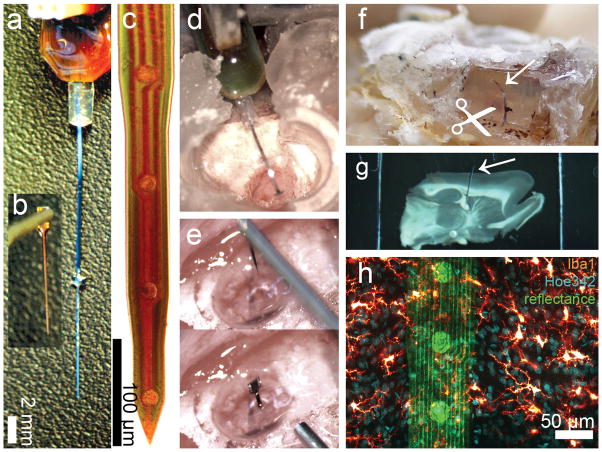
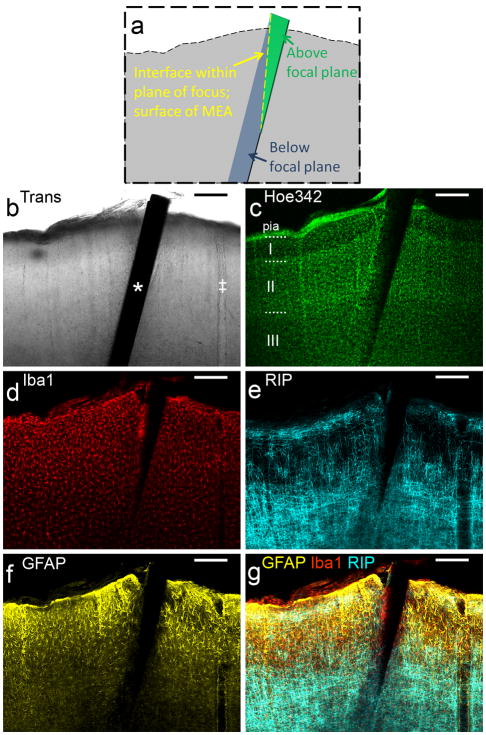
Figure 1.
DCHist method overview. A chronic single-shank, silicon MEA (a), along with an unmounted MEA (b) held by vacuum tweezers. (c) A magnified bright field view of a NeuroNexus MEA tip; circular iridium electrode sites and polysilicon tracings are visible. (d) A cortically-implanted, chronic MEA is shown during surgery, prior to covering the ribbon cable with protective silicone elastomer and dental acrylic. (e) Insertion of an unmounted MEA into rat motor cortex; a micromanipulator positioned vacuum tweezers which hold and release the MEA. (f) A soldering iron was used to expose the silicon elastomer; the chronic MEA ribbon cable (arrow) was then cut at the surface of the fixed brain. (g) A brain section with in situ microdevice (arrow) is shown, mounted within a two-sided slide after histological labeling. (h) In this image fluorescent chemical (Hoe342) and antibody (Iba1) labels were collected, along with laser reflectance outlining the microdevice surface and reflective components. (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
Unmounted MEAs, held by the bond pad using vacuum tweezers (Pro-craft Pick-Up Set, Grobet USA, Carlstadt, NJ), were positioned perpendicular to the dural incision. Care was taken to avoid positioning the MEA over visually identifiable surface vasculature. Each device was rapidly driven into the tissue through the dural incision. A cut from micro scissors removed the exposed shank and bond pad above the dura. Autoclaved, low-melting point agarose gel (1% in MilliQ water, wt/vol, type III agarose from Sigma-Aldrich Inc., St. Louis, MO) was tested for temperature on the back of the hand and then pushed through a 0.2 μm syringe filter (Nalgen Co., Rochester, NY) over the exposed craniotomy. Finally, the adhesive and steel bar were removed and nylon sutures (Ethicon Inc., Somerville, NJ) were used to close the incision.
When implanting active MEAs (Figs. 1a, 1d), multiple screws were driven into burr holes along the dorsal surface of the skull to help secure a permanent dental acrylic cap. Muscle was removed to expose the right temporal region of skull, where a circular craniotomy was made to expose auditory cortex. A chronic MEA was mounted to a magnetically held micromanipulator over the subjects exposed skull. A dural slit was created and the MEA was carefully inserted by hand into the cortex, avoiding visible vasculature elements. A sterile plastic piece was placed below the craniotomy, above which silicon elastomer (Kwik-Sil, World Precision Instrument Inc., Sarasota, FL) was applied. Dental cement (Lang Dental Mfg. Co., Inc. Wheeling, IL) was used to cover the elastomer and exposed bone, and secure the chronic MEA connector package on the head.
After each surgery, triple antibiotic ointment was applied around the outside of the surgical site, and subjects received 0.04 ml of Baytril (Bayer HealthCare LLC Animal Health Division, Shawnee Mission, KS) in 3 ml sterile saline by subcutaneous injections. Subjects were monitored until ambulatory, and returned to their home cage.
2.2. Tissue collection
The tissue collection was based on previous published methods (Bjornsson et al., 2004). Prior to tissue collection, rats were injected with Ketamine (75–95 mg/kg) and Xylazine (5 mg/kg) via intraperitoneal injection. Additional Ketamine was administered until no foot pressure reflex was evident. Subjects were then placed in a surgical tray partially filled with ice, and transcardially perfused through the aorta using a 15 gauge, olive-tipped needle (Popper and Sons, INC, New Hyde Park, NY) with room temperature phosphate buffered saline (PBS: In g/L; 9 g NaCl, 1.44 g KH2PO4, 7.95 g Na2HPO4, at pH 7.4) followed by room temperature 4% buffered formaldehyde (in ml/L; 202 ml 0.4M Na2HPO4 solution, 48 ml 0.4M NaH2PO4 solution, 500 ml 8% formaldehyde, 250 ml MilliQ H2O, at pH 7.4). The PBS was passed through the hemostat-clamped needle at 80 mmHg, delivering ~300 ml of solution; 4% buffered formaldehyde fixative was then delivered through the same needle, increasing the perfusion pressure from 80 to 130 mmHg over several minutes and delivering ~300 ml. The head was removed following perfusion, and the ventral brain was carefully exposed by removal of jaw and hard palate. The head was then placed in the formaldehyde solution overnight at 4°C. The tissue was washed three times in PBS over the following day, returning the container to 4°C each time.
