Abstract
The HIV/AIDS pandemic is primarily caused by HIV-1. Another virus type, HIV-2, is found mainly in West African countries. We hypothesized that population migration and mobility in Africa may have facilitated the introduction and spreading of HIV-2 in Mozambique. The presence of HIV-2 has important implications for diagnosis and choice of treatment of HIV infection. Hence, the aim of this study was to estimate the prevalence of HIV-2 infection and its genotype in Maputo, Mozambique.
HIV-infected individuals (N = 1,200) were consecutively enrolled and screened for IgG antibodies against HIV-1 gp41 and HIV-2 gp36 using peptide-based enzyme immunoassays (pepEIA). Specimens showing reactivity on the HIV-2 pepEIA were further tested using the INNO-LIA immunoblot assay and HIV-2 PCR targeting RT and PR genes. Subtype analysis of HIV-2 was based on the protease gene.
After screening with HIV-2 pepEIA 1,168 were non-reactive and 32 were reactive to HIV-2 gp36 peptide. Of this total, 30 specimens were simultaneously reactive to gp41 and gp36 pepEIA while two samples reacted solely to gp36 peptide. Only three specimens containing antibodies against gp36 and gp105 on the INNO-LIA immunoblot assay were found to be positive by PCR to HIV-2 subtype A.
The proportion of HIV-2 in Maputo City was 0.25% (90%CI 0.01-0.49). The HIV epidemic in Southern Mozambique is driven by HIV-1, with HIV-2 also circulating at a marginal rate. Surveillance program need to improve HIV-2 diagnosis and consider periodical survey aiming to monitor HIV-2 prevalence in the country.
Keywords: HIV-2, laboratory diagnosis, sub-Saharan Africa, Mozambique
Introduction
Human immunodeficiency virus (HIV) type 2 (HIV-2) is a related but distinct virus from HIV-1, in spite of having similar gene products, genetic organization and cell tropism in vitro. HIV-2 also shows decreased cell killing, syncytium formation, and replication rates when compared with HIV-1. These features may justify the lower transmissibility of HIV-2, as well as its reduced pathogenicity and longer clinical course. Nevertheless, HIV-2 can cause immunosuppression and progression to AIDS [1-3]. HIV-2 was first isolated in West Africa in 1986 and has been reported in many countries of Western Africa such as the Gambia, Guinea, Ghana, Cape Verde, Guinea-Bissau, Cote d'Ivoire, Liberia, Senegal, and Niger [4,5]. Outside Africa, HIV-2 infection is sporadically detected in countries with socioeconomic relations with West Africa such as Portugal, Spain, France, United Kingdom and India [6-9]. The presence of HIV-2 is not regularly surveyed and generally assumed to be low in other African countries outside West Africa. However, in the last three decades there has been an increase in population migration within Africa, mainly attributed to regional wars, political upraises and even to increased international travel inside Africa [3,10].
The laboratory diagnosis of HIV-2 infection is challenged by the high cross-reactivity rate between the two viruses. Overall, HIV diagnosis is performed by a combination of one screening assay, often an Enzyme Immunoassay (EIA), followed by a confirmatory Western blot assay. However, the use of HIV-1 Western blots may lead to misclassification of HIV-2- infected individuals since HIV-1 and HIV-2 Western blots lack specificity for type-specific diagnosis because there is significant cross-reactivity to gag, pol, and env oligomeric proteins. PCR has been used as gold standard for HIV-2 diagnosis but it still remains a research tool and a commercial assay is not yet available [3,11,12].
The diagnosis of HIV-2 has important implications for the choice of antiretroviral treatment (ART) regimens, as HIV-2 strains are naturally resistant to non-nucleoside reverse transcriptase and fusion inhibitors and are, at least in vitro, less sensitive to some protease inhibitors [13]. Consequently, accurate diagnosis of HIV-2 infection is critical for clinical management of patients [14,15].
Mozambique is situated in southeastern Africa and is bordered by six countries: Tanzania, Malawi, Zambia, Zimbabwe, Swaziland and South Africa and gained independence from Portugal in 1975, after nearly five centuries of Portuguese occupation. Furthermore, in the last two decades, there have been a considerable number of people from West Africa, Europe, and Asia visiting and living in Mozambique. These migratory movements may have facilitated the introduction and spreading of HIV-2 in Mozambique. Despite the existence of sporadic reports, the prevalence of HIV-2 in Mozambique is largely unknown [16]. In this work, we utilized a combination of serological and molecular techniques to estimate the prevalence of HIV-2 infection in Maputo City, Mozambique.
Materials and methods
Study design and subjects
This was a cross-sectional study that enrolled individuals with a recent HIV diagnosis attending the Centro de Saúde do Alto Maé outpatient clinic in Maputo City, Mozambique. A total of 1,200 consecutive individuals were recruited to participate at the time of routine CD4+ T-lymphocyte (CD4+) count. After written informed consent, two EDTA tubes were collected from each volunteer for CD4+ count and HIV-2 screening test, both performed at the Instituto Nacional de Saúde (INS), Mozambique. Confirmatory serological testing and HIV-2 molecular characterization was conducted at the Federal University of Rio de Janeiro, Brazil. The study protocol was approved in 2009 by the Mozambican National Health Bioethics Committee under Protocol number 129/CNBS.
