Abstract
Summary: Identifying mentions of named entities, such as genes or diseases, and normalizing them to database identifiers have become an important step in many text and data mining pipelines. Despite this need, very few entity normalization systems are publicly available as source code or web services for biomedical text mining. Here we present the Gnat Java library for text retrieval, named entity recognition, and normalization of gene and protein mentions in biomedical text. The library can be used as a component to be integrated with other text-mining systems, as a framework to add user-specific extensions, and as an efficient stand-alone application for the identification of gene and protein names for data analysis. On the BioCreative III test data, the current version of Gnat achieves a Tap-20 score of 0.1987.
Availability: The library and web services are implemented in Java and the sources are available from http://gnat.sourceforge.net.
Contact: jorg.hakenberg@roche.com
1 INTRODUCTION
The extremely rapid growth of published literature in the biological sciences necessitates the constant improvement of automated text-mining tools to extract relevant information and convert it into structured formats. Terms for the same entities used in biomedical articles can vary widely between authors and across time (Tamames and Valencia, 2006). Thus, two key tasks in biomedical text analysis are named entity recognition (NER; finding names of genes, cell lines, drugs, etc.) and entity mention normalization (EMN; mapping a recognized entity to a repository, such as Entrez Gene or PubChem). Both tasks enable indexing, retrieval and integration of literature with other resources. Gene and protein names in particular represent central entities that are discussed in biomedical texts. While a growing number of tools for gene NER are freely available e.g.(Leaman and Gonzalez, 2008), only a limited number of tools provide gene normalization capabilities that can be used off-the-shelf by end users e.g.(Huang et al., 2011).
In this article, we present a new version of the Gnat system for gene mention recognition and normalization (Hakenberg et al., 2008) and make it available as an open-source Java library and as a remote web service. Gnat now relies on a modular architecture, allowing integration of new components by implementing relatively simple HTTP interfaces and allows its components to be distributed on servers (local or remote; public or private). The framework allows end users to send PubMed or PubMed Central document identifiers as well as free text to our server, returning lists of gene mentions with Entrez Gene IDs. Text mining application developers can make use of the same service by using Gnat as a component in their own processing pipelines or by customizing Gnat toward their requirements.
Here we present the major components in Gnat, demonstrate how they interact and how they can be exchanged and extended by developers. We present an overview of the web services provided, which can be used remotely from our server or set up by users at their local sites. Finally, we discuss the performance of gene mention normalization provided by Gnat.
2 APPROACH
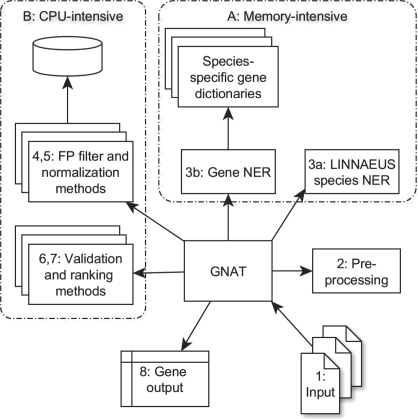
Gnat consists of a set of modules to handle all steps required in a text processing pipeline, from document retrieval to named entity normalization. A general Gnat processing pipeline (Fig. 1) consists of modules that perform the following steps:
Retrieve documents,
Pre-process each text,
Perform named entity recognition for genes and species,
Remove likely false positive gene mentions,
Assign candidate identifiers to genes,
Validate identifiers, and
Rank candidate gene identifiers.
Fig. 1.
Overview of the Gnat processing pipeline with typical components [(1) through (7); see text] and final output (8). Gnat is designed in a modular manner, where data exchange is performed using the HTTP protocol. It allows memory- and CPU-intensive components (A and B) to be run separately on appropriate hardware. Memory-intensive components typically run as (remote or local) services, as they require longer startup times less suited for small queries. The Gnat client (center) manages which components to invoke in which manner, and sends data to the components for annotation. Some components rely on annotations provided by other components, such as the assignment of candidate identifiers during step (5), which requires species annotations from step (3a).
Steps (1) and (2) comprise essential text retrieval and pre-processing tasks. Document retrieval uses NCBI e-utils to obtain records from PubMed and PubMed Central when such identifiers are given. Pre-processing of texts includes, for instance, a name range expansion that replaces mentions such as ‘freac1-3’ with ‘freac1, freac2, and freac3’, to aid subsequent gene NER.
In step (3), Gnat recognizes names referring to both genes and species using a dictionary-based approach. A set of candidate Entrez Gene identifiers is assigned to each gene mention in this step as well, comprising all potential matches based on the gene's name alone. The NER modules available in the current default version of Gnat include the species-dependent dictionary lookups present in previous versions (Hakenberg et al., 2008) for 20 common model organisms including human, mouse, rat and Escherichia coli. In addition to the dictionary-based gene NER taggers, we now provide an interface to Banner (Leaman and Gonzalez, 2008), which uses conditional random fields to recognize candidate gene names. Users can select either of these NER modules, the joint results of both methods or implement their own NER component (3b in Fig. 1). To identify species names, we incorporated Linnaeus (Gerner et al., 2010) (3a in Fig. 1), whose output determines which dictionary-based gene taggers to run, and to narrow down identifiers for ambiguous gene names later in the pipeline [step (6)].