The skull was then carefully removed using bone ronguers to expose the brain of the subjects with unmounted, inactive MEAs. In subjects with MEAs anchored to the skull cap, a soldering iron was used in a fume hood to carefully remove the dental cement and expose the silicon elastomer. Fine surgical scissors were then used, with the aid of a surgical microscope, to cut through the silicon elastomer and through the device shank at the surface of the brain (Fig. 1f). This separating cut was repeated multiple times to insure the implanted portion of the device was detached from the elastomer before the brain was subsequently removed. The tissue was frequently returned to PBS during this process, to prevent drying. This step required slow, methodical dissection (>30 minutes per subject) of the silastic and electrode cabling down to the brain surface. Care was taken to avoid dislodging the entire silastic block from the dental cement cavity while silastic pieces were being removed. Though performing this step under a surgical microscope significantly lessons the difficulty, this step was consistently performed very slowly. Finally, all extracted brains were placed in HEPES Buffered Hank’s saline (HBHS) containing sodium azide (in g/L; 7.5 g NaCl, 0.3 g KCl, 0.06 g KH2PO4, 0.13 g Na2HPO4, 2 g Glucose, 2.4 g HEPES, 0.05 g MgCl2:6H2O, 0.05 g MgSO4:7H2O, 0.165 g CaCl2, 90 mg NaN3, at pH 7.4) and stored at 4°C until slicing.
To obtain a tissue section with the implanted device intact, brains were first oriented under a surgical microscope while resting in a glass petri dish containing HBHS, and cut parallel to both the implanted device angle and the wider diameter dimension of the implanted MEA. Producing a sectioning plane which closely matches the implantation plane is of key importance to later capturing the device. This was performed using a razor blade held by a micromanipulator arm, and created a flat surface matching the in situ microdevice plane which could then be mounted on a vibratome plate. These cuts were informed by both the carefully documented surgical insertion of the device and the microscope view of the exposed end of the device at the surface of the cortex. This plane-creation process may prove technically challenging without well documented insertion information. Blade-guides, such as a ‘Rodent Brain Matrix’ (ASI Instruments, Warren, MI), may also be used to match sagittal, coronal or dorsal/ventral implant angles to collect a tissue block for sectioning. The flat tissue surface was then glued (Instant Krazy Glue Pen, Elmer’s Products Inc., Columbus, OH) to the stage of a vibratome (Model VT1000S, Leica Microsystems GmbH, Wetzlar, Germany). Ice was then placed under the vibratome stage. After the glue cured, cold PBS was poured around the tissue to cool it. Sections of the brain were taken until the device was visible through the tissue, after which thinner sections (50–100 μm) were taken until the device was easily visible through the surface. Single sections, 150 to 400 μm thick, were then collected, capturing the MEAs in situ. Variability in collection thicknesses reflects variability in MEA orientation with respect to the plane of sectioning. Previously mounted DCHist tissue slices were held alongside the vibratome to help to visually inform this collection step. Sections containing devices were lifted with a small scoop-spatula and placed into a 24-well plate containing HBHS. Control sections were taken matching the thickness of the device-containing sections. With the sectioning plane closely matching the device implantation plane, individuals with limited vibratome experience were able to estimate and perform this capturing method.
2.3. Previous Tissue Slicing/Immunohistochemistry Methods
To compare with previously reported histological methods, rats were implanted with unmounted MEAs and perfused as described above. After perfusion, MEAs were carefully explanted from the brain under a surgical scope using tweezers, sliced perpendicular to the implanted device angle and processed as previously reported (Turner et al., 1999); (Bjornsson et al., 2008); images from these 100 μm thick slices are shown in Figure 2.
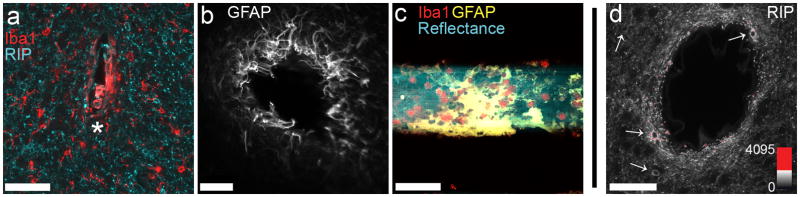
Figure 2.
Explanting MEAs produces LSCM histology data with variable morphological distortion (a, b), including formerly interfacing tissue now collected in the hole (*) or removed and remaining on the explanted device (c). Fluorescence intensity of an applied marker can be affected by distance from a surface contacting the labeling solution, as shown in (d) where oligodendrocyte cell bodies (arrows) and myelinated branches are more intensely labeled given closer proximity to a large blood vessel (centered) within a 100 μm thick tissue slice. These confounding factors associated with explanting can limit the detailed study of microdevice integration in the CNS. Images in (a, b, d) are single optical sections; image in (c) is a 90 μm highest-intensity z-projection. Microdevices in (a) and (c) were implanted 2 weeks, while device in (b) was implanted 4 weeks. Scale bars are 50 μm (a, c, d), 100 μm (b). (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
2.4. Tissue processing and mounting
A list of antibody and chemical labels shown in the data presented, as well as their sources and item numbers, is presented in Supplementary Table. An example immunohistochemistry labeling protocol sheet can be found in Supplementary Note. Slices were processed at room temperature (unless noted) in 24-well plates wrapped in aluminum foil. Solutions were added using adjustable pipettes and removed by vacuum suction, through disposable tips. The tissue was carefully flipped halfway through many of the processing steps to enable a more uniform treatment. Slices were first washed three times with 400 μl of HBHS, with 5 minutes between rinses, then were incubated in 400 μl of fresh NaBH4 in HBHS (5 mg/ml) for 30 minutes, and washed again three times, 5 minutes each, in 400 μl of HBHS. Slices were next incubated in 400 μl of fresh Wash Solution (WS; 1% vol/vol Normal Goat Serum, 0.3% Triton X-100 in HBHS, vol/vol; WS was refrigerated at 4°C for use in subsequent steps) for 1 hour. After this step, WS with diluted primary antibodies (all used at a 1:100 dilution) was applied, and the well plate was set on a plate rotator for 2 hours at room temperature. The plates were then placed in 4°C for an additional 24 to 48 hour incubation. Primary antibody solution was removed after the 24 to 48 hour interval, and the tissue was washed rapidly five times with WS. A 30 minute wash interval and six, 60 minute washes, were then performed in WS at 4°C, to thoroughly remove unbound or non-specific primary antibodies from the tissue (plates may be left overnight in 4°C during this step). Fluorescent secondary antibodies, at a concentration of 1:100 in 400 μl WS, were next applied, along with fluorescent chemical labels. The same incubation and washing steps performed with the primary antibodies were repeated.
