Abstract
Background
Host-microbe interactions at the intestinal mucosal-luminal interface (MLI) are critical factors in the biology of inflammatory bowel disease (IBD).
Methods
To address this issue, we performed a series of investigations integrating analysis of the bacteria and metaproteome at the MLI of Crohn’s disease, ulcerative colitis, and healthy human subjects. After quantifying these variables in mucosal specimens from a first sample set, we searched for bacteria exhibiting strong correlations with host proteins. This assessment identified a small subset of bacterial phylotypes possessing this host interaction property. Using a second and independent sample set, we tested the association of disease state with levels of these 14 “host interaction” bacterial phylotypes.
Results
A high frequency of these bacteria (35%) significantly differentiated human subjects by disease type. Analysis of the MLI metaproteomes also yielded disease classification with exceptional confidence levels. Examination of the relationships between the bacteria and proteins, using regularized canonical correlation analysis (RCCA), sorted most subjects by disease type, supporting the concept that host-microbe interactions are involved in the biology underlying IBD. Moreover, this correlation analysis identified bacteria and proteins that were undetected by standard means-based methods such as ANOVA, and identified associations of specific bacterial phylotypes with particular protein features of the innate immune response, some of which have been documented in model systems.
Conclusions
These findings suggest that computational mining of mucosa-associated bacteria for host interaction provides an unsupervised strategy to uncover networks of bacterial taxa and host processes relevant to normal and disease states.
Keywords: metaproteome, microbiome, mucosal luminal interface, rRNA genes
INTRODUCTION
Inflammatory bowel disease (IBD) is a group of remittent intestinal diseases, the two most common forms being Crohn’s disease (CD) and ulcerative colitis (UC). Inflammation in UC is typically restricted to the colonic mucosa while damage in CD can occur throughout the gastrointestinal tract and penetrate deeper into the bowel wall. Although a precise understanding remains elusive, IBD etiology appears to be complex, including both heritable and environmental factors.1
Genome-wide association studies (GWAS) have identified as many as 71 putative susceptibility loci,2 several of which are components of the immune system involved in managing microbial infections. For example, variants of the NOD2 gene have been linked to Crohn’s disease susceptibility,3–5 and one key function of NOD2 appears to involve clearing intracellular infections. In studies that challenged mice (intragastrically) or cell cultures with living intracellular pathogens, NOD2 variants/knockouts exhibited a diminished ability to kill or clear the microorganisms.6–10 Similarly, GWAS studies have also discovered CD-associated polymorphisms in ATG16L1 and IRGM, which are genes involved in autophagy.11–15 Given that an important role of autophagy is in host cell elimination of intracellular pathogens,16,17 these associations combined with the aforementioned NOD2 results and the functional linkages between these two groups of molecules18,19 point toward intracellular infections playing a role in IBD etiology.
While results from both human and animal studies suggest a microbial role in IBD etiology, the most compelling evidence comes from investigations with animals. In numerous rodent models, colitis is absent in germ-free conditions but rapidly develops when standard intestinal microorganisms are introduced.20 In addition, monoassociation studies have shown that colitis phenotype is dependent upon the type of bacterial species,21,22 suggesting disease etiology involves specific microorganisms.
In this study, we describe a series of investigations that endeavored to develop a greater understanding of host-microbe interactions in human IBD. We hypothesize that bacteria that play a functional role in IBD will be those that interact with the host. To identify such bacteria, we collected and examined samples at the intestinal mucosal-luminal interface (MLI), a critical locale for disease biology, but one not often studied. In addition to cataloging the variables and relating them to disease type, we also searched for bacteria exhibiting strong and numerous correlations with host proteins, as we posit that such relationships are indicators of microorganisms that interact with the host.
MATERIALS AND METHODS
Patients
Two patient cohorts were examined in this study (Table S1). The first was comprised of nine patients: UC (n = 6) and HC (n = 3) subjects. The second was comprised of forty-two patients: CD (n = 14) and UC (n = 15), and HC (n = 13) subjects. This study was performed in accord with human subjects protocols approved by the institutional review boards of UCLA and Cedars-Sinai Medical Center.
