Abstract
Motivation: The ability to detect copy-number variation (CNV) and loss of heterozygosity (LOH) from exome sequencing data extends the utility of this powerful approach that has mainly been used for point or small insertion/deletion detection.
Results: We present ExomeCNV, a statistical method to detect CNV and LOH using depth-of-coverage and B-allele frequencies, from mapped short sequence reads, and we assess both the method's power and the effects of confounding variables. We apply our method to a cancer exome resequencing dataset. As expected, accuracy and resolution are dependent on depth-of-coverage and capture probe design.
Availability: CRAN package ‘ExomeCNV’.
Contact: fsathira@fas.harvard.edu; snelson@ucla.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The development of next-generation sequencing has enabled routine large-scale resequencing projects, permitting us to perform increasingly more comprehensive DNA variant analysis. However, the cost and analytical complexity of sequencing still limit the number of whole genomes that can be sequenced in any single project (Teer and Mullikin, 2010). In fact, the analysis of complete human genome sequence often interprets DNA alterations in protein coding regions primarily. This is in practice a reasonable strategy since ~85% of the disease-causing mutations are found in the coding regions or canonical splice sites (Choi et al., 2009). Thus, whole-exome sequencing presents an effective alternative to whole-genome sequencing and provides an unbiased, cost-effective and time-efficient tool for the study of the genetic basis for disease. Following the first successful application of whole-exome sequencing in re-discovering the cause of a dominantly inherited rare Mendelian disorder Freeman–Sheldon syndrome (Ng et al., 2009), a number of studies have reported similar successes (Chou et al., 2010; Hoischen et al., 2010; Johnston et al., 2010; Raca et al., 2010; Rehman et al., 2010; Volpi et al., 2010). Although the cost of both genome and exome sequencing continues to fall at a rapid pace, whole-exome sequencing has a number of advantages, including a lower cost, more straightforward data analysis and interpretation and significantly greater depth of coverage with a corresponding overall improvement in data quality. Exome sequencing is rapidly becoming a fundamental tool for genetic and functional genomic research laboratories and a diagnostic tool in clinics. At present the main applications of targeted exonic sequencing is for the determination of single nucleotide variants (SNVs) or small indel variants but not structural variation.
Structural variation, especially copy-number variation (CNV) and loss of heterozygosity (LOH), is an important class of genetic variability in Mendelian, common inherited diseases and cancer (Choi et al., 2007; Deng et al., 2010; Fong et al., 1989; Jankowska et al., 2009; Sha et al., 2009; Shuin et al., 1994; Stankiewicz and Lupski, 2010; Wain et al., 2009). As is true of SNVs, there are population-specific, common CNVs and rare, disease-causing CNVs (Kato et al., 2010; Sudmant et al., 2010). Many large-scale projects (Conrad et al., 2010a,b; McCarroll, 2010) and technological platforms (Pinkel et al., 1998; Urban et al., 2006) have been devised to estimate the prevalence and impact of CNV. Array Comparative Genomic Hybridization (CGH) (Pinkel et al., 1998) and SNP genotyping arrays have been widely used as standard methods to detect CNV and LOH. However, with the rapid increase in genomic and exomic sequence, there is growing interest in the use of these data to detect CNVs.
While methods have been developed for CNV estimation in whole-genome sequencing (Chiang et al., 2009; Yoon et al., 2009), these methods make key assumptions that fail to hold in the exome sequencing setting. For example, Yoon et al. (2009) assumes random, unbiased distribution of sequence reads, such that read depth can be modeled as a normal distribution across the genome, and deviation from the background indicates the presence of CNV. This random read distribution assumption breaks down in the context of exome capture as the probes have variable specificity and efficiency for the targeted exonic regions. The discrete nature of exome sequences also presents problems to existing methods. Many whole-genome CNV detection tools use segmentation algorithms that assume continuity of search space and do not function properly when given discontinuous and variable length exome sequencing data. SegSeq (Chiang et al., 2009), for example, merges windows of a fixed length based on a log-ratio difference statistic. Lastly, because exons are generally smaller than insert sizes for paired-end sequencing (200–500 bp), paired-end based CNV detection methods are not generally applicable to exome data.