Slices were mounted in custom (thick-tissue) slides (Supplementary Fig. 1) to avoid compressing the tissue and to allow imaging into both sides of the specimen. An approximately 10 × 20 mm rectangle was cut from a plastic slide (Rinzle Plastic Micro-Slides, Electron Microscopy Sciences, Hatfield, PA). A coverglass slip (No. 1, Corning Incorporated, Corning, NY) was attached to one side of this slide using quick drying nail polish (01 Invisible, Sally Hansen Inc., New York, NY) applied in multiple layers under and around the coverglass edges. Thick tissue sections were carefully placed in the slide well, to which Prolong Gold (Invitrogen Inc., Eugene, OR) mounting media was added (Fig. 1g). Small, manually shaped coverglass ‘shim’ pieces were sometimes added over thinner tissue slices to help keep them flat. A second coverglass slip was secured down on the other side using nail polish, sealing the slice within. Slides were stored at 4°C away from light.
2.5. Tissue imaging, device imaging, and data quantification
All images were collected on an Olympus IX81 microscope equipped with an Olympus FV1000 laser scanning confocal system as well as Olympus 10x/0.40 air and 40x/0.80 water emersion objectives (Olympus America Inc., Center Valley, PA). Laser light sources at 488 nm, 543 nm, and 633 nm were used, along with a tunable Ti:Sapphire laser (MaiTai, Spectra-Physics Inc., Mountain View, CA) set at 750 nm for two-photon excitation of a UV label. An external photomultiplier (R3896, Hamamatsu Corp., Bridgewater, NJ) equipped with a 405/40 nm filter (Chroma Technology Corp., Bellows Falls, VT) was used for UV dye fluorescence detection. Laser reflectance from devices was collected on internal detectors by setting the wavelength detection window to a ± 3 nm width centered on the laser light wavelength being scanned (Supplementary Figs. 2b, 2f). A transmission light (Trans) setting on the Olympus IX81 was used to collect laser light that had passed through the sample and an aligned condenser to a forward detector (Supplementary Fig. 2a). Coherent anti-Stokes Raman scattering (CARS) microscopy images of MEAs (Supplementary Figs. 2c-d) were taken on the Olympus IX81 microscope using 790 nm output from the Ti:Sapphire laser as a pump beam and frequency-doubled idler at 1018 nm from optical parametric oscillator (Opal, Spectra-Physics Inc., Mountain View, CA) as Stokes beam, as previously described (Chen et al., 2009). When necessary, neighboring channels were imaged in succession to avoid fluorophore signal overlap. Image averaging of 2–4 frames was frequently performed during data collection, to increase image clarity while using low laser power settings. Images were leveled and pseudo-colored in Olympus Fluoview V1.7. Figure labels, scale bars, and figure layouts were created in Photoshop CS2 (Adobe Systems Inc., San Jose, CA).
Cell nuclei were counted using ‘Cell Count’ plugin in ImageJ (National Institute of Health, USA) by manually marking nuclei every 15 μm within z-stacks (Figs. 6b-c). Fluorescence intensity measurements (Figs. 7a-b) were gathered by averaging multiple line scans, which were evenly spaced and perpendicular to the implanted device, using Olympus Fluoview V1.7 (Olympus America). To investigate changes in directional orientation of cortical myelination near implanted devices, Fourier component analysis for directionality (Fig. 9e) was performed on data detailed in Supplementary Figure 3 using the ImageJ plug-ins ‘Directionality’ created by Jean-Yves Tinevez (http://pacific.mpi-cbg.de/wiki/index.php/Directionality) and ‘OrientationJ’ created by Daniel Sage (http://bigwww.epfl.ch/demo/orientation/), following their respective instructions. Graphs were created in Excel 2007 (Microsoft Co., Redmond, WA).
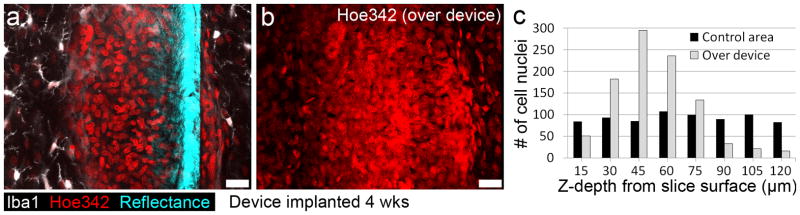
Figure 6.
Example quantification allowed by collection of intact interface. (a) A single optical section, 40 μm into slice, shows many tightly packed nuclei (Hoe342) surrounded by a cloud of microglia marker (Iba1) along the face of the microdevice. (b) A 3D rendering showing all nuclei in the full 120 μm image stack. (c) Cell nuclei were counted in 15 μm steps over the device and a control area in the same cortical layer. Scale bars are 20 μm. (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
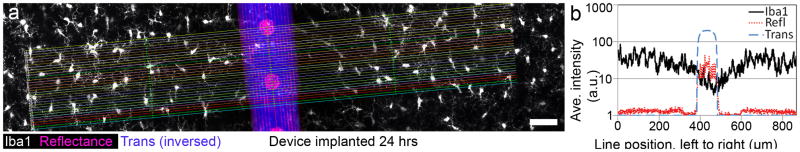
Figure 7.
Example quantification allowed by collection of intact interface. (a) A z-stack through 20 μm of tissue around an implanted device, coated with an experimental anti-inflammatory peptide, was quantified by averaging horizontal line scans. (b) Microglial signal changes (Iba1, black line) are compared with device location by graphing laser reflectance (Refl, red line) and the inversed transmission light (Trans, blue line) shadow; a dip occurs in Iba1 fluorescence centered over the right edge of the device. Scale bar is 50 μm. (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
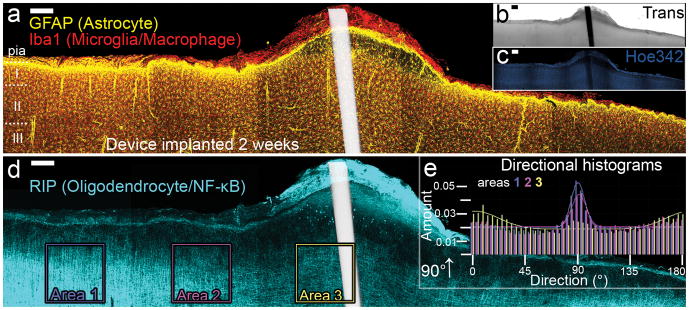
Figure 9.