HIV serological assays
All individuals enrolled in this study had been previously diagnosed as HIV infected according to the Mozambican algorithm for HIV diagnosis which employs two sequential HIV-1/2 rapid tests: the Determine HIV 1/2 test (Abbott Laboratories, Tokyo, Japan), used for screening and UniGold HIV test (Trinity Biotech, Ireland), used to confirm initial reactivity on the Determine assay. HIV infection is ascertained only if both assays are reactive. Of note, both assays have the HIV-2 gp36 antigen on the solid phase, and are able to detect HIV-2 specific antibodies.
In addition to the HIV-1/HIV-2 specific peptide-EIA (pepEIA) assay described below, two confirmatory serological assays were used in this study: INNO-LIA HIV I/II Score (Innogenetics, Gent, Belgium) and Bio-Rad New Lav Blot II (Bio-Rad, Marnes-la-Conquette, France).
In order to initially differentiate the reactivity against HIV-1 and HIV-2 we used the same assay described by Qiu et all [17]. Briefly, synthetic peptides, carrying 14 amino acid residues from the immunodominant regions of transmembrane proteins of HIV-1 (gp41) or HIV-2 (gp36) were synthesized with the addition of 7 non-specific residues at the amino-terminus (RGDKDGG) to increase the solubility of both peptides. HIV-1 or HIV-2 peptides were coated onto wells on separate microplates (Nalge Nunc International, Rochester, NY) at 5 μg/mL in 100 μL of 10mM phosphate buffer saline (PBS), pH 7.4 for 2 hr at 37°C. Following five washes with PBS, wells were blocked with 100 μL of BSA buffer (PBS with 0.05% Tween-20 and 0.5%BSA). Serum or plasma samples (diluted 1:100 in BSA buffer) were added to the coated wells and incubated for 1 hr at 37°C. After the wash, bound antibodies were detected by goat anti-human IgG peroxidase conjugate (30 min at 37°C, diluted in BSA buffer), followed by substrate (TMB) incubation for 25 min at room temperature.
The cutoff values were arbitrarily set at 0.5 OD for both EIAs; all negative specimens were below the cutoff value. The criteria for interpreting results were as follows: 1) HIV-1 OD ≥ 0.5 and HIV-2 OD < 0.5 were considered HIV-1 reactive only; (2) HIV-2 OD ≥ 0.5 and HIV-1 OD < 0.5 were considered HIV-2 reactive only; 3) HIV-1 OD and HIV-2 OD ≥ 0.5 were considered HIV-1 and HIV-2 dually reactive and; 4) HIV-1 OD and HIV-2 OD < 0.5 were considered non reactive. Samples dually reactive were further diluted 1:10 (to a final serum dilution of 1:1,000) and retested on the pepEIA, and the results were interpreted as above.
Molecular detection and characterization
Buffy coats from samples suspected of having HIV-2 were used for DNA extraction using a blood DNA extraction kit (QIAamp, Qiagen, Hilden, Germany), according to the manufacturer's instructions. Detection of HIV-2 proviral DNA was carried out using nested polymerase chain reaction (PCR) of region pol (protease and transcriptase reverse) with some modifications to the protocol described by Colson et al [14]. Nested PCR was performed using 5 μL of extracted genomic DNAThe PCR conditions were 2,5U of Taq Platinum DNA polymerase (Invitrogen Life Technologies, Carlsbad, Calif), 1× polymerase buffer, 0.4 mM of each deoxynucleoside triphosphate, 3mM of Mgcl2, and 25 pmol of the outer primers (forward: H2Mp1, 5'-GGGAAAGAAGCCCCGCAACTTC-3';reverse: H2Mp2, 5'-GGGATCCATGTCACTTGCCA-3') in the first round. This amplification was carried out in a 50 μl reaction mixture to obtain a genomic fragment of 1,753 bp. The cycling conditions were 94°C for 2 min, followed by 40 cycles of 94°C for 30sec, 56°C for 45 sec, and at 72°C for 2 min) with one final cycle of 72°C for 7 min. The 2nd round was performed using 5 μL of the previous amplification product and 25 pmol of the inner primers (forward: H2Mp3, 5'-ACTTACTGCACCTCGAGCA-3'; reverse: H2Mp4, 5'-CCCAAATGACTAGTGCTTCTT-3'). Five microliters of the amplicon of the first-round were added in the second-round of PCR and performed under the same conditions of the 1st PCR to obtain a genomic fragment of 1,507 bp. Amplicons were detected by electrophoresis on 1,5% agarose gel and visualized under UV light after ethidium bromide staining.