Here we present ExomeCNV, which uses depth-of-coverage and B-allele frequencies from mapped short sequence reads to estimate CNV (Fig. 1, left side) and LOH (Fig. 1, right side). We describe an assessment of its validity, sensitivity, specificity and limitations through an analysis of a melanoma tumor and a matched normal sample. Important model assumption and the effect of important confounding factor such as sample admixture rate are also considered.
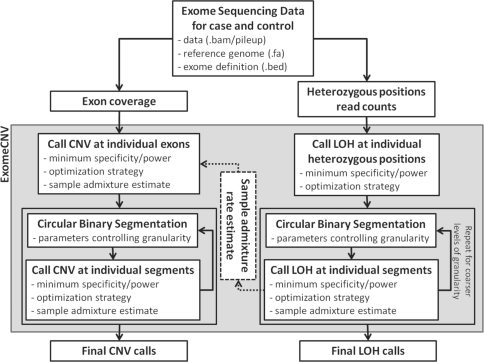
Fig. 1.
Overview of ExomeCNV analysis workflows. Two workflows are present: CNV detection and LOH detection. Each involves similar steps of exon/position/segment-wise CNV/LOH calling, Circular Binary Segmentation, and interval merging. User inputs and parameters are listed at each step.
2 METHODS
2.1 Correlation of depth-of-coverage
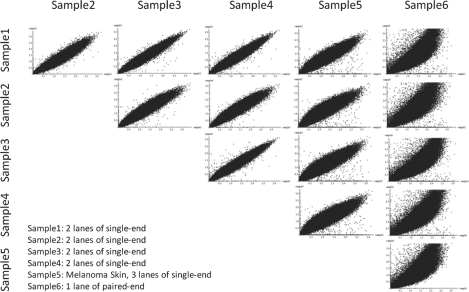
To plot the correlation of depth-of-coverage (Fig. 2), we used the internal exome data that were captured by the same Agilent SureSelect Human All Exon Kit and sequenced on the Illumina GAIIx. Samples 1 through 4 were generated by two lanes of 76 bp single-end sequencing, Sample 5 was generated by three lanes of 76 bp single-end sequencing and Sample 6 was generated by one lane of 76 + 76 bp paired-end sequencing. For each sample, the average depth-of-coverage per exon was normalized by dividing the average coverage by the overall exome average coverage and then, the normalized depth-of-coverage were compared between 15 pairs of samples.
Fig. 2.
Correlation of depth-of-coverage across exome sequencing samples. To demonstrate the consistency of capture and sequencing efficiency of individual exons represented by the depth-of-coverage per exon, the normalized individual exon coverage in all pairs of six-independent exomes were plotted. All 6 samples were captured using the Agilent SureSelect Human All Exon G3362. Samples 1–5 had mean base coverages of 36~39× as a result of 2 (Samples 1–4) or 3 (Sample 5) lanes of GAIIx single-end sequencing per sample. Sample 6 had mean base coverage of 60× as a result of 1 lane of GAIIx paired-end sequencing and demonstrates substantially different biases in individual exonic depth-of-coverage.
2.2 The CNV detection algorithm
2.2.1 Power analysis of CNV Detection
Consider an exon of length L, let X and Y denote the numbers of reads, each of length w, mapped within the exon in question in case (e.g. tumor) and control (e.g. matched normal), respectively. The depth-of-coverage is then Xw/L and Yw/L for case and control, respectively. Although we discuss our method in terms of depth-of-coverage, our method is developed in terms of the count statistics X and Y. Let NX and NY be the total numbers of aligned reads in case and control, respectively. Define the read count ratio:
| (1) |
We divide the raw counts X and Y by the total number of reads NX and NY to mitigate the effect of overall increase in local counts due to the increase in total depth-of-coverage. Finally, we adjust the ratio so that the exome-wide median is 1. Without lost of generality, we assume NX=NY and reduce R=X/Y. Because X and Y follow Poisson distributions with parameters λX and λY, respectively, with sufficient depth-of-coverage the Poisson distributions converge to normals with equal means and variances: N(λX, λX) and N(λY, λY). Under the null hypothesis of no CNV, λX=λY, and under the alternative hypothesis, λX=ρλY=ρλ. ρ indicates the copy-number ratio; for example, ρ=0.5 for deletion and ρ=1.5 for duplication. By Geary–Hinkley transformation (Geary, 1930,1944; Hinkley, 1969), let
| (2) |
and t(ρ) follows the standard normal distribution. Thus, the specificity and sensitivity are 1 − α and 1 − β where
| (3a) |
| (3b) |
The formulas above describe the achievable specificity and sensitivity of a given cutoff ratio R. Considering values of R from zero to infinity, we can plot the receiver operating characteristic (ROC) curve (Supplementary Materials). In practice, we may wish to identify a cutoff r(ρ) which yields the desired minimum specificity and/or sensitivity for testing a particular copy-number ratio ρ at an exon with certain depth-of-coverage and length. This can be achieved by solving above equations, and an appropriate cutoff r(ρ) can be chosen from a set of solutions to maximize user-selected quantity metrics such as specificity, sensitivity or area under curve.