Panoramas of five highest-intensity projection images (200 μm z-stacks) around a 2-week- implanted MEA, show a complex immune response to the implanted MEA. A superficial tissue bump (b), containing many macrophages surround a weak glia limitans (a), which rests over a swollen, poorly- nucleated area of CNS tissue (c). Large-scale changes in typically columnar myelination directionality were observed (d) and quantified using Fourier component analysis of the boxed Areas 1, 2, and 3. A comparative loss in columnar orientation under the atypical cortical surface was measured (e) (see Supplementary Fig. 3 for graph source data, Supplementary Fig. 4 for magnified images). Scale bars 200 μm (a-d). Device location is shown white in (a,d) as overlay of inverted-Trans channel. (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
3. Results
3.1. Explanting devices limits current histological analysis around CNS implanted microdevices
Figure 2 presents histological images gathered after the post-mortem explantation of CNS-implanted MEAs in accordance with commonly performed histological methods used to analyze the biological impact of these devices (see section 2.3). Relative to native morphology with the MEA in place, explanting MEAs from fixed tissue produced visibly disrupted tissue morphologies at the former MEA location (Figs. 2a, 2b), including distorted extraction-hole shapes and lumping of tissue formerly adhering to the microdevice’s surface (Fig. 2c). As labeling intensity can be affected by the tissue-penetration depth of the label, another limitation of the Device Explanting Method is observed in Figure 2d, where the relative location of antigen binding sites to the labeling solution contained in a CNS blood vessel hole produced a higher fluorescent intensity in tissue closer to the vessel surface. This result demonstrates how histological characterization around holes in brain tissue sections can be confounded by factors related to the diffusion of the label.
3.2. In Situ collection captures intact tissue interface
Example image results from using the DCHist method, taken under laser scanning confocal microscopy (LSCM), are presented in Figure 3. An illustrative image (Fig. 3a) and transmission light microscopy (Trans) image scan intersecting the 1 week implanted MEA are shown (Fig. 3b). Cell nuclei (Fig. 3c) are illuminated by chemical marker, while cell-specific antibodies mark microglia, oligodendrocytes, and astrocytes (Figs. 3d-f). Of note in this example DCHist data set is the appearance of a depth-dependent microglial response around the cortical layer I/II boundary. More generally, this example data illustrates that an MEA microdevice can be captured within its surrounding tissue, labeled, mounted and imaged to reveal the expected morphologies associated with the labels with low background fluorescence. Throughout the development of the DCHist method all single-shank implants were successfully captured in separate histological slices.
Figure 3.
In situ labeling of a thick tissue slice around a captured microdevice. Images of a single optical section taken sequentially using 488, 546, 633 and 750 nm laser light at approximately 50 μm into a 300 μm thick sagittal section through motor cortex at the site of a 1 week implanted MEA. (a) Orientation of the device and optical section is illustrated. (b) An implanted MEA is visible as a shadow (*) in Trans image. A large cortical blood vessel is also visible (‡). (c) Two-photon excitation of a cell nuclei label (Hoe342) provides an idea of the total cell density, and aids in delineating cortical layers. Antibodies to cell-type-specific proteins were used to label (d) microglia/macrophages, (e) oligodendrocytes, and (f) astrocytes in the interfacing tissue. (g) In the overlay of the three antibody labels, a dense microglial signal is visible along the device at approximately the cortical layer I/II border. Each cell-type label displays expected morphological structures over a low background fluorescence. Scale bars are 200 μm.
3.3 Fluorescent label penetration depth assessment
Passive diffusion from solution into tissue drives typical immunological and non-immunological labeling. Assessment of the penetration of applied fluorescent labels into the thicker DCHist tissue specimens can be performed using LSCM along with a long working-distance (≥2mm) but higher-magnification ( ≥40x) microscope objective. Figure 4 presents example data obtained in order to characterize the variation in penetration depth of a handful of markers. These results demonstrate penetration depth variability between different fluorescent markers in this tissue slice; the fluorescently labeled antibody against a protein expressed in microglia (Iba1) (Fig. 4a) and the DNA/cell nuclei label (Hoe342) (Fig. 4b) were detected above background fluorescence throughout this slice, while detection of the fluorescent marker against a protein expressed in neurons (β3tub) (Fig. 4c) was limited to ~100 μm into the slice. A graph comparing the relative fluorescent intensities of these three labels as a function of depth into tissue is shown in Figure 4d, further illustrating labeling variability assessment in a tissue slice.
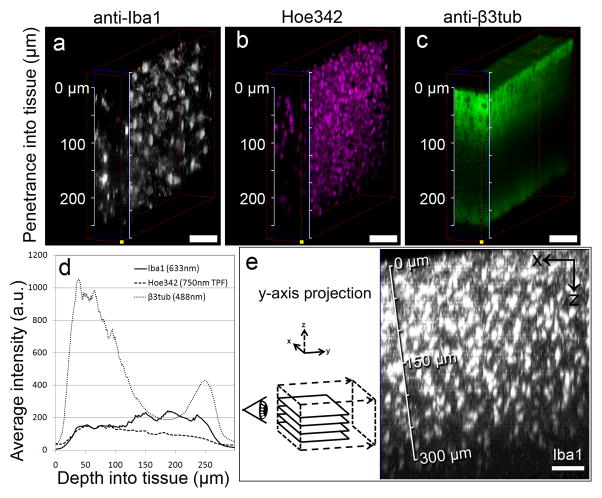
Figure 4.
Assessing penetration of tissue labels. (a-c) Confocal image stacks, taken into a ~250 μm thick tissue slice near an implanted microdevice left in situ (implanted 2 weeks). Fluorescent markers were excited with a 488 nm (anti-β3tub; neurons) and a 633 nm laser (anti-Iba1; microglia), as well as a 750 nm laser for two-photon fluorescence of Hoe342 (nuclei). Although low non-specific labeling is seen in each, Iba1 and Hoe342 show consistent labeling and imaging throughout the slice, while β3tub labeling is limited beyond 100 μm. (d) A comparison of average fluorescent intensity data in a-c is graphed, showing relatively stable fluorescence intensities for Iba1 and Hoe342 and a bimodal distribution for β3tub as the imaging depth approaches the surface of the slice distal to the microscope objective. These results were repeated in nearby tissue areas, with comparable results (data not shown). (e) A y-axis project of a LSCM z-stack into a labeled, 800 μm-thick slice demonstrates an immuno-label (Iba1) imaged down to 300 μm. Detector sensitivity in (a),(c) and e) was adjusted while z-scanning to match the range of emitted fluorescence; laser power, dwell time and all other settings were unchanged during scanning. Scale bars are 50 μm.
Figure 4e shows data collected from an 800 μm thick slice labeled with anti-Iba1. A depth-detectable limit of ~300 μm from the surface of this slice was determined. Contrast was not sufficient to clearly visualize microglial morphology above background fluorescence and detector noise beyond 300 μm. Determining imaging limitations like this for particular labels informs us as to how thick we can expect to image down to tissue around collected devices.