Steps (4) to (7) comprise the actual gene mention normalization task, for which we have implemented a range of filters to remove likely false positive gene mentions as well as candidate IDs. Removal of false positives (FPs) uses information in the gene name itself, the surrounding text, as well as entire paragraphs or full text to ensure that a found name refers to a specific gene, and not another non-gene term. Likely FPs are further removed if not also recognized by BANNER. Note that in contrast to most gene name identification tools, mentions that refer to gene families are considered FPs in the current version of Gnat, since the aim is to find gene mentions that can be mapped to a specific entry in Entrez Gene. Thus, one of the filters removes mentions such as ‘G proteins’, although this step can be tailored to specific needs.
Candidate identifiers can then be further filtered or validated, for example, by removing genes from species not mentioned in the text, or by other user-defined methods [step (6)]. In step (7), the remaining ambiguous cases (gene mentions with more than one potential Entrez Gene ID) are ranked by comparing contextual information found in the text surrounding the mention with knowledge about each gene. For example, known Gene Ontology annotations for a gene increase its rank when that GO term is found in the nearby text, and similar methods are used for chromosomal locations, associated diseases, enzymatic activity, tissue specificity, etc. More details on the individual components, especially for disambiguation and normalization, can be found in (Hakenberg et al., 2008) and (Solt et al., 2010), which discuss specific implementations for BioCreative II and III, respectively (also see Section 5).
3 USING THE GNAT JAVA LIBRARY
For each of the aforementioned steps, we provide implementations as Gnat components that can be used as they are, extended or replaced by developers within their own pipelines. Most components can run either locally within the client (for instance, during development) or as remote services (with public or restricted access). For example, users might want to implement different NER strategies or supply custom dictionaries for species currently not provided in the default version. Users might alternatively want to include non-specific gene mentions that could be mapped to structured vocabularies such as MeSH that include gene families, or to include information from DNA or protein sequences in the text to improve gene mention normalization (Haeussler et al., 2011). Likewise, the final ranking methods can be adapted, or different input/output formats could be defined.
4 USING THE GNAT WEB SERVICE
The Gnat system also implements web services using HTTP POST and GET requests that can be used by both end users and developers. To submit a text for annotation, the following three URL parameters can be used: pmid, pmc and text (combinations are allowed). The pmid parameter takes one or more comma-separated PubMed IDs as values, pmc takes PubMed Central ID(s) and text takes a text (sentence, paragraph, full document) in plain ASCII format. Note that for large requests (especially when submitting full-text articles), an HTTP POST request should be used instead of GET.
Users can also modify the default behavior of the web service to specify the particular tasks to perform with the parameter task, which can take gner (gene NER), sner (species NER) or gnorm (normalization to Entrez Gene IDs) as arguments. Specifying tasks can be useful when application developers want to take Gnat's NER results as input for their own pipelines, for instance. Finally, the user can specify the format of the returned results (parameter: returntype), either as a tab-separated list (value: tsv, which is the default) or XML (xml). A help page listing all current parameters and valid values is available by calling the service without parameters. In addition to making these web services available as source code, we also host a remote service for a set of 10 common model organisms (available at http://gnat.sourceforge.net).
5 DISCUSSION
Large-scale community challenges to assess the status and compare methods for gene mention normalization have been ongoing since 2003 see the overview of BioCreative I, Task 1B, in Hirschman el al., 2005). Gnat has been evaluated on three BioCreative datasets: BioCreative I is composed of abstracts from papers on mouse, fruit fly, and yeast genes, BioCreative II is composed of abstracts from papers on only human genes, and BioCreative III is composed of full-text articles with no restriction on species. For human genes only, an earlier version of Gnat was ranked first among the participants in BioCreative II (Morgan et al., 2008), achieving a precision and recall of 82.1 and 81.6%, respectively, on a test set of 262 abstracts. Subsequent modifications to Gnat improved precision to 90.1% and with recall at 81.1 (Hakenberg et al., 2008). On a dataset derived from BioCreative I+II, covering genes from 13 species in 100 abstracts (Hakenberg et al., 2008), the provided default processing pipelines achieves 79% precision at 65% recall. For BioCreative III, performance was evaluated using the TAP-k metric (Lu et al., 2010), which is based on a ranked list of predictions (Carroll et al., 2010). The 50 manually annotated full-text articles chosen for maximal variability among submissions served as the gold standard for BioCreative III, on which Gnat achieves a TAP-20 score of 0.1987, with the highest ranking method yielding only 0.3466 (Lu et al., 2010). Due to the difference in the scoring metrics, results are not easy to compare directly between BioCreative challenges; our own experiments show precision and recall values for the current system of 53.6 and 47.4%, respectively, on the manually curated training data (see Lu et al., 2010).