In the presence of sample admixture, the ‘true’ copy-number ratio will tend to 1. In particular, if a fraction c of the tumor sample has a normal copy-number (either by contamination of normal tissue or heterogeneity within the tumor), the copy-number ratio of this admixed sample will be ρ′=c+ρ(1−c). Thus, in heterogeneous samples, the only change to the method described above is the replacement of ρ by ρ′. The admixture rate c can be estimated from data by back-calculating c from empirical ρ′ in LOH regions (see Supplementary Materials).
2.3 Segmentation and sequential merging
We used the circular binary segmentation (CBS) algorithm (Olshen et al., 2004), as implemented in the R package DNAcopy (Venkatraman and Olshen, 2007), to subdivide the genome (exome). For each segment we combined the coverage by direct sum, and used mean coverage log ratio as the segment's log ratio log(R). Log ratio is used here to satisfy the input requirement of CBS algorithm. CNV call proceeds on each segment in the same manner as described above.
In order to achieve the most sensitive segmentation, we chose to start CBS with parameters that produce a large number of small segments, call CNV on the segments, and repeat the process with a smaller number of larger segments. We then merge the CNV segments sequentially, from finest segmentation to coarsest. By nature of CBS, finer segments are contained within coarser segments, and in merging step, we need to resolve conflicting CNV calls between finer segments within a larger segment. If a finer (smaller) segment has sufficient coverage to call a CNV event, the call persists. However, if it does not have sufficient coverage to reject the null hypothesis (i.e. not being called, or being called as copy-number neutral), a positive CNV call in a larger segment overrides the negative calls. An illustration of this sequential merging algorithm is given in the Supplementary Materials.
2.4 LOH detection
First, we consider all polymorphic positions in the exome of the control sample, and for each of the positions, the B-allele count B is the number of reads with non-reference- or B-allele at that position. For a polymorphic position i, let Ni be the total number of reads mapped to that position (i.e. depth-of-coverage); thus the B-allele count Bi follows a binomial distribution Binomial (pi, Ni). A binomial that rejects the null hypothesis: pi=0.5 can be used to detect LOH at each polymorphic position.
Segmentation is done using CBS algorithm based on the absolute difference in B-allele frequencies (BAFs) |BAFi,case − BAFi,control|, where BAFi=Bi/Ni. Within each segment, based on the realization that B-allele frequencies deviate from the null value of 0.5 under LOH, an F-test for equality of variance is used to detect significance increase in variance of BAFcase from that of BAFcontrol (other statistics were also considered, see Supplementary Materials). Finally, the LOH calls are merged sequentially as described above.
2.5 Exome sequencing and data analysis
2.5.1 Exome sequencing
High molecular weight whole genomic DNA from matched skin and tumor pair samples of a melanoma patient were sequenced on the Illumina Genome Analyzer (GAIIx). The UCLA Institutional Review Board (IRB) approved the collections of the DNA samples. The libraries were generated following the Agilent SureSelect Human All Exon Kit version 1.0.1 protocol, and the Illumina Genome Analyzer (GAIIx) flowcell was prepared according to the manufacturer's protocol. We performed three and four lanes of single-end sequencing for each of the skin and tumor samples, respectively, within the UCLA Center of High-throughput Biology (CHTB). The base-calling was performed by the real time analysis (RTA) software (version 1.6) provided by Illumina.