Intestinal MLI Sample Collection
MLI samples were obtained from various regions of the intestine using an endoscopic saline-lavage sampling technique. Subjects were prepared for colonoscopy by taking Golytely the day before the procedure. For IBD subjects, colonoscopy procedures were performed during periods of inactive disease. During the colonoscopy procedure, 30 ml of sterile 0.9% saline was injected to the surface of each of the six different locations of the colon. The mucosal lavage samples were collected by vacuum suction with a Fujinon magnifying colonoscope. Typically, 20 ml of saline was recovered for each region. Lavage samples were kept on ice immediately after collection and processed as described below.
MLI Sample Processing for Metaproteomic Analyses
MLI samples were centrifuged at 4,200 × g for 30 minutes to pellet particulate matter (samples were stored on ice between collection and centrifugation). Proteins in the supernatant were precipitated by adding three volumes of acetone, incubating these mixtures overnight at −80°C, and centrifuging at 4,200 × g for 30 minutes at 4°C. Pellets were washed with 10 ml of ice-cold 70% acetone and then dried at room temperature for 1 to 2 hours.
Metaproteomic Analyses
Surface-enhanced laser desorption/ionization time of flight (SELDI-TOF) mass spectrometry analysis of the intestinal MLI samples from the first cohort of subjects was performed as described below. Pellets were resuspended by vortex mixing with 2 to 3 mL of PBS containing 1% Triton X-100. These samples were centrifuged at 17,000 × g for 15 minutes to precipitate and remove the insoluble material. The resulting detergent-solubilized protein solutions were examined by metaproteomic analyses. A small aliquot of the protein detergent-soluble supernatant (10–20 μL) was added to 200 μL of binding buffer (0.2 M ammonium acetate (pH 4) containing 0.1% Triton X-100) and shaken for 30 to 45 minutes at room temperature on weak cation exchange (WCX) Protein Chip arrays (CM10, a carboxymethyl WCX surface). These samples were then removed from the arrays and the arrays were washed 3 times for 5 minutes with 250 μl of binding buffer and vigorous agitation to remove non-specifically bound proteins. The arrays were rinsed with deionized water to remove buffer salts and detergents and then twice spotted with matrix (2 μL of a saturated solution of sinapinic acid (SPA) in 50% acetonitrile with 0.5% TFA) to allow laser desorption and ionization. The arrays were read using a Ciphergen PBS-IIc instrument at a laser setting of 195 to detect proteins between the masses of 2–30 kDa.
High-resolution reflectron matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry analysis of the intestinal MLI samples from the second cohort of subjects was performed as described below. To each precipitated protein sample, 500 μl of PBS with 1% Triton-X was added and thoroughly mixed, and then transferred to 2 ml microcentrifuge tubes. Samples were centrifuged at 10,000 × g and the supernatant was collected. A BCA assay was carried out for each supernatant sample to determine the protein concentration. 300 μg of total protein from each sample was diluted in PBS and applied to subsequent analyses. Samples were then passed through a 1-μm filter plate separately. 10 μl of the extracts are mixed with 200 μg of weak cation exchange (WCX) magnetic beads (S-COOH MoBiTec, Goettingen, Germany) with 90 μl of 0.2M ammonium acetate pH 4.0 with 0.01% TX-100. The process has been automated in a 96-well format with a Hamilton Starlet robot (Reno, NV) where the beads are pelleted on a strong plate magnet and washed 3 times with 100 μl of binding buffer. The beads were then desalted with 5 mM ammonium acetate and extracted with 15 μl of 1% trifluoroacetic acid. 10 μl of the extracts are removed and mixed with an equal volume of 5 mg/mL α-cyano-hydroxycinnamic acid matrix (CHCA, LaserBio Labs, France) dissolved in 90% acetonitrile. 2 μl each of the extract-matrix mixture is then applied to a 96-well MALDI target in triplicates. After drying, the plate is read in a Perkin-Elmer Sciex (San Jose, CA) prOTOF2000 reflectron mass spectrometer with settings for optimal detection of peptides and small proteins between 2 and 20 kDa.