3.5 Imaging both sides of a DCHist tissue slice
Mounting tissue slices within chambers enabled imaging into the specimens around in situ microdevices (Fig. 5). Imaging both sides of an antibody-labeled-slice using LSCM revealed the applied neuron and microglia-specific markers around an MEA 24 hours after implantation (Figs. 5a, 5b). Imaging this 300 μm slice from both sides helped overcome the light-scattering induced depth limitation of LSCM, and allowed detailed imaging of areas which would otherwise be shadowed by the opaque MEA microdevice (Fig. 5c). Preserving the tissue in a sealed chamber also prevented unwanted tissue compression, as might occur if mounted by tightly sandwiching between two glass pieces.
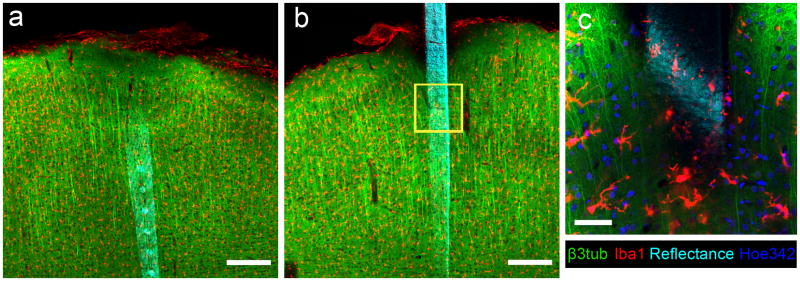
Figure 5.
Both sides of a tissue slice can be imaged around DCHist collected device. (a) Reflectance (cyan) from the traces and electrode sites of an implanted MEA are visible, along with markers for β3tub and Iba1. (b) The backside of this silicon device is visible after flipping the tissue section. (c) A magnified view of the yellow box in (b) shows the morphological characteristics of the labeled microglia (Iba1) and neurons (β3tub), and also nuclei (Hoe342) along with the device surface. Data taken under LSCM; (a, b) are highest intensity projections 150 μm thick; (c) is a single optical section. Device implanted into rat motor cortex for 24 hours. Scale bars are 200 μm (a,b), 50 μm (c). (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
3.6 DCHist allows detailed quantification of peri-microdevice tissue composition
Collecting the intact device/tissue interface allowed detailed quantification of structural details up to the surface of the in situ device. Figure 6a presents a single-optical-scan of the bio-integration of a microdevice 4 weeks post-implantation. A dense, encapsulating response is visible in this position in cerebral cortex, consisting primarily of closely packed cell nuclei surrounded by a cloud of microglia-specific labeling (Iba1). A highest-intensity projection of nuclei labeled through 120 μm, shown in Figure 6b, outlines the shape of this highly nucleated response around the microdevice. Quantification of nuclei at this position over the device relative to a position in the same cortical layer (but ~2 mm from this implant) enabled comparison of the atypical cell nuclei response with a control region (Fig. 6c). The abnormal aggregate of nuclei and the abundance of Iba1 fluorescence indicate a multinucleated giant cell immune response in the CNS (Edell et al., 1992) responding to the chronic presence of the microdevice.
To further evaluate the capture of immunogenic conditions around an implanted microdevice, another subject was implanted for 24 hours with a device coated with an experimental anti-inflammatory peptide (Ward et al., 2009; see Panitch, et al., 2011, for details related to this peptide) (Fig. 7a). The impact of this recently implanted microdevice was again inferred from Iba1 labeling. To quantify this response, the fluorescent intensity with respect to device location was investigated; intensity data along lines centered over and aligned perpendicular to the device were collected. Once averaged and graphed (Fig. 7b), a dip in biomarker intensity near the device appears to describe a subtle but likely physiologically relevant anti-inflammatory effect in the closely interfacing tissue along one edge of this microdevice. This preliminary finding demonstrates the potential of the DCHist method to aid in characterizing the biological impact of bio-active coatings applied onto the microdevice.
3.7 DCHist method provides device location and implantation impact data
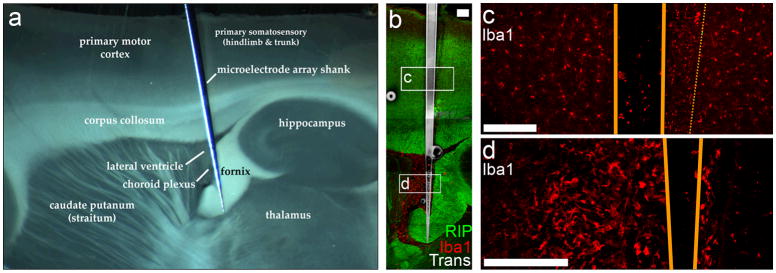
As seen in Figure 8, capturing the device and surrounding tissue allowed detailed microdevice localization through cortical and subcortical tissue. A subject with a deeply implanted microdevice was sacrificed 6 hours post-surgery. This subject was not perfused before brain extraction, in order to avoid clearing blood-borne responses associated with the recent surgery. Viewing the mounted, DCHist processed tissue slice under a surgical microscope (Fig. 8a), the surrounding brain features were clearly visible in relation to the in situ microdevice (Paxinos and Watson, 2007). The fornix has been ripped and deformed by the implantation, and the typically empty lateral ventricle appears clouded with tissue related to the injury. The collected microdevice along with markers for oligodendrocytes (RIP) and microglia/macrophages (Iba1) were imaged under LSCM (Fig. 8b), showing their precise locations in relation to one another. Closer inspection of the intact interface revealed an asymmetric activation of microglia/macrophages to one side of the implanted device (Fig. 8c), likely indicating the device was implanted at a slight angle with respect to inserter’s direction of travel, displacing tissue predominantly to the highlighted side. Evidence of the microdevice insertion impact is further presented in Figure 8d, where a closer view of the ventricle adjacent to the torn fornix reveals it to contain a high concentration of the labeled microglia/macrophage protein Iba1. Recent vascular damage proximal to the lateral ventricle could explain the significant presence of this marker.
Figure 8.
In situ collection provides precise device location and implantation impact data. (a) A midbrain-implanted MEA device, collected in non-perfused tissue 6 hours after surgery, is imaged under dissection microscope and shown relative to neighboring anatomical brain structures. (b) Under LSCM, immunohistochemical data for oligodendrocyte (RIP) and microglia (Iba1) can be taken along the entire device. (c) An asymmetric activation of microglia to one side of the implanted device (highlighted with thin, dashed line) indicates device-implantation at a slight angle with respect to the inserter’s direction of travel, displacing tissue to one side. (d) Near the highly deformed fornix, a macrophage-filled ventricle was captured, preserving indications that a significant amount of damage and bleeding likely occurred in the lateral ventricle subsequent to the implantation. Device edges outline in orange in (c,d). Scale bars are 200 μm (b), and 50 μm (c, d). (For interpretation of the references to color in this gure legend, the reader is referred to the web version of the article.)