One drawback of the current default processing pipeline of Gnat relative with the BioCreative III test set comes from limiting our predictions to genes from 20 model organisms. The manually curated gold standard for 50 full-text documents includes an unusual composition of species compared with the training set: for example, 23% of all genes in the gold standard belong to Enterobacter sp. 638. This species and three more that contribute an additional 15% to the gold standard genes are not currently supported by the default dictionary-based NER in Gnat, but user-specific dictionaries could be added quickly when new species are anticipated, a procedure for which we provide detailed instructions in the documentation. Future extensions of the Banner library within Gnat to map any recognized gene name to species and candidate IDs should also help to make up for the low recall introduced by the current species limitation in species supported.
The current version of Gnat implements a client–server architecture, where individual modules can be set up to run within the client or as servers. Typically, a module would run as a server if it performs a memory-intensive processing step, requires a certain time for startup or is a finalized component; modules run as client are those which are suited to individual users' needs or those undergoing development. Using the default pipeline, it takes an average of 5 s to annotate a PubMed abstract; however, this number clearly depends on the underlying hardware and usage of remote services and can thus serve only as a rough estimate. Given the modular architecture, Gnat's modules can be easily tailored or replaced. For example, Gnat currently relies on Linnaeus for species NER and provides an interface to Banner for gene NER, demonstrating the ability to easily incorporate external tools, especially if they provide a Java API.
6 CONCLUSION
Here we presented the Gnat library for gene name recognition and normalization in biomedical text, now freely available from SourceForge at http://gnat.sourceforge.net. Gnat is written in a modular fashion to allow end users to annotate their textual data using the public web services, as well as text-mining developers to customize Gnat and host their own remote services, either public or private. Gnat provides many individual components of a typical text processing and gene name normalization pipeline, which can be extended or swapped by developers where necessary. As such, Gnat adds to the set of open-source tools now available for researchers to use for large-scale gene name normalization studies, providing a variety of access points to different users, from end users submitting text to a web service and treating Gnat's processing pipeline as a ‘black box’, to developers who use only some of Gnat's modules and replace others.
Precision and recall of Gnat can range from 54/47% on full-text documents that do not match main model organisms, to 82/82% on abstracts that reflect the species composition of the majority of PubMed. For the latter, consisting of an ensemble of ten common model organisms, we host web services that accept PubMed and PubMed Central IDs and free text as input, and return mentions and/or EntrezGene identifiers, which we hope will provide an opportunity to enhance research across many domains of bioinformatics.
Funding: Biotechnology and Biological Sciences Research Council (CASE studentship to M.G., grant BB/G000093/1 to C.M.B., G.N.); the European Commission (grant HEALTH-F4-2008-223210 to C.M.B.); German Academic Exchange Service (DAAD) to I.S.
Conflict of Interest: none declared.
REFERENCES
- Carroll H.D., et al. TAP-k: a measure of retrieval designed for bioinformatics. Bioinformatics. 2010;26:1708–1713. doi: 10.1093/bioinformatics/btq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner M., et al. Linnaeus: a species name identification system for biomedical literature. BMC Bioinformatics. 2010;11:85. doi: 10.1186/1471-2105-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M., et al. Annotating genes and genomes with DNA sequences extracted from biomedical articles. Bioinformatics. 2011;27:980–986. doi: 10.1093/bioinformatics/btr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenberg J., et al. Inter–species normalization of gene mentions with GNAT. Bioinformatics. 2008;24:i126–i132. doi: 10.1093/bioinformatics/btn299. [DOI] [PubMed] [Google Scholar]
- Hirschman L., et al. Overview of BioCreAtIvE task 1B: normalized gene lists. BMC Bioinformatics. 2005;6:S11. doi: 10.1186/1471-2105-6-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., et al. GeneTUKit: a software for document-level gene normalization. Bioinformatics. 2011;27:1032–1033. doi: 10.1093/bioinformatics/btr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman R., Gonzalez G. BANNER: An executable survey of advances in biomedical named entity recognition. Pac. Symp. Biocomput. 2008;13:652–663. [PubMed] [Google Scholar]
- Lu Z., Wilbur J. Proceedings of the BioCreative III. USA: Bethesda/MD; 2010. Overview of BioCreative III Gene Normalization. 20-35, September 13-15. [Google Scholar]
- Morgan A., et al. Overview of BioCreative II Gene Normalization. Genome Biol. 2008;9:S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt I., et al. Proceedings of the BioCreative III. USA: Bethesda/MA; 2010. Gene mention normalization in full texts using Gnatand Linnaeus. 134-139, September 13-15. [Google Scholar]
- Tamames J., Valencia A. The success (or not) of HUGO nomenclature. Genome Biol. 2006;7:402. doi: 10.1186/gb-2006-7-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]