To detect HIV-1 nested PCR was designed to amplify a 1000-bp fragment encompassing 297 bp of the protease gene and 703 bp of the RT gene. The conditions were 2.5 mM MgCl2, 0.4 mM of each deoxynucleotide triphosphate, 25 pmol of each primer (forward: K1F, 5'-5' CAGAGCCAACAGCCCCACCA -3'; reverse: K2R, 5'- TTTTCCCACTAACTTCTGTCATTGAC-3'), and 1.25U of Taq Platinum DNA polymerase (Invitrogen, CA, USA), 1x polymerase buffer and 5 μL of DNA genomic extracted in a final volume of 50 μL. The thermal conditions of amplification in the thermocycler device (ABI 9700, Applied Biosystems, USA) were: 94°C for 3 min, 5 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 45 sec. and extension at 72°C for 2 min followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 45 sec. and extension at 72°C for 2 min. and incubation at 72°C for 7 min. The secondround was done with 3 μL of the amplicon of the first-round and 25 pmol of inner primers (forward: DP10, 5'- 5'CAACTCCCTCTCAGAAGCAGGAGCCG 3'; reverse: RT12 5'- ATCAGGATGGAGTTCATAACCCATCCA-3') in the same conditions of the first-round. Amplicons of 1000bp were detected by electrophoresis on 1% agarose gel and visualized under UV light after ethidium bromide staining [17-19].
For each run, HIV-1 and HIV-2 positive samples, in addition to a genomic DNA from an individual seronegative for HIV-1/2, were used as controls. The gp41 immunodominant region (IDR) was sequenced following the protocol previously described [20].
For sequencing analysis, PCR products corresponding to 1000 bp and 1507 bp of HIV-1 and HIV-2 pol region respectively were purified using QIAamp PCR purification kit (Qiagen) according to the manufacturer's instructions. The amplicons were directly sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit, with a 3730 Automated DNA Sequencer (Applied Biosystems, USA).
Sequences were edited and aligned using BioEdit program v5.0.9. (Department of Microbiology, North Carolina State University, USA), and Clustal W algorithm, respectively. Neighbor-joining (NJ) tree was built on Mega 4.0 software. The NJ tree was evaluated by bootstrap analysis of 1,000 replicates. The mean genetic distance between the Mozambican HIV-2 samples was determined using substitution model on Mega 4.0 software.
Statistical analysis
Confidence Interval (CI) was used to address precision of the proportion estimates and the degree of confidence was set to 90% [21].
Results
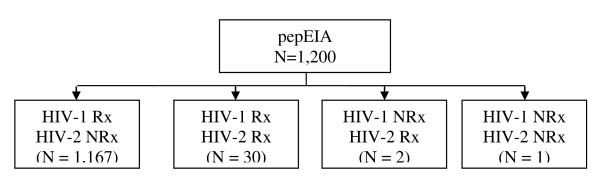
We initially screened 1,200 specimens by parallel testing with HIV-1 and HIV-2 pepEIA (Figure 1). One sample (FEH0045) was non-reactive to the gp41 and gp36 derived peptide. Further testing with the Vironostika HIV Uni-Form II plus O and the HIV-1 Western blot was also negative. Consistently with this serological finding, a PCR reaction set to amplify the reverse trasncriptase and protease regions from HIV-1 and HIV-2 was negative.. This sample was removed from analysis.
Figure 1.
Results of the pepEIA screening test for HIV types 1 and 2 in 1,200 HIV infected patients followed-up at the Alto Maé outpatient clinic. Rx = pepEIA reactive and NRx = pepEIA non-reactive.
A total of 1,167 specimens were reactive only on the HIV-1 gp41 pepEIA after the initial screening. Thirty samples were simultaneously reactive to gp41 and gp36 peptides and after re-testing these specimens with a 1:10 dilution, 17 became reactive to HIV-1 peptide only. The remaining 13 samples remained reactive to both HIV-1 and HIV-2 peptides (Table 1). Finally, two samples were reactive with the gp36 peptide only after the initial screening. For these last two samples, HIV-2 reactivity persisted after 1:10 specimen dilution.
Table 1.
Serological and genomic tests results of 32 patients reactive on the pepEIA HIV-2 screening assay
| Result of pepEIA screeninga | Result of confirmatory assay | Result of nested PCR (Subtyping) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INNO-LIA HIV 1/2b | ||||||||||||
|
Patient code |
HIV-1 | HIV-2 | HIV type | |||||||||
| HIV-1 OD | HIV-2 OD | HIV-2 OD (1/10)d | p24 | gp41 | gp120 | gp36 | gp105 |
New LAV Blotc II (HIV-2) |
HIV-1 | HIV-2 | ||
| FEH0251 | 2578 | 563 | 155 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0049 | 2886 | 592 | 356 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0220 | 2815 | 624 | 196 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0105 | 2820 | 694 | 283 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0345 | 2804 | 713 | 230 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0047 | 3024 | 718 | 310 | 3 | 3 | 3 | 2 | neg | 1-2 | nd | POS (C) | Neg |