3.8 DCHist method capturing tissue architectural reorganization
Because the outlined method is capable of capturing the tissue interface along the length of an implanted microdevice, the often variable and complex tissue responses induced by indwelling microdevices can be examined through multiple intact layers. In Figure 9, a surface bump on the cortex seen in transmission light (Fig. 9b), is found to be composed of layers of macrophages surrounding a poorly formed glia limitans, the astrocyte-composed barrier designating CNS from non-CNS (Fig. 9a). The atypical tissue swelling continues internal to the glia limitans, and contains both elongated astrocytes and microglia, but few nuclei (Supplementary Fig. 3); numerous RIP+ cell bodies are visible just internal to this atypical tissue (Fig. 9d). Typically, a highly myelinated area of cortex known as the plexus of Exner runs parallel to the surface in superficial layer I, however, this structure is lost within the swollen tissue. Looking deeper into the cortex, the normally column-oriented myelination pattern of oligodendrocytes is lost directly below the inflamed, bulging cortical surface; this atypical structural directionality was quantified using Fourier components analysis comparing three 0.5 mm2 squares of data centered above the device (Area 3), 0.5 mm away (Area 2) and 0.5 mm further away (Area 1) from the previous area (Fig. 9e). The 90° columnar orientation is lost under the atypical cortical layer 1 structure. This complex tissue-altering response is observable due to DCHist collection of the undisturbed tissue near a brain implanted MEA, and would likely go unnoticed using histological methods requiring device removal.
3.9 Imaging the microdevices with LSCM microscopy
Using the DCHist method allows capture of the microdevice within a thick section of surrounding tissue. Methods to allow imaging the captured microdevice itself under LSCM are presented in Supplementary Figure 2. Collecting transmission light (Trans) images of the microdevices, created with LSCM by detecting laser light that has passed through the sample and an aligned condenser onto a forward detector, shows contrast where light is absorbed or reflected away by semi-transparent and non-transparent structures. Supplementary Figure 2a presents a Trans image outlining a semi-transparent silicon microdevice. The surface of items may be imaged under LSCM by detecting the reflected laser light scanned across the sample. Supplementary Figure 2b shows laser reflectance collected from a microdevice surface, with electrical traces and electrode sites visible. Other, more selective imaging may be performed to further investigate specific elements of a microdevice; a previously unreported example is using Coherent anti-Stokes Raman Scattering (CARS) microscopy (Cheng and Xie, 2004) to image polysilicon traces within microdevices (Supplementary Figs 2c, 2d), potentially revealing sources of device malfunction upon histological analysis. Fluorescent coatings may also be applied to outline the device (Supplementary Fig. 2e-g) against a non-fluorescent background.
4.0 Discussion
In addition to enabling imaging of the device/tissue interface across multiple cortical strata (Fig. 3) and providing highly quantifiable data of the immediately surrounding tissue (Figs. 6, 7), we demonstrate how these methods can be used to analyze implantation-related tissue changes (Fig. 8) and atypical tissue organization (Fig. 9). We also detailed how the device itself may be imaged under LSCM (Supplementary Fig. 2) and demonstrated how imaging into both sides of a slice may be performed (Fig. 6, Supplemental Fig. 1). Finally, we have outlined techniques to assess the limitations of the labeling and imaging strategies (Fig. 4), which are individual to a researcher’s exact tissue processing methods and available microscopy tools.
The methods presented in this article improve upon prevailing histological methods for capturing the biological impact of brain-implanted microdevices with light microscopy. The DCHist method achieves this by avoiding device explantation during histological processing. Capturing the interfacing tissue and in situ device in a single section of tissue collects the tissue most impacted by the device’s presence — tissue which would likely have been lost or highly distorted using current extraction-based histological methods (Fig. 2). The data presented, capturing intact tissue and cellular morphologies (Figs. 6, 7, 8, 9), would have been difficult to interpret if processed using the prevailing Device-Extraction Histology method. As shown in Figure 2, explanting the device typically produces unpredictably distorted holes, either missing formerly interfacing tissue (Fig. 2b) or partially filled with formerly device-adherent tissue (Fig. 2a) scraped from the surface of the device during the explanting step. The hole left after MEA extraction also creates a surface through which labeling solution may passively diffuse into the tissue section; misleading label intensities relative to this solution-contacting surface can result, especially with poorly penetrating labels. This problem is exemplified here in the labeling of tissue around a large vascular element in Figure 2d, imaged ~ 50 μm into a 100 μm thick tissue section.
Our results suggest that the DCHist method typically requires thicker (>100 um) tissue sections. Our findings demonstrate that many labels can successfully penetrate through thick tissue by 1) using a dilute blocking and detergent solution in all processing steps (Hamam and Kennedy, 2003), 2) applying each label at a high concentration for a long period of time, and 3) washing thoroughly to avoid background labeling. The method used to fix the tissue (Leong et al., 1988) and the degree of tissue fixation (Pikkarainen et al., 2010) can be important variables in determining accessibility of antigen binding sites to antibody labels. However, antigen retrieval steps may be employed to improve antigen accessibility (D’Amico et al., 2009). With genetic expression of fluorescent markers (Shaner et al., 2005) the limitations of label penetration can be avoided entirely. Detailed imaging of thick tissue sections through LSCM is limited by scattering of light through the tissue. As demonstrated in Figure 5, imaging into both sides of a histology specimen helps to avoid imaging depth limitations (Glaser and Vanderloos, 1981). Depth imaging limitations of LSCM can be overcome by using high sensitivity detectors and longer wavelength laser light, such as the near-infrared light used for two-photon fluorescent excitation (Helmchen and Denk, 2005) in this publication to image the UV-fluorescent Hoechst 33342. Mounting tissue in a ‘clearing solution’, which closely matches the optical index of molecular components, can also promote deeper histological imaging (Tuchin, 2005; Dodt et al., 2007; Tsai et al., 2009), but at the cost of the fixed tissue shrinking somewhat in these solutions.
Although previous studies have shown that it is possible to collect CNS implanted microdevices within fixed tissue sections (Turner et al., 1999; Bragin, 2000; Holecko et al., 2005), the presented work formalizes the consistent capture of brain-implanted microdevices towards improved characterization of the interfacing tissue. Future studies applying this novel histological method to larger numbers of subjects will likely improve our understanding of the inter-subject and intra-subject biological response variability, a subject not addressed in this method report. By also presenting detailed labeling and imaging methods, we hope to encourage the adoption of the resulting DCHist method as a means to closely characterize biological changes around CNS implants.