2.5.2 Sequence data analysis
Novoalign from Novocraft Short Read Alignment Package (http://www.novocraft.com/index.html) was used to align each lane's QSEQ file to the reference genome. Human Genome reference sequence (hg18, March 2006, build 36.1), downloaded from the UCSC genome database located at http://genome.ucsc.edu and mirrored locally, was indexed using novoindex program (-k 14 -s 3). The output format was set to SAM and default settings were used for all options. Using SAMtools (http://samtools.sourceforge.net/), the SAM files of each lane were converted to BAM files, sorted and merged for each sample and potential PCR duplicates were removed using Picard (http://picard.sourceforge.net/) (Li et al., 2009). To retrieve the depth of coverage information of each base, we generated a PILEUP file for each sample using SAMtools and calculated the average coverage per capture interval using a custom script. Here, we used processed BAM files that were used to call the SNVs while reducing the likelihood of using spuriously mismapped reads to call the variants: the last 5 bases were trimmed and only the reads lacking indels were retained (Clark et al., 2009). The detailed description of the mutational landscape of this tumor sample is in preparation.
2.6 Genome-wide SNP genotyping
Both the skin and the tumor samples were submitted to the Southern California Genotyping Consortium (SCGC) at UCLA for genotyping on the Illumina Omni-1 Quad BeadChip, which consists of 1 140 419 SNPs (1 016 423 genotyping probes and 123 996 CNV probes) distributed across the genome. The Illumina GenomeStudio V2010.1 Genotyping Module version 1.6.3 was used to calculate the BAF values and the log R ratio (LRR) for each probe and the copy number aberration (CNA) and LOH of the autosomes were inferred from these values using the genoCN R package (Sun et al., 2009). The genoCNA function was used with the default parameters.
2.7 Copy-number analysis using ERDS
The same PILEUP file we used to generate the average coverage per capture interval was used to run Estimation by Read Depth with SNVs (ERDS) to demonstrate the need to control for the variability of capture of exons observed in exome sequencing (M.Zhu et al., manuscript in preparation). The SNV file that is required by ERDS was generated by using SAMtools varFilter tool default parameters and SVA (Sequence Variant Analyzer) snp_filter.pl script. The result file generated by ERDS was summarized using the SVA software (http://people.genome.duke.edu/~dg48/sva/index.php).
2.8 Comparison between CNV calling methods
In assessing performance of ExomeCNV, we used all mapped exons as the sample space. CNV calls on other platforms were mapped to the exons and compared to calls by ExomeCNV. Thus specificity is the proportion of copy-neutral exons correctly identified by ExomeCNV, while sensitivity is the proportion of amplified (or deleted) exons correctly identified. Similarly, LOH performance is assessed using all polymorphic positions as the sample space.
3 RESULTS
3.1 ExomeCNV for CNV and LOH detection
ExomeCNV uses a normalized depth-of-coverage ratio approach to identify CNV and LOH from exome sequencing information of paired case/control samples (for example, paired tumor/normal) in a way that optimizes sensitivity and specificity. We begin by assuming that although there are potentially exon-specific biases due to laboratory capture methods and sequence-specific biases, these are independent of sample and so are nearly uniform for a particular exon across samples. As a result, simply assessing the ratio of depth-of-coverage of each exon reduces such bias (see Supplementary Materials).
3.1.1 Correlation of depth-of-coverage across exome sequencing samples
To establish validity of this fundamental assumption, we compared depth-of-coverage of exons across five independent samples from five different subjects (Samples 1–5 in Fig 2). All samples were captured using the same probe set (Agilent SureSelect Human All Exon G3362) and sequenced at mean base coverages of 36–39× as a result of two (Samples 1–4) or three (Sample 5) lanes of GAIIx single-end sequencing per sample (see Section 2). As shown in Figure 2, a high correlation was observed among the five samples (Pearson correlation 0.908–0.975, mean=0.947, SD=0.027), arguing for the validity of our assumption.
The same level of consistency was not observed when single-end data were compared with paired-end data (Sample 6; Pearson correlation 0.855–0.877, mean=0.871, SD=0.009) due to the lack of independence between pairs of reads in paired-end data. Thus, care must be taken to ensure consistency of library preparation and sequencing method between samples used in analysis. Here, all of our analyses used exome sequencing data from a melanoma (Sample 5) and a matched normal skin, both processed and sequenced in the same manner (see Section 2).