| FEH0190 | 2972 | 937 | 144 | 3 | 3 | 3 | 1 | neg | 1-2 | nd | POS (C) | Neg |
| FEH0027 | 2979 | 947 | 273 | 2 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0596 | 2892 | 962 | 160 | 3 | 3 | 3 | neg | 1 | 1-2 | Indeterm | POS (C) | Neg |
| FEH0281 | 2539 | 1086 | 296 | 3 | 3 | 3 | 2 | neg | 1-2 | POS | POS (C) | Neg |
| FEH0226 | 3008 | 1275 | 416 | 3 | 3 | 2 | 2 | neg | 1-2 | POS | POS (C) | Neg |
| FEH0210 | 2820 | 1619 | 393 | 3 | 3 | 3 | 1 | neg | 1-2 | POS | POS (C) | Neg |
| FEH0244 | 2805 | 1695 | 291 | 2 | 3 | +/- | 2 | neg | 1-2 | Indeterm | POS (C) | Neg |
| FEH1044 | 2645 | 1764 | 446 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0327 | 2885 | 2156 | 389 | 3 | 3 | 2 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0700 | 2405 | 699 | 116 | +/- | 4 | 2 | neg | 2 | 1-2 | nd | POS (C) | Neg |
| FEH0687 | 2799 | 1485 | 279 | 1 | 4 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0086 | 2959 | 1426 | 755 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0394 | 2794 | 1794 | 864 | 3 | 3 | 3 | 1 | neg | 1-2 | POS | POS (C) | Neg |
| FEH1049 | 3010 | 2146 | 729 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0875 | 2859 | 2222 | 702 | 2 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0982 | 2412 | 2261 | 1425 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0404 | 3031 | 2262 | 1054 | 3 | 3 | 3 | neg | neg | 1 | POS | POS (C) | Neg |
| FEH0857 | 2837 | 2272 | 1481 | 3 | 3 | 2 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0646 | 3008 | 2364 | 573 | 3 | 3 | 3 | neg | neg | 1 | POS | POS (C) | Neg |
| FEH1196 | 3017 | 2620 | 1145 | 2 | 3 | 3 | neg | neg | 1 | Indeterm | POS (C) | Neg |
| FEH1097 | 2996 | 2662 | 547 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH1040 | 2996 | 2697 | 672 | 3 | 3 | 3 | neg | neg | 1 | nd | POS (C) | Neg |
| FEH0681 | 2882 | 2069 | 2988 | 2 | 4 | 3 | 1 | neg | 1-2 | nd | POS (C) | Neg |
| FEH0089 | 2858 | 2809 | 1600 | 3 | 3 | 3 | 3 | 3 | 1-2 | POS | POS (C) | POS (A) |
| FEH0726 | 197 | 2051 | 922 | neg | neg | neg | 3 | 1 | 2 | POS | Neg | POS (A) |
| FEH0924 | 194 | 2779 | 2080 | +/- | neg | neg | 2 | 3 | 2 | POS | Neg | POS (A) |
aUnderlined numbers denotes reactivity on the pepEIA;
bOnly reactivity to antigens of interest is reported;
c nd, not tested; indeterm, indeterminate;
d assay performed with sample further diluted 1/10
The 30 initially dually reactive samples and the two HIV-2 reactive samples were further tested on the INNO-LIA immunoblot assay (Table 1). All 30 pepEIA dually reactive samples reacted against all five HIV-1 specific proteins present on the INNO-LIA (gp120, gp41, p31, p24 and p17), except for sample FEH1196 that was negative for p17. The analysis of reactivity against HIV-2 proteins (gp105 and gp36) revealed that among the 17 samples that became HIV-2 negative after re-testing with 1:10 sample dilution in the pepEIA assay, gp36 alone was present on six samples, gp105 alone was present on two samples and both bands were absent for nine samples. Of the 13 samples that remained HIV-2 positive after pepEIA re-testing with 1:10 sample dilution, 10 did not react to gp36 or gp105 while two reacted against gp36 only. Solely one sample (FEH0089) reacted to both gp36 and gp105 HIV-2 proteins. The two samples reactive on gp36 pepEIA only, reacted to both HIV-2 gp36 and gp105 proteins. One of these specimens (FEH0726) did not react to any HIV-1 proteins present on the INNO-LIA assay while the other (FEH0924) showed weak reactivity to the HIV-1 p24 protein (Table 1).
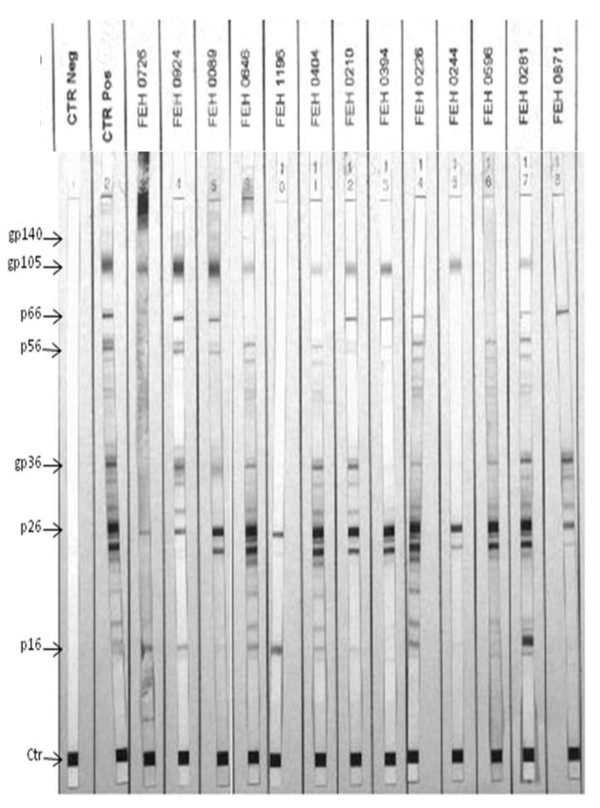
In order to explore the discriminatory power of HIV-2 Western blot we have analyzed the capacity of 9 HIV-1/HIV-2 dually reactive samples in recognizing HIV-2 proteins (Figure 2). The data indicated no discriminatory pattern and, on the basis of the manufacturer's WB interpretation criteria, all samples could be considered as HIV-2 positive. Likewise, the reactive on the HIV-1 Western blot from the 2 solely HIV-2 pepEIA reactive samples were evaluated. Sample FEH0726 did not react to any HIV-1 proteins; however, sample FEH0924 showed a strong reactivity to gp120 and weaker reactivity to p24, p31, p51 and p66 (data not shown).