Identifying the Index Bacterial Phylotypes
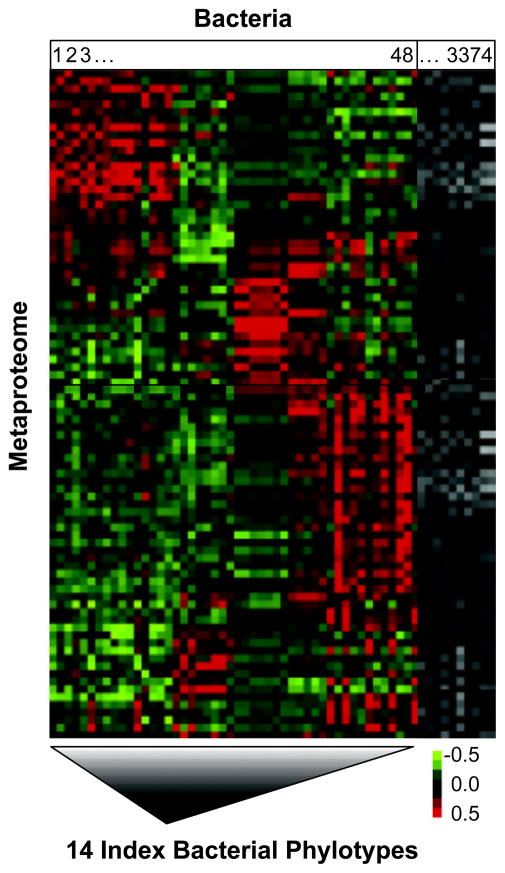
The index phylotypes were identified as follows. Starting with the entire complement of 3,374 phylotypes from the aforementioned OFRG analysis of the first cohort, we identified the 48 phylotypes with the highest numbers of clones per phylotype and removed the rest from further analysis. Correlation analyses were then performed on the numbers of clones per phylotype for each of the remaining 48 phylotypes and the quantitative values of the proteins from each of the 87 peaks. The resulting Pearson’s correlation coefficients were subjected to a cluster analysis and then depicted in a heat map (Figure 1). Ten of these remaining 48 phylotypes were included in our final set of 14 index phylotypes; they were chosen because they (i) exhibited strong relationships with the proteins, (ii) represented different regions of the heat map and (iii) possessed properties facilitating the development of sequence-selective qPCR assays. The final set of 14 index phylotypes also contained 4 phylotypes exhibiting high abundance in several IBD subjects.
FIGURE 1.
Self-organizing map (SOM) cluster analysis of the relationships between the amounts of the bacterial phylotypes and proteins in MLI samples from the first cohort of IBD and healthy subjects. Bacterial rRNA gene and metaproteome composition were examined by OFRG and SELDI-MS analyses, respectively. Pearson correlation coefficients for each phylotype-protein pair are depicted by the color in each cell; stronger correlations have brighter colors (see scale bar). Bacterial phylotypes are on the horizontal axis while the proteins are on the vertical axis (see Supporting Information for details). The heat map contains a subset of the bacterial phylotypes; for details on how this subset was selected, see Identifying the Index Bacterial Phylotypes in Materials and Methods.
DNA Extraction of Intestinal MLI Samples
MLI samples were centrifuged for 1 minute at 14,000 × g. Pellets were resuspended with 1 ml CLS-Y buffer and transferred to Lysis Tubes from a FastDNA Spin Kit (Qbiogene, Carlsbad, CA). DNA was extracted using the FastDNA Spin Kit (Qbiogene, Carlsbad, CA) as described by the manufacturer, with a 30-second bead-beading step at a FastPrep instrument setting of 5.5. DNA was further purified by subjecting it to electrophoresis in a 1% agarose gel and then recovering it from the gel using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) as described by the manufacturer, but without exposing the DNA to UV or ethidium bromide and excluding the heating step to dissolve the gel in Buffer QG.
Additional Materials and Methods are provided in the Supporting Information.
RESULTS
The design of this study consisted of two phases. In phase one, we obtained the bacterial and metaproteomic compositions of MLI samples from a first cohort of IBD and healthy subjects. Analysis of these results led to the identification of a reduced set of bacterial phylotypes, termed index phylotypes, which included those exhibiting strong correlations (or relationships) with the proteins. In phase two, we tested the index phylotypes in a series of experiments examining an independent set of subjects (second cohort).
Phase One: Phylotype Reduction
Twenty-four MLI samples were obtained from various regions of the intestine from 9 subjects comprising the first cohort: 6 UC and 3 healthy controls (HC) (Table S1). These MLI samples were collected using an endoscopic saline-lavage technique that samples discreet sites (1 cm diameter). Each sample was examined for its bacterial rRNA gene and metaproteomic composition using oligonucleotide fingerprinting of ribosomal RNA genes (OFRG) and surface-enhanced laser desorption/ionization mass spectrometry (SELDI-MS) analyses, respectively. These analyses identified 87 proteins and 3,374 bacterial phylotypes. The relationships between the amounts of the proteins and the most abundant 48 phylotypes were examined using a correlation analysis (Figure 1). The resulting heat map shows numerous relationships among these bacteria and proteins, reflecting a myriad of putative host-microbe interactions occurring in this habitat. Based on this analysis, we derived a reduced set of phylotypes that were tested in the second phase of these investigations; the resulting 14 index phylotypes included those exhibiting high population densities and strong relationships with the proteins; for additional details on the selection process, see Identifying the Index Bacterial Phylotypes in Materials and Methods. Sequence-selective qPCR assays were developed for all 14 of the index phylotypes. The phylogenetic relationships among the phylotypes (Figure S1) along with pertinent primer design information are provided (Table S2).