3.1.2 Analytic power calculation of exonic CNV and LOH detection
For each exon, the number of sequencing reads aligning within it appears to follow a Poisson with mean directly proportional to the size of the exon and the copy-number (see Supplementary Materials), but assuming that we have sufficiently deep coverage, we can approximate this by a normal distribution with mean equal to variance. We apply the Geary–Hinkley transformation (Geary, 1930,1944; Hinkley, 1969), which converts a ratio of two normally distributed variables to a standard normal distribution, and a CNV is identified by a significant deviation of the transformed ratio from the null, standard normal distribution (see Section 2). Allowing only one false positive per genome, we analytically determine the statistical power of this CNV detection approach for different depth-of-coverage, and the results are shown in Figure 3a–b. For detecting deletions, 95% power is achieved for segments of size 500 bp or more (Fig. 3a), while detection of a single copy duplication is achieved with 95% power for segments of size 1,000 bp or more (Fig. 3b) with a mean segmental base coverage of 35×. We note that the power of the method improves substantially with higher depth-of-coverage, and an individual exons deletion/duplication status would be more powerfully observed by including additional flanking intronic sequence in the capture probe design. Genomic DNA admixture, as expected, diminishes power, but even with 35× coverage of a given exon, a length of greater than 1000 bp is observed over 95% of the time. Exons or segments captured with 500 bp at target sequence are observed at 95% power only with greater than 55× base coverage. A more thorough consideration of specificity and ROC curves are produced in the Supplementary Materials.
Fig. 3.
Examples of the power of ExomeCNV to detect segmental duplication, deletion and LOH based on an analytical calculation. Power is plotted relative to mean depth-of-coverage in the genomic segment, setting false positive to 1 per genome based on an analytical model of genome-wide power of detection at different window sizes (inset, a–d). Windows are the total length of a given sequence at a given exon or the sum of length of exons adjacent to each other in the genome. The effect of admixture (rate of 30%) on the power to detect deletions and single copy duplications are shown in (c) and (d), respectively. (e) plots the power of LOH detection versus depth-of-coverage of individual polymorphic position (single base pair) with variable admixture rates (inset). The periodicity of the power curve is due to discrete nature of the binomial test. The 35× depth of coverage is chosen because it is a typical minimal average depth of coverage for exome sequencing and is thus a conservative view of power within typical exome sequencing datasets.
To estimate LOH, we focused on the non-reference-allele or BAF of polymorphic positions in the sequenced regions. The observed B-allele count at a polymorphic position can be modeled by a binomial distribution with depth-of-coverage as sample size and the probability of observing a B-allele proportional to the B-allele copy-number, which is equivalent to the LOH state. Because the expected value of BAF at a normal (non-LOH) polymorphic position is 0.5, a significant deviation of BAF from 0.5 identifies LOH. With sufficient depth-of-coverage, LOH can be detected at a single polymorphic position.
3.1.3 The effect of sample admixture
The specificity and sensitivity of this CNV detection method depends not only on the depth-of-coverage but also the rate of admixture, whereby non-mutated genomes contaminate mutated genomes in the sampled tissue/cells. In the absence of admixture, the average depth-of-coverage ratio is 0.5 for deletion, 1.5 for one-copy duplication and the BAF at an LOH site is either 0 or 1; however, in cancer biopsy sequencing, this is rarely observed in practice due to admixture with normal or non-mutated tumor cells. Thus, we assess the effect of admixture ranging from 10% to 70%, which is frequently observed in tumor samples (Fig. 3e). With admixture, the depth-of-coverage ratio and the BAF will tend to the null values of 1 and 0.5, respectively, making the CNV and LOH detection harder. Figure 3c and d show a reduction of power in detecting deletion (Fig. 3c) and one-copy duplication (Fig. 3d) as a result of 30% admixture. There is an approximate two-fold increase in the size of the exonic sequence detectable in the presence of 30% non-mutated genomic DNA. Capping the false positive rate at 0.001 and assuming 35× mean depth-of-coverage, a power curve (Fig. 3e) shows 0.95 sensitivity of detecting LOH at a single polymorphic position with admixture up to 30%.