Figure 2.
Evaluation of HIV-2 Western blot profile (New Lav Blot II). The first two strips are negative (CTR Neg) and positive (CTR Pos) controls. The other lanes show samples from patients infected with HIV-2 (FEH 00726 and 0924), dually infected (FEH 0089) and dually reactive to HIV-1 and HIV-2 gp41/36 reactive, except for sample FEH0871 which is reactive to gp41 peptide only. The arrow indicates the molecular weights of viral proteins in kDa.
All 32 samples showing reactivity to the HIV-2 gp36 peptide on the pepEIA were submitted to HIV-2 specific PCR. Only three samples were positive by PCR: one double- reactive on the pepEIA (FEH0089) and the two samples that reacted with the gp36 peptide only (FEH0726 and FEH0924). With the exception of these last two samples, all other 30 samples were also amplified using HIV-1 specific primers and the majority of those analyzed grouped with subtype C based on Stanford HIV Database subtyping tool (Table 1). Interestingly, the three HIV-2 PCR positive samples were the only samples reactive to both gp36 and gp105 on the INNO-LIA immunoblot. All HIV-2 positive samples (FEH0089, FEH0726 and FEH0924) pol gene (1507 bp fragment) were analyzed and sequences clustered with HIV-2 subtype A (Table 1) sequences after phylogenetic analysis.
In order to explore the cross-reactivity between HIV-1 and HIV-2 antibodies, a sub-group of HIV-1 positive samples (n = 22), some displaying cross-reactivity to HIV-2 (n = 16), were sequenced at the gp41 gene which included the 14 amino acid present at the gp41 immunodominant peptide used in the pepEIA. The gp41/36 IDR aa alignment showed that consensus sequence of 5 HIV-1 samples displaying no cross-reactivity to HIV-2 were not drastically different from 16 samples displaying cross-reactivity to HIV-2 IDR peptide. Of note, we found samples like FEH0871 and FEH0281 from both groups showing some similar aa signatures in IDR peptide to HIV-2 consensus such as A27 and R29 (Table 2).
Table 2.
Analysis of amino acid sequence of the variable region of gp41 IDR peptide at positions 21 to 34 from 22 samples showing mono and dually reaction to HIV-1/HIV-2 gp41 IDR peptide
| HIV-2 peptide | 21L | N | S | W | G | C | A | F | R | Q | V | C | H | T34 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 peptide | L | G | I | W | G | C | S | G | K | L | I | C | T | T | |
| FEH0856 | non-xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0871 | non-xR | . | . | M | . | . | . | . | . | R | . | . | . | . | . |
| FEH0882 | non-xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH1107 | non-xR | . | . | M | . | . | . | . | . | . | . | . | . | . | . |
| FEH1117 | non-xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0049 | xR | . | . | L | . | . | . | . | . | . | . | . | . | . | . |
| FEH0105 | xR | . | . | L | . | . | . | . | . | . | . | . | . | . | . |
| FEH0210 | xR | . | . | . | . | . | . | . | N | . | . | . | . | . | . |
| FEH0327 | xR | . | . | M | . | . | . | . | . | . | . | . | . | . | . |
| FEH0027 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0047 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0281 | xR | . | . | V | . | . | . | A | . | . | H | . | . | . | . |
| FEH0596 | xR | . | . | L | . | . | . | . | A | . | . | . | . | . | . |
| FEH0086 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0394 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0404 | xR | . | . | M | . | . | . | . | . | . | . | . | . | . | . |
| FEH0646 | xR | . | . | L | . | . | . | . | . | . | . | . | . | . | . |
| FEH0875 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0982 | xR | . | . | M | . | . | . | . | . | . | F | . | . | . | . |
| FEH1049 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH1097 | xR | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| FEH0089 | DI | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
non-xR - HIV-1 positive samples that did not show cross-reactivity to gp36 peptide on pepEIA; xR - HIV-1 positive samples that showed cross-reactivity to gp36 peptide on pepEIA; DI - HIV-1 and HIV-2 dually infected sample. Dots mean similarity between sequences and bold underlined letters correspond to positions with HIV-1 genetic divergence but similar amino acid to HIV-2.