Phase Two: Testing the Index Bacterial Phylotypes
To test the index bacterial phylotypes, we performed a series of experiments examining MLI samples from a larger and independent cohort comprised of 42 subjects (second cohort): 14 CD, 13 HC, and 15 UC (Table S1). These experiments endeavored to (i) determine if there were significant differences in the abundance of the index phylotypes between IBD and healthy subjects, (ii) determine whether these phylotypes exhibited significant relationships with host proteins and, (iii) assess if and how those relationships (and/or the variables involved in those relationships) were linked to disease. These experiments also examined the ability of the MLI metaproteomes to classify subjects by disease type.
Associations Between Disease and the Bacterial Phylotypes or Metaproteomes
Associations between the 14 index bacterial phylotypes and disease type were examined in the cecum and sigmoid colon, using ANOVA of qPCR data from the second cohort. Five of the 14 phylotypes exhibited significant differences (P < 0.05) between disease types, with the population densities being lower in either CD or UC or both compared to HC (Figure S2). The population densities of 2 phylotypes were different in both intestinal regions while 3 were different only in the sigmoid colon, the latter of which suggests that specific host responses can vary by compartment. Within each disease type there were no differences in the population densities of the phylotypes between the cecum and sigmoid colon, suggesting that the disease-associated changes can be broadly distributed through the intestine. A similar analysis was performed on the metaproteomic data. Concordant with the bacterial results, a larger number of proteins were significantly different (P < 0.05) between disease types in the sigmoid colon (96/589) than in the cecum (36/589) (see Supporting Information for details).
Classification of Disease Type by Bacterial Phylotypes or Metaproteomes
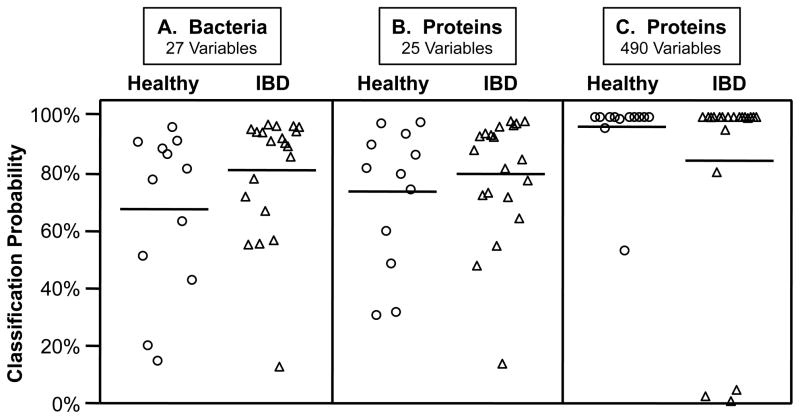
To determine if the levels of the index phylotypes or proteins in the MLI samples could classify subjects by disease type, we performed nearest shrunken centroids analyses.23 This method was originally developed to identify subsets of expressed genes that can classify subjects by host phenotype. Applying this method to our data, optimal classification came from inclusion of 27 of the 30 phylotype-region variables (see Figure 2 legend for more details). The posterior probabilities generated by this analysis showed that these MLI-inhabiting bacteria were able to correctly classify (probability > 50%) almost all subjects (28/32) and that classification was better in the category of IBD than healthy (Figure 2a). Classification using a similar number of protein-region variables produced comparable posterior probabilities (Figure 2b), suggesting that both types of variables (bacteria and proteins) possess similar power to sort these biologic states. Optimal classification of the metaproteomic data utilized 490 of the 1,178 protein-region variables, resulting in very high probability values for almost all of the subjects (Figure 2c).
FIGURE 2.