3.1.4 Using circular binary segmentation to merge exonic CNV/LOH
Because CNV and LOH can, and usually do, span multiple exons, we extended our method above to call CNV/LOH on larger segments derived from summing data of sequentially spaced exons in the human genome. We apply circular binary segmentation (CBS) (Olshen et al., 2004; Venkatraman and Olshen, 2007) to subdivide the genome and then combine depth-of-coverage of exons and BAF of polymorphic positions within each segment, composed of arbitrary number of individual exons, to search for larger CNV and LOH. In the case of CNV, since reads are independent of each other, the sum of depth-of-coverage of all exons in a segment constitutes the segment's depth-of-coverage, and the CNV test can be performed as described above. In the case of LOH, since B-alleles are not always on the same chromosome, BAF cannot be combined by direct summation. Instead, since BAF deviates from the null value 0.5 under LOH, a significance increase in variance of BAF from control (F-test for equality of variances) indicates LOH (several other statistics were also considered, see Supplementary Materials). Finally, we repeated the process of CBS and CNV/LOH-calling, ranging granularity of segmentation from finest to coarsest, and merged the CNV/LOH calls by prioritizing positive calls of finer segments over coarser ones (see Section 2 for details). In the case of our melanoma sample, we performed CBS/sequential merging at five levels of granularity and observed 165 130 merging events in the first iteration followed by 121, 79, 105 and 66 in the subsequent iterations for a total of 165 501 merging events.
3.2 Validation
To test the performance of ExomeCNV we analyzed exome sequencing data from a melanoma and a matched normal skin (Supplementary Materials); the average depth-of-coverage of the data is 42.8× for the tumor and 37.5× for the normal sample, which are sufficient to achieve at least 90% sensitivity and specificity based on the power calculation above.
3.2.1 Validating false positive and false negative rates
We first estimated the false positive rate of the algorithm by calling CNVs on two sequencing lanes of the same normal tissue library, treating one as case and the other as control; any CNV call from this would be false positive. Our method correctly called most exons as non-CNV. In particular, setting P-value thresholds to ensure minimum specificity of 0.9, 0.99 and 0.999, we observed specificity of 0.916, 0.995 and 1.0, respectively (Supplementary Materials). Furthermore, we tested the sensitivity of ExomeCNV by analyzing copy-number of sex chromosomes in a pair of male and female exome data that were available internally (see Section 2). Using the male exome as control, ExomeCNV correctly identified female chromosome X as being ‘duplicated’ and chromosome Y as being ‘deleted’ (Supplementary Materials) with no false negatives.
3.2.2 Comparison with SNP genotyping array
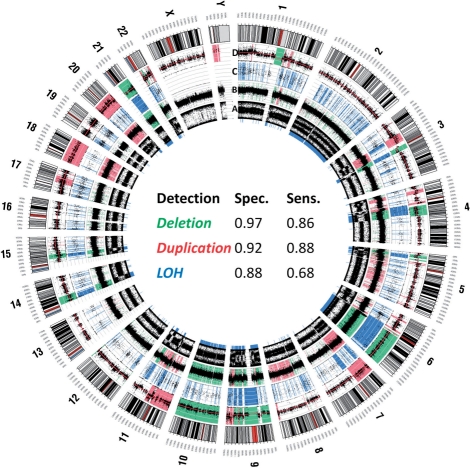
We then used ExomeCNV to predict CNV and LOH in the melanoma samples and compared our results to those obtained from Illumina Omni-1 Quad Beadchip genotyping array assessment of the same samples (Fig. 4 and Supplementary Materials). The sizes of the CNV segments from ExomeCNV range from single exon (120 bp) to whole chromosome (chr 10 and 18) (size distribution of CNV calls is presented in the Supplementary Materials). Treating calls from the genotyping array experiment as a standard, ExomeCNV had 97% specificity and 86% sensitivity for detecting deletions, 92% specificity and 88% sensitivity for detecting amplifications, and 88% specificity and 68% sensitivity for detecting LOH even though there is substantial variability across the genome. Higher depth-of-coverage from the sequence data for each exome would likely further improve concordance. We note that this is a dramatic improvement of ExomeCNV relative to the inappropriate application of ERDS (M.Zhu et al., manuscript in preparation) CNV caller which, when applied to these data, achieves only 16% sensitivity and 83% specificity for deletion and 50% sensitivity and 56% specificity for amplification (see Circo plot showing results from the three methods in the Supplementary Materials). This is due to the fact that there is substantial variability of the efficiency of capture of exons in exome sequencing not accounted for in ERDS. For CNV segments called by ExomeCNV but not by the genotyping arrays, we found that most lie within regions in which there is a low density of genotyping markers; thus the false positive rates (and the associated specificity) for ExomeCNV here may in fact be lower.