Discussion
This study found a low prevalence (0.25%, CI 90% 0.01-0.49) of HIV-2 in Maputo City, Southern Mozambique among HIV positive population attending ARV clinics. Additionally, of the three patients with HIV-2 infection, one was co-infected with HIV-1 subtype C. The number of dually infected individuals was lower than the HIV-2 infected subjects (one versus two). Barreto et al (1993) described the presence of individuals infected with HIV-2 in the city of Maputo since the late 80's using serological testing, and claimed that the entry of this virus was due to post war movement of population growth especially among countries of Portuguese language in this continent [22]. The overall prevalence of HIV-2 infected individuals found in our study is very low when compared to figures from West Africa where HIV-2 and dual infections account for 1 to 10%. This is similar to what is shown in West Africa where HIV-1 epidemic is increasing and overtaking HIV-2 infections [3,23-26].
Phylogenetically, all three HIV-2 specimens segregated as subtype A and due to a large (> 10%) genetic distance between them it is likely that these patients represent three different independent transmission events of HIV-2 in country. The presence of subtype A isolates in Mozambique fits with the hypothesis of introduction of HIV-2 from West Africa, where this variant dominates the epidemic. In fact, subtype A is the major HIV-2 variant representing the epidemic in Portuguese Speaking countries in West Africa such as Guine Bissau and Cape Verde [27-30,12].
On the other hand, EIA using IDR peptides were able to distinguish HIV-1 and HIV-2 infection with a sensitivity of 100% and specificity of 97.6% (90% CI 99.7% to 99.9%). There was a high proportion of cross-reactivity between HIV-1 and HIV-2 IDR peptides from sera of individuals infected with HIV-1. It seems that sera of HIV-2 mono-infected individuals solely recognize HIV-2 related IDR peptide in our pepEIA assay. The cross-reactivity of some HIV-1 infected individuals can be a source of false "dual infection" diagnosed in many publication based solely in IDR peptides EIA screening. In this sense, our data are in agreement with Ciccaglione et al [31]. This cross-reactivity can be minimized by 1:10 serum dilution in pepEIA which is an alternative strategy to identify false-reactive HIV-2 samples. Our data clearly demonstrated that the presence of gp105 and gp36 bands in INNO-LIA assay was strongly associated with HIV-2 infection. However, the same is not true when solely one of these bands is recognized. In our study the PCR reaction targeting HIV-2 pol region was very sensitive and specific to identify truly HIV-2 infected individuals. This fact led us to conclude that gp41/36 IDR pepEIA can be a good screening method followed by INNO-LIA and PCR as confirmatory tests for HIV-2 infection. This algorithm of test seems to be very robust and enable us to identify 3 HIV-2 infections among 1199 HIV positive sera.
The prevalence of HIV-2 in Mozambique and the boundary countries need to be monitored throughout the time to have a better understanding of its trend and geographical distribution. Limited information exists regarding commercial serological assays for HIV-2 diagnosis [32,33]. Therefore, the continuous survey for HIV-2 need to be incorporated at serological and molecular level in HIV surveillance programs in sub-Saharan countries. Guidelines for ART and algorithms for HIV diagnosis must take into account the prevalence of HIV-2 infection in a country. The new testing algorithm proposed here can be implemented in regions where HIV-2 is reported to circulate in order to optimize the 1st line ARV regimens as well PMTCT interventions.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CA, and RC carried out the molecular genetic studies, AT participated in the sequence alignment and drafted the manuscript. CM, and DC carried out the immunoassays. NB, and CW participated in clinical sites selecting patients. OCFJr participated in the design of the study and performed the statistical analysis. IJ and At FG conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript
Contributor Information
Cremildo Maueia, Email: cregoma@yahoo.com.br.
Deise Costa, Email: dacosta@biologia.ufrj.br.
Bindiya Meggi, Email: bindiyameggi@gmail.com.
Nalia Ismael, Email: nalusha180@hotmail.com.
Carla Walle, Email: carla_wale@yahoo.com.br.
Raphael Curvo, Email: raphaelpmc@terra.com.br.
Celina Abreu, Email: celina@biologia.ufrj.br.
Nilesh Bhatt, Email: nbhatt.mz@gmail.com.
Amilcar Tanuri, Email: atanuri1@gmail.com.
Ilesh V Jani, Email: ivjani@email.com.
Orlando C Ferreira, Jr, Email: orlandocfj@gmail.com.
Acknowledgements
We are thankful to the laboratory and Hospital staff of the Centro de Saúde do Alto Maé and to Dr Ramadan Sultane from the Hospital Militar de Maputo. Special thank to Adolfo Vubil, Dulce Bila, Thebora Sultane, Cynthia Semá, Cidia Francisco and to the staff of the Departamento de Imunologia of the INS for their collaboration to this project. We are also in debt to Miss Tasmiya Ira for her valuable work on patient recruitment.
The study was partially supported by the Brazilian National Research Council, CNPq (grant # 490.439/2007-1).