Classification of healthy and IBD subjects by nearest shrunken centroids analyses of the amounts of the index bacterial phylotypes or proteins from intestinal MLI samples. Values are posterior probabilities: healthy (circles) and IBD (triangles). Means are solid horizontal lines and standard deviations are dashed lines (only the lower value is shown). Only second cohort subjects sampled in both cecum (CE) and sigmoid colon (SIG) were used in this analysis (n = 32), and both regions were analyzed separately. A: Bacterial population density values were generated using 15 qPCR assays: 14 index phylotypes (Table S2 and Figure S1) and one targeting all bacterial rRNA genes; the optimal classification solution (threshold = 0.175, error = 4/32) included all bacteria-region variables except Eubacterium 2766 from the cecum and Clostridium 12 from the cecum and sigmoid colon (27/30 phylotype-region variables). B and C: Proteins were enumerated by MALDI-MS. B: Classification from 25 protein-region variables (threshold = 1.788; error = 5/32) produced similar posterior probabilities as those from the bacterial analysis (A); C: The optimal classification solution (threshold = 0.670; error = 3/32) included 490 protein-variables (see Supporting Information for protein lists).
Relationships Among Bacteria and Proteins at the Intestinal MLI
To test the index phylotypes for their putative abilities to interact with the host, we determined whether they exhibited significant relationships with proteins in MLI samples from a second cohort of subjects. Sequence-selective qPCR assays were used to enumerate the phylotypes while quantitative high-dimensional matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) was used to examine the metaproteomes. To analyze these data, we used regularized canonical correlation analysis (RCCA), which is a method that endeavors to identify linear relationships between two sets of variables.24
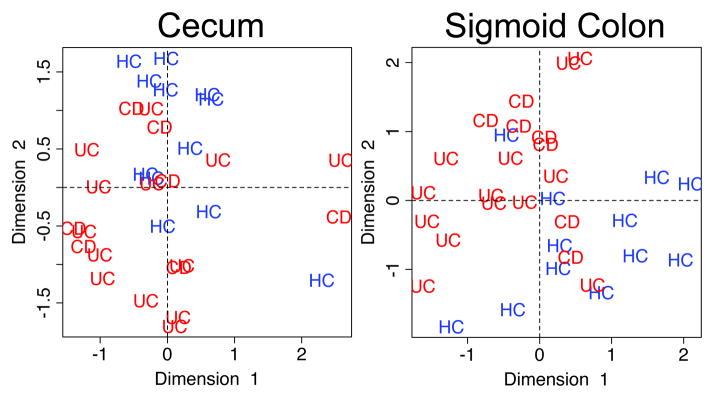
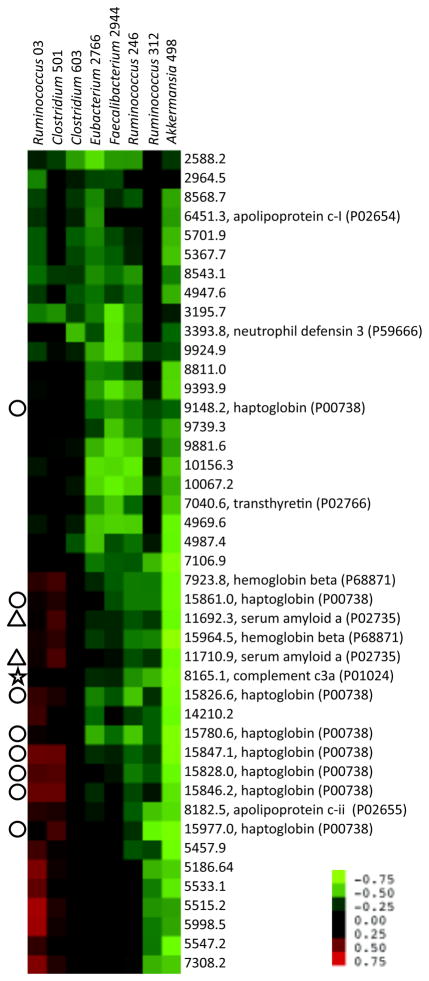
The RCCA demonstrated that the relationships between the index phylotypes and the metaproteomes were able to sort most of the subjects by disease type (Figure 3). Patterns of subject sorting were distinct between the sigmoid colon and cecum. The ability of the RCCA to sort the subjects suggests that these bacteria-protein relationships may be contributing factors in host-microbe interactions underlying IBD. To examine these relationships in greater detail, correlations between the levels of 8 bacteria and 43 proteins, which were the strongest contributors to the inter-relationships identified by the RCCA, were depicted using a cluster analysis (Figure 4). In the Discussion, we describe how these relationships provide new insights into IBD etiology.