With the exacting depth and trajectory with which brain implants are typically placed in humans, application of DCHist collection methods could translate well to the analysis of post-mortem human tissue, potentially improving our understanding of implant impact on human tissue. Although we did not attempt to collect multi-shank microdevices, 3D microdevice arrays, or larger CNS implants during this study, we are highly optimistic that their intact collection and analysis is accomplishable. With increased tissue processing time allowing deeper label penetration and the utilization of optical clearing solutions, substantial portions of larger brain-implants and multi-shank implants may be collected, labeled and imaged with only slight modification of the methods presented here. Similarly, we are optimistic that close analysis of data derived from the DCHist method will help to further the optimization of microdevice structure material (Williams, 2008; Winslow et al., 2010) and microdevice shape (Ward et al., 2009; Bjornsson et al., 2006) for use in future brain-implanted microdevices. Coupled with detailed quantification methods (Bjornsson et al., 2008; Kopec et al., 2010), we expect the outlined tissue characterization methods to significantly impact the development of future neural interfacing strategies.
Supplementary Material
Acknowledgments
We thank; M. Slipchenko for technical microscopy assistance; S. Vaidyanathan for thin-film coating expertise; Y. Venkataraman and E. Bartlett for vibratome assistance; A. Panitch and N. Onunkwo for sharing their expertise in utilizing the experimental anti-inflammatory peptide; W. Shain, K. Smith, and C. Bjornsson for previous histological instruction through the Center for Neural Communication Technology, and the Center for Neural Communication Technology for their donation of unmounted microelectrode arrays. We also thank M. Ward, S. Wilks, and A. Koivuniemi for their valuable manuscipt critiques.
Source of funding
This work was supported by the National Institutes of Health (R03DC009339-02(NIDCD)) and the Purdue Research Foundation.
Abbreviations
- DCHist
Device Capture Histology
- LSCM
Laser Scanning Confocal Microscopy
- Trans
Transmission light microscopy
- MEAs
Microelectrode Arrays
Footnotes
Conflict of Interest
The authors declare no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew J. Woolley, Email: awoolley@purdue.edu.
Himanshi A. Desai, Email: hdesai@purdue.edu.
Mitchell A. Steckbeck, Email: msteckbe@purdue.edu.
Neil K. Patel, Email: patel2@purdue.edu.
Citations
- Azemi E, Lagenaur CF, Cui XT. The surface immobilization of the neural adhesion molecule L1 on neural probes and its effect on neuronal density and gliosis at the probe/tissue interface. Biomaterials. 2011;32:681–692. doi: 10.1016/j.biomaterials.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T. Brain-computer interfaces: an international assessment of research and development trends. Springer; Dordrecht: 2008. [Google Scholar]
- Biran R, Martin DC, Tresco PA. Neuronal cell loss accompanies the brain tissue response to chronically implanted silicon microelectrode arrays. Experimental Neurology. 2005;195:115–126. doi: 10.1016/j.expneurol.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, Lin G, Abdul-Karim MA, Szarowski DH, Smith KL, LeBlanc D, Roysam B, Shain W, Turner JN. Brain Vascular Damage Due to Neuroprosthetic Insertion:Casting and Quantitative Analysis. MAM. 2004:10. [Google Scholar]
- Bjornsson CS, Oh SJ, Al-Kofahi YA, Lim YJ, Smith KL, Turner JN, De S, Roysam B, Shain W, Kim SJ. Effects of insertion conditions on tissue strain and vascular damage during neuroprosthetic device insertion. J Neural Eng. 2006;3:196–207. doi: 10.1088/1741-2560/3/3/002. [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, Lin G, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B. Associative image analysis: A method for automated quantification of 3D multi-parameter images of brain tissue. Journal of Neuroscience Methods. 2008;170:165–178. doi: 10.1016/j.jneumeth.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragin A, Hetke J, Wilson CL, Anderson DJ, Engel J, Jr, Buzsáki G. Multiple site silicon-based probes for chronic recordings in freely moving rats: implantation, recording and histological verification. Journal of Neuroscience Methods. 2000;98:77–82. doi: 10.1016/s0165-0270(00)00193-x. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang H, Slipchenko MN, Jung Y, Shi Y, Zhu J, Buhman KK, Cheng J. A multimodal platform for nonlinear optical microscopy and microspectroscopy. Opt Express. 2009;17:1282. doi: 10.1364/oe.17.001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Xie XS. Coherent Anti-Stokes Raman Scattering Microscopy: Instrumentation, Theory, and Applications. J Phys Chem B. 2004;108:827–840. [Google Scholar]
- D’Amico F, Skarmoutsou E, Stivala F. State of the art in antigen retrieval for immunohistochemistry. Journal of Immunological Methods. 2009;341:1–18. doi: 10.1016/j.jim.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Dodt H, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Meth. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Drake K, Wise K, Farraye J, Anderson D, BeMent S. Performance of planar multisite microprobes in recordingextracellular single-unit intracortical activity. IEEE Transactions on Biomedical Engineering. 1988;35:719–732. doi: 10.1109/10.7273. [DOI] [PubMed] [Google Scholar]
- Edell D, Toi V, McNeil V, Clark L. Factors influencing the biocompatibility of insertable silicon microshafts in cerebral cortex. IEEE Trans Biomed Eng. 1992;39:635–643. doi: 10.1109/10.141202. [DOI] [PubMed] [Google Scholar]
- Glaser E, Vanderloos H. Analysis of thick brain sections by obverse—Reverse computer microscopy: Application of a new, high clarity Golgi—Nissl stain. Journal of Neuroscience Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical Deconstruction of Parkinsonian Neural Circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamam B, Kennedy TE. Visualization of the dendritic arbor of neurons in intact 500 μm thick brain slices. Journal of Neuroscience Methods. 2003;123:61–67. doi: 10.1016/s0165-0270(02)00341-2. [DOI] [PubMed] [Google Scholar]
- Hascup ER, af Bjerkén S, Hascup KN, Pomerleau F, Huettl P, Strömberg I, Gerhardt GA. Histological studies of the effects of chronic implantation of ceramic-based microelectrode arrays and microdialysis probes in rat prefrontal cortex. Brain Research. 2009;1291:12–20. doi: 10.1016/j.brainres.2009.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Meth. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hirakawa K, Hashizume K, Hayashi T. Viscoelastic property of human brain -for the analysis of impact injury (author’s transl) No To Shinkei. 1981;33:1057–1065. [PubMed] [Google Scholar]