References
- Lemey P, Oliver G, Pybus OG, Bin W, Saksena NK, Salemi M, Vandamme AM. Tracing the origin and history of the HIV-2 epidemic. PNAS. 2003;100(11):6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso H, Borrego P, Bártolo I, Marcelino JM, Família C, Quintas A, Taveira N. Evolutionary and Structural Features of the C2, V3 and C3 Envelope Regions Underlying the Differences in HIV-1 and HIV-2 Biology and Infection. Plos One. 2011;6(1) doi: 10.1371/journal.pone.0014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omobolaji T, Campbell-Yesufu, Rajesh TG. Update on Human Immunodeficiency Virus (HIV)-2 Infection. Clin Infect Dis. 2011;51:780–787. doi: 10.1093/cid/ciq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F. HIV-2, the West African AIDS virus. AIDS. 1987;1(3):135–40. [PubMed] [Google Scholar]
- Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey MA, Santos-Ferreira MO, Laurent AG, Dauguet C, Katlama C, Rouzioux C, Klatzman D, Champalimaud JL, Montagnier L. Isolation of a new human retrovirus from West African patients with AIDS Science. A. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- Soriano V, Gomes P, Heneine W, Holguín A, Doruana M, Antunes R, Mansinho K, Switzer WM, Araujo C, Shanmugam V, Lourenço H, González-Lahoz J, Antunes F. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J Med Virol. 2000;61:111–116. doi: 10.1002/(SICI)1096-9071(200005)61:1<111::AID-JMV18>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Valadas E, França L, Sousa S, Antunes F. 20 Years of HIV-2 Infection in Portugal: Trends and Changes in Epidemiology. Clin Infect Dis. 2009;48:1166–1167. doi: 10.1086/597504. [DOI] [PubMed] [Google Scholar]
- Dougan S, Patel B, Tosswill JH, Sinka K. Diagnoses of HIV-1 and HIV-2 in England, Wales, and Northern Ireland associated with West Africa 1. Sex Transm Infect. 2005;81:338–341. doi: 10.1136/sti.2004.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangai R, Nair SC, Sridharan G, Prasannakumar S, Daniel D. Frequency of HIV type 2 infections among blood donor population from India: A 10-year experience. Indian J Med Microbiol. 2010;28(2):111–113. doi: 10.4103/0255-0857.62485. [DOI] [PubMed] [Google Scholar]
- De Silva T, Cotton M, Rowland-Jones. HIV-2: The forgotten AIDS virus. Trends Microbiol. 2008;16:588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Torian LV, Eavey JJ, Punsalang AP, Pirillo RE, Forgione LA, Kent SA, Oleszko WR. HIV Type 2 in New York City, 2000-2008. Clin Infect Dis. 2010;51(11):1334–1342. doi: 10.1086/657117. [DOI] [PubMed] [Google Scholar]
- Zacarias JS, Oliveira I, Andersen A, Dias F, Rodrigues A, Holmgren B, Andersson S, Aaby P. Changes in prevalence and incidence of HIV-1, HIV-2 and dual infections in urban areas of Bissau, Guinea-Bissau: is HIV-2 disappearing? AIDS. 2008;22:1195–1202. doi: 10.1097/QAD.0b013e328300a33d. [DOI] [PubMed] [Google Scholar]
- Damond F, Brun-Vezinet F, Matheron S, Peytavin G, Campa P, Pueyo S, Mammano F, Lastere S, Farfara I, Simon F, Chene G, Descamps D. Polimorphism of the Human Immunodeficiency Virus Type 2 (HIV-2) Protease Gene and selection of Drug Resistence Mutations in HIV-2 patients treated with Protease inhibitors. J Clin Microbiol. 2006. pp. 484–487. [DOI] [PMC free article] [PubMed]
- Colson P, Henry M, Tourres C, Lozachmeur D, Gallais H, Gastaut JA, Moreau J, Tamalet C. Polymorphism and Drug-Selected Mutations in the Protease Gene of Human Immunodefiency Virus type 2 from Patients living in Southern France. J Clin Microbiol. 2004;42(2):570–5. doi: 10.1128/JCM.42.2.570-577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelle J, Sanou M, Liu H-F, Vandenbroucke AT, Duquenne A, Goubau P. Genetic Polymorphisms and Resistance Mutations of HIV Type 2 in Antiretroviral-Naive Patients in Burkina Faso. AIDS Res and Hum Retroviruses. 2007;23(8):955–964. doi: 10.1089/aid.2007.0034. [DOI] [PubMed] [Google Scholar]
- Renny G. HIV-2 infectionthroughout the World. A geographical perspective Sante. 1998;6:440–446. [Google Scholar]
- Qiu M, Liu Xin, Yan Jiang, Nkengasong JN, Xing W, Pei L, Parekh BS. Current HIV-2 Diagnostic Strategy OverestimatesHIV-2 Prevalence in China. J Med Virol. 2009;81:790–797. doi: 10.1002/jmv.21441. [DOI] [PubMed] [Google Scholar]
- Abreu CM, Brindeiro PA, Martins NA, Arruda MB, Bule E, Stakteas S, Tanuri A, Brindeiro RM. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 isolates circulating in pregnant women from Mozambique. Arch Virol. 2008;153:2013–2017. doi: 10.1007/s00705-008-0215-6. [DOI] [PubMed] [Google Scholar]
- Dumans AT, Soares MA, Pieniazek D, Kalish ML, De Vroey V, Hertogs K, Tanuri A. Prevalence of protease and reverse transcriptase drug resistance mutations over time in drug-naiive human immunodeficiency virus type 1-positive individuals in Rio de Janeiro, Brazil. Antimicrob Agents Chemother. 2002;46(9):3075–3079. doi: 10.1128/AAC.46.9.3075-3079.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanuri A, Swanson P, Devare S, Berro OJ, Savedra A, Costa LJ, Telles JG, Brindeiro R, Schable C, Pieniazek D, Rayfield M. HIV-1 subtypes among blood donors from Rio de Janeiro, Brazil. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20(1):60–6. doi: 10.1097/00042560-199901010-00009. [DOI] [PubMed] [Google Scholar]
- Altman DG, Gore SM, Gardner MJ, Pocock SJ. Statistical guidelines for contributors to medical journals. Br Med J (Clin Res Ed) 1983;286:1489–1493. doi: 10.1136/bmj.286.6376.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto J, Liljestrand J, Palha de Sousa C, Bergström S, Böttiger B, Biberfeld G, De la Cruz F. HIV-1 and HIV-2 antibodies in pregnant women in the city of Maputo, Mozambique. A comparative study between 1982/1983 and 1990. Scand J infect Dis. 1993;25(6):685–688. doi: 10.3109/00365549309008563. [DOI] [PubMed] [Google Scholar]
- Ghys PD, Fransen K, Diallo MO, Ettiègne-Traoré V, Coulibaly IM, Yeboué KM, Kalish ML, Maurice C, Whitaker JP, Greenberg AE, Laga M. The associations between cervicovaginal HIV shedding, sexually transmitted diseases and immunosuppression in female sex workers in Abidjan, Cote d'Ivoire. AIDS. 1997;11:85–93. doi: 10.1097/00002030-199712000-00001. [DOI] [PubMed] [Google Scholar]
- Langley CL, Benga-De E, Critchlow CW, Ndoye I, Mbengue-Ly MD, Kuypers J, Woto-Gaye G, Mboup S, Bergeron C, Holmes KK, Kiviat NB. HIV-1, HIV-2, human papillomavirus infection and cervical neoplasia in high-risk African women. AIDS. 1996;10:413–417. doi: 10.1097/00002030-199604000-00010. [DOI] [PubMed] [Google Scholar]
- Poulsen AG, Kvinesdal B, Aaby P, Mølbak K, Frederiksen K, Dias F, Lauritzen E. Prevalence of and mortality from human immunodeficiency virus type 2 in Bissau, West Africa. Lancet. 1989. pp. 827–831. [DOI] [PubMed]
- Wilkins A, Ricard D, Todd J, Whittle H, Dias F, Da Silva PA. The epidemiology of HIV infection in a rural area of Guinea-Bissau. AIDS. 1993;7:1119–1122. doi: 10.1097/00002030-199308000-00015. [DOI] [PubMed] [Google Scholar]
- Chen Z, Luckay A, Sodora DL, Telfer P, Reed P, Gettie A, Kanu JM, Sade RF, Yee J, Ho DD, Zhang L, Marx PA. Human Immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, Neequaye AE, Whelan TM, Ho DD, Shaw GM. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Koumare B, Mulanga C, Brengues C, Mounirou B, Bougoudogo F, Ravel S, Bibollet-Ruche F, Delaporte E. Genetic subtypes of HIV type 1 and HIV type 2 strains in commercial sex workers from Bamako, Mali. AIDS Res Hum Retroviruses. 1998;14:51–58. doi: 10.1089/aid.1998.14.51. [DOI] [PubMed] [Google Scholar]
- Takehisa J, Osei-Kwasi M, Ayisi NK, Hishida O, Miura T, Igarashi T, Brandful J, Ampofo W, Netty VB, Mensah M, Yamashita M, Ido E, Hayami M. Phylogenetic analysis of HIV type 2 in Ghana and intrasubtype recombination in HIV type 2. AIDS Res Hum Retroviruses. 1997;13:621–623. doi: 10.1089/aid.1997.13.621. [DOI] [PubMed] [Google Scholar]
- Ciccaglione AR, Miceli M, Pisani G, Bruni R, Iudicone P, Costantino A, Equestre M, Tritarelli E, Marcantonio C, Tataseo P, Marazzi MC, Ceffa S, Paturzo G, Altan AM, San Lio MM, Mancinelli S, Ciccozzi M, Lo Presti A, Rezza G, Palombi L. Improving HIV-2 Detection by a combination of Serological and Nucleic acid amplification Test Assays. J Clin Microbiol. 2010. pp. 2902–2908. [DOI] [PMC free article] [PubMed]
- Van der Ende ME, Guillon C, Boers PH, Ly TD, Gruters RA, Osterhaus AD, Schutten M. Antiviral resistence of biologic HIV-2 clones obtained from individuals on nucleoside reverse transcriptase inhibitor therapy. JAIDS. 2000;25:11–18. doi: 10.1097/00126334-200009010-00002. [DOI] [PubMed] [Google Scholar]
- Ntemgwa ML, Toni TD, Brenner BG, Camacho RJ, Wainberg WA. Antiretroviral Drug Resistance in Human Immunodeficiency Virus Type 2. Antimicrob Agents Chemother. 2009. pp. 3611–3619. [DOI] [PMC free article] [PubMed]