FIGURE 3.
Regularized canonical correlation analysis of bacteria and proteins from MLI samples of IBD and healthy subjects. Canonical variates 1 and 2, based on correlation analysis of the levels of bacteria and proteins, are plotted for samples from the cecum and sigmoid colon. CD, HC and UC indicate Crohn’s disease, healthy controls and ulcerative colitis. n = 13 (CD-SIG), 7 (CD-CE), 12 (HC-SIG), 13 (UC-HE), 14 (UC-SIG) and 14 (UC-CE).
FIGURE 4.
Self-organizing map (SOM) cluster analysis of the correlations between the levels of the 8 bacteria and 43 proteins that were strongest contributors to the interrelationships identified by the RCCA. Bacterial rRNA gene and metaproteome composition were examined by sequence-selective qPCR and MALDI-MS analyses, respectively. Pearson correlation coefficients for each phylotype-protein pair are depicted by the color in each cell; stronger correlations have brighter colors (see scale bar). Bacterial phylotypes are on the horizontal axis while the proteins are on the vertical axis. The selected bacteria and proteins had the largest coefficients for canonical variates 1 and 2.
DISCUSSION
Examining distinct and biologically important habitats will be a key factor in understanding host-microbe interactions involved in disease and physiological processes. Biogeographic variation in microbial communities can be striking, reflecting differences in host responses as well as physical and chemical features of the habitats. A recent examination of 27 different locations from healthy humans provided a clear depiction of habitat selection.25 More pertinent to IBD, several studies have shown that fecal and mucosal communities are highly divergent.26,27
In this study, we examined the interface between the intestinal mucosa and lumen (MLI). The intestinal mucosa is a barrier layer that keeps microorganisms from entering host tissue. Most interactions between the host and its resident luminal microorganisms therefore occur at this interface. In this study, we used a saline-lavage technique to collect MLI samples, which are comprised of a particulate component (diverse bacterial morphotypes, with rare epithelial and hemopoeitic cells), and a soluble component (63% human, 30% bacterial, 7% other eukaryote and viral, by protein identification). This technique therefore collects mostly microorganisms and soluble proteins at this interface. Alternative approaches for examining this region, such as endoscopically obtained mucosal biopsies, produce samples that are less regionally defined, as they are comprised of contents from both the MLI as well as numerous intestinal layers including the lamina propria. Analysis of biopsy samples by methods including rRNA gene analysis could therefore produce results that are confounded by aggregation of functionally distinct regions or by DNA from dead organisms that are being processed in phagocytic cells. In future work, we will also endeavor to collect materials from the epithelium, as such samples would include important host-associated proteins such as receptors, and because prior work investigating such samples in a mouse model showed that most of the associations between bacteria and immunoregulatory cells were found in that habitat.28
Our investigations showed that specific bacteria and proteins in the MLI can be used to differentiate and classify IBD and healthy human subjects. Five of the 14 index phylotypes exhibited differential abundance between disease types (Figure S2). We posit that this high rate of success is primarily due to the biological importance of the MLI habitat. In addition, nearest shrunken centroids analyses of the bacteria and proteins from this habitat showed that their amounts could be used to classify most subjects at relatively high confidence levels, even when small numbers of variables were utilized (Figure 2a,b). As was shown by the classification analyses of the metaproteomic data (Figure 2b,c), we anticipate that utilization of greater numbers of bacterial phylotypes will also considerably enhance the classification results. The fact that the optimal classification solutions used almost all of the bacterial variables (27/30) and approximately half of the protein variables (490/1,178) provides further support for the importance of this habitat in both understanding host-microbe interactions associated with IBD etiology and for developing methodologies for stratifying disease subtypes. Indeed, to our knowledge, no other classification analyses have sorted IBD subjects at the high confidence levels produced by our MLI analysis (Figure 2c).
To identify putative host-microbe interactions associated with IBD, we performed a correlation analysis (RCCA) on the levels of bacteria and proteins collected at the MLI of IBD and healthy subjects. The results from this analysis suggests that host-microbe interactions in IBD involve a self-sustaining cycle whereby inflammation causes shifts in bacterial composition, including an increase in the relative proportion of proinflammatory bacteria, which causes an increase in local host inflammatory products.