Fig. 4.
Analysis of melanoma and paired normal samples. Interpretation of deletion, duplication and LOH from exonic sequence data using ExomeCNV and plotted with Circos. The most outer ring shows the chromosome ideograms in a pter–qter orientation, clockwise with the centromeres in red. From inside to outside, each data track represents (A) B-allele-frequency (BAF) from Omni-1 genotyping array with the region of LOH highlighted in blue underneath the track; (B) Log R Ratio (LRR) from genotyping array with the region of gain highlighted in red and the region of loss highlighted in green; (C) BAF from ExomeCNV output from ~40× depth-of-coverage exome sequencing with the region of LOH highlighted in blue; (D) log ratio of tumor and normal depth-of-coverage with the segment mean in red line, the region of gain highlighted in red and the region of loss highlighted in green. The LOH and CNV for the chromosome X and Y were not called for the genotyping data as genoCN (the algorithm used to call CNV from Omni-1) is not designed to analyze chromosome X and Y. The table in the middle summarizes best achievable specificity and sensitivity of ExomeCNV in detecting CNV and LOH relative to CNV/LOH calls from Omni-1 array assessment.
4 DISCUSSION AND CONCLUSION
The resolution of CNV detection with ExomeCNV is limited largely by the probe design. The CNV segments identified by our method range from 120 bp (single exons with higher than average coverage) to 240 Mb in size (whole chromosomes); however, the true breakpoint can be anywhere in the space between the terminal exon called within a CNV region and the adjacent exon in a non-CNV region. Hence, although a given CNV event can be detected at a single exon in some instances, the absolute resolution of our method is in fact limited to the inter-exon distance around an exon, which can be as small as 125 bp or as large as 22.8 Mb with the median of 5 kb (statistics based on SureSelect Human All Exon Kit G3362).
Although ExomeCNV relies on the availability of matched control samples, we can also derive a matched control sample from a pool of other samples, which then serves as an effective control. This is useful for the identification of germline inherited or de novo CNVs in an individual. Because the expected copy number in the reference population is constant (usually two), by the law of large numbers, averaging depth-of-coverage from sufficiently many samples yields a good control set, assuming that they are all captured using the same probe set and capture method and sequenced in the same manner. This may limit the application of ExomeCNV to data generated at a given site with a given protocol. Calling CNVs using this pooled sample as background will generate CNV calls that are present in the case sample but not the control population. Also, by the central limit theorem, pooling independent samples helps reduce variance in depth-of-coverage and increases precision of our method. We have pooled as few as eight samples and have observed that this is indeed the case (Supplementary Materials). However, it is important to note that using pooled sample as control imposes a strong assumption that the samples do not share common CNV regions and that the population has an average genomic copy number of two. Other potential challenges of using the pooled sample as control are discussed in the Supplementary Materials.
Because ExomeCNV depends on an estimate of the admixture rate c, misspecification of c would affect its performance. We performed sensitivity analysis and found that misestimating c would have a strong effect on sensitivity and specificity of CNV detection. Fortunately, LOH detection provides some data to directly estimate c, as LOH detection does not depend on a prior knowledge of c (Supplementary Materials). For the melanoma sample, our estimate of 30% admixture rate matches that from genotyping arrays, confirming the validity of this approach. However, there are advantages to slightly overestimating c as it makes the method more conservative and reduces false positives.
As we have shown, CNV and LOH detection is readily possible from exome sequencing data, extending the utility of this powerful approach. The fundamental basis that makes this approach possible is the consistency of depth-of-coverage of each exon (and BAF by extension) across multiple samples for each individual exon, as demonstrated in five samples performed in our laboratory (Supplementary Materials, Fig. 2). This consistency permits reliable parametric modeling of the shift in depth-of-coverage and BAF distributions, hence accurate identification of CNV and LOH. However, we do not observe the same level of consistency when comparing depth-of-coverage across different library types. For instance, a sixth sample was performed using a paired-end approach that results in very different coverage of each exon (Fig. 2), and as a result, ExomeCNV does not perform well when the control sample library is of one type and the case is of another, or when the case and control have significantly different coverage levels. Resolving these issues is a work-in-progress.