- Holecko MM, Williams JC, Massia SP. Visualization of the intact interface between neural tissue and implanted microelectrode arrays. J Neural Eng. 2005;2:97–102. doi: 10.1088/1741-2560/2/4/005. [DOI] [PubMed] [Google Scholar]
- Jackson N, Sridharan A, Anand S, Baker M, Okandan M, Muthuswamy J. Long-term neural recordings using MEMS based moveable microelectrodes in the brain. FrontNeuroeng. 2010 doi: 10.3389/fneng.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroch DB, Ward MP, Chow EY, Rickus JL, Irazoqui PP. Magnetic insertion system for flexible electrode implantation. Journal of Neuroscience Methods. 2009;183:213–222. doi: 10.1016/j.jneumeth.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Franklin RK, Gibson MD, Brown RB, Kipke DR. Implantable microelectrode arrays for simultaneous electrophysiological and neurochemical recordings. Journal of Neuroscience Methods. 2008;174:62–70. doi: 10.1016/j.jneumeth.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Martin DC. Sustained release of dexamethasone from hydrophilic matrices using PLGA nanoparticles for neural drug delivery. Biomaterials. 2006;27:3031–3037. doi: 10.1016/j.biomaterials.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Kopec CD, Bowers AC, Pai S, Brody CD. Semi-automated atlas-based analysis of brain histological sections. Journal of Neuroscience Methods. 2010 doi: 10.1016/j.jneumeth.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong AS, Milios J, Duncis CG. Antigen preservation in microwave-irradiated tissues: A comparison with formaldehyde fixation. J Pathol. 1988;156:275–282. doi: 10.1002/path.1711560402. [DOI] [PubMed] [Google Scholar]
- McConnell GC, Rees HD, Levey AI, Gutekunst C, Gross RE, Bellamkonda RV. Implanted neural electrodes cause chronic, local inflammation that is correlated with local neurodegeneration. J Neural Eng. 2009;6:056003. doi: 10.1088/1741-2560/6/5/056003. [DOI] [PubMed] [Google Scholar]
- Moxon K, Leiser S, Gerhardt G, Barbee K, Chapin J. Ceramic-Based Multisite Electrode Arrays for Chronic Single-Neuron Recording. IEEE Trans Biomed Eng. 2004;51:647–656. doi: 10.1109/TBME.2003.821037. [DOI] [PubMed] [Google Scholar]
- Otto KJ, Rousche PJ, Kipke DR. Microstimulation in auditory cortex provides a substrate for detailed behaviors. Hearing Research. 2005;210:112–117. doi: 10.1016/j.heares.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 6. Academic; Oxford: 2007. [DOI] [PubMed] [Google Scholar]
- Panitch A, Otto KJ, Woolley A, Onunkwo N. MK2 Inhibitor compositions and methods to enhance neurite outgrowth, neuroprotection, and nerve regeneration. 2011 http://www.freepatentsonline.com/y2011/0052658.html.
- Pikkarainen M, Martikainen P, Alafuzoff I. The Effect of Prolonged Fixation Time on Immunohistochemical Staining of Common Neurodegenerative Disease Markers. Journal of Neuropathology and Experimental Neurology. 2010;69:40–52. doi: 10.1097/NEN.0b013e3181c6c13d. [DOI] [PubMed] [Google Scholar]
- Rousche P, Pellinen D, Pivin D, Williams J, Vetter R, Kipke D. Flexible polyimide-based intracortical electrode arrays with bioactive capability. IEEE Trans Biomed Eng. 2001;48:361–371. doi: 10.1109/10.914800. [DOI] [PubMed] [Google Scholar]
- Rousche PJ, Normann RA. A method for pneumatically inserting an array of penetrating electrodes into cortical tissue. Ann Biomed Eng. 1992;20:413–422. doi: 10.1007/BF02368133. [DOI] [PubMed] [Google Scholar]
- Seymour JP, Kipke DR. Neural probe design for reduced tissue encapsulation in CNS. Biomaterials. 2007;28:3594–3607. doi: 10.1016/j.biomaterials.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, Lyden PD, Kleinfeld D. Correlations of Neuronal and Microvascular Densities in Murine Cortex Revealed by Direct Counting and Colocalization of Nuclei and Vessels. Journal of Neuroscience. 2009;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchin VV. Optical clearing of tissues and blood using the immersion method. J Phys D: Appl Phys. 2005;38:2497–2518. [Google Scholar]
- Turner JN, Shain W, Szarowski DH, Andersen M, Martins S, Isaacson M, Craighead H. Cerebral Astrocyte Response to Micromachined Silicon Implants. Experimental Neurology. 1999;156:33–49. doi: 10.1006/exnr.1998.6983. [DOI] [PubMed] [Google Scholar]
- Velliste M, Perel S, Spalding MC, Whitford AS, Schwartz AB. Cortical control of a prosthetic arm for self-feeding. Nature. 2008;453:1098–1101. doi: 10.1038/nature06996. [DOI] [PubMed] [Google Scholar]
- Ward B, Seal BL, Brophy CM, Panitch A. Design of a bioactive cell-penetrating peptide: when a transduction domain does more than transduce. J Pept Sci. 2009;15:668–674. doi: 10.1002/psc.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Rajdev P, Ellison C, Irazoqui PP. Toward a comparison of microelectrodes for acute and chronic recordings. Brain Res. 2009;1282:183–200. doi: 10.1016/j.brainres.2009.05.052. [DOI] [PubMed] [Google Scholar]
- Weppelmann E, Field J, Swain M. Observation, analysis, and simulation of the hysteresis of silicon using ultra-micro-indentation with spherical indenters. J Mater Res. 1993;8:830–840. [Google Scholar]
- Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29:2941–2953. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- Williams JC, Holecko MM, Massia SP, Rousche P, Kipke DR. Multi-site incorporation of bioactive matrices into MEMS-based neural probes. J Neural Eng. 2005;2:L23–L28. doi: 10.1088/1741-2560/2/4/L03. [DOI] [PubMed] [Google Scholar]
- Winslow BD, Christensen MB, Yang W, Solzbacher F, Tresco PA. A comparison of the tissue response to chronically implanted Parylene-C-coated and uncoated planar silicon microelectrode arrays in rat cortex. Biomaterials. 2010;31:9163–9172. doi: 10.1016/j.biomaterials.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JO, Cogan SF, Rizzo JF. Neurotrophin-eluting hydrogel coatings for neural stimulating electrodes. J Biomed Mater Res. 2007;81B:551–563. doi: 10.1002/jbm.b.30696. [DOI] [PubMed] [Google Scholar]
- Zhang J, Laiwalla F, Kim JA, Urabe H, Van Wagenen R, Song Y, Connors BW, Zhang F, Deisseroth K, Nurmikko AV. Integrated device for optical stimulation and spatiotemporal electrical recording of neural activity in light-sensitized brain tissue. J Neural Eng. 2009;6:055007. doi: 10.1088/1741-2560/6/5/055007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.