From the analytical power calculations, assuming 35× coverage (which is the lower end of a reasonable amount of sequence for variant calling and easy to generate with a variety of technologies), CNV detection has a limit of about 500 bp (in transcript coordinates), which is typically equivalent to 2–3 exons and spans about 10 kb of genomic space on average. Increased depth-of-coverage, which is likely to become the norm as sequencing costs decrease, reduces the interval size that is reliably detectable and should push the method to single exonic deletion resolution. Currently, CNV and LOH information should be detectable in whole-exome sequencing data at a resolution that is almost equivalent to what one can obtain from a dense SNP genotyping array.
ExomeCNV is available as a CRAN package ‘ExomeCNV’.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Kevin Squire and the members of Nelson Lab for thoughtful discussion and suggestion, Markus Schröder for help with R package and Bret Harry, Zugen Chen and Traci Toy for technical support.
Funding: National Heart, Lung, Blood Institute of the US National Institutes of Health grant (1RC2 HL101715, to J.Q.); National Institute for Mental Health (R01 MH071852); National Institute of Arthritis, Musculoskeletal and Skin Disorders (P30 AR057230); National Cancer Institute (P30 CA16042).
Conflict of Interest: none declared.
REFERENCES
- Chiang D.Y., et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.H., et al. Hypermethylation and loss of heterozygosity of tumor suppressor genes on chromosome 3p in cervical cancer. Cancer Lett. 2007;255:26–33. doi: 10.1016/j.canlet.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Choi M., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou L.S., et al. DNA sequence capture and enrichment by microarray followed by next-generation sequencing for targeted resequencing: neurofibromatosis type 1 gene as a model. Clin. Chem. 2010;56:62–72. doi: 10.1373/clinchem.2009.132639. [DOI] [PubMed] [Google Scholar]
- Clark M.J., et al. U87MG Decoded: The Genomic Sequence of a Cytogenetically Aberrant Human Cancer Cell Line. PloS Genet. 2009;6 doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D.F., et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010a;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D.F., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010b;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F.Y., et al. Genome-wide copy number variation association study suggested VPS13B gene for osteoporosis in Caucasians. Osteoporos. Int. 2010;21:579–587. doi: 10.1007/s00198-009-0998-7. [DOI] [PubMed] [Google Scholar]
- Fong C.T., et al. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: correlation with N-myc amplification. Proc. Natl Acad. Sci. USA. 1989;86:3753–3757. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary R.C. The frequency distribution of the quotient of two normal variates. J. R. Stat. Soc. 1930;93:442–446. [Google Scholar]
- Geary R.C. Extension of a theorem by Harald Cramer on the frequency distribution of the quotient of two variables. J. R. Stat. Soc. 1944;107:56–57. [Google Scholar]
- Hinkley D.V. On ratio of 2 correlated normal random variables. Biometrika. 1969;56:635–639. [Google Scholar]
- Hoischen A., et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum. Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- Jankowska A.M., et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.J., et al. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., et al. Population-genetic nature of copy number variations in the human genome. Hum. Mol. Genet. 2010;19:761–773. doi: 10.1093/hmg/ddp541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A. Copy number variation and human genome maps. Nat. Genet. 2010;42:365–366. doi: 10.1038/ng0510-365. [DOI] [PubMed] [Google Scholar]
- Ng S.B., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B., et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Pinkel D., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Raca G., et al. Next generation sequencing in research and diagnostics of ocular birth defects. Mol. Genet. Metab. 2010;100:184–192. doi: 10.1016/j.ymgme.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A.U., et al. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am. J. Hum. Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B.Y., et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J. Hum. Genet. 2009;54:199–202. doi: 10.1038/jhg.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuin T., et al. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Sudmant P.H., et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer J.K., Mullikin J.C. Exome sequencing: the sweet spot before whole genomes. Hum. Mol. Genet. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A.E., et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman E.S., Olshen A.B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- Volpi L., et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain L.V., et al. The role of copy number variation in susceptibility to amyotrophic lateral sclerosis: genome-wide association study and comparison with published loci. PLoS One. 2009;4:e8175. doi: 10.1371/journal.pone.0008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., et al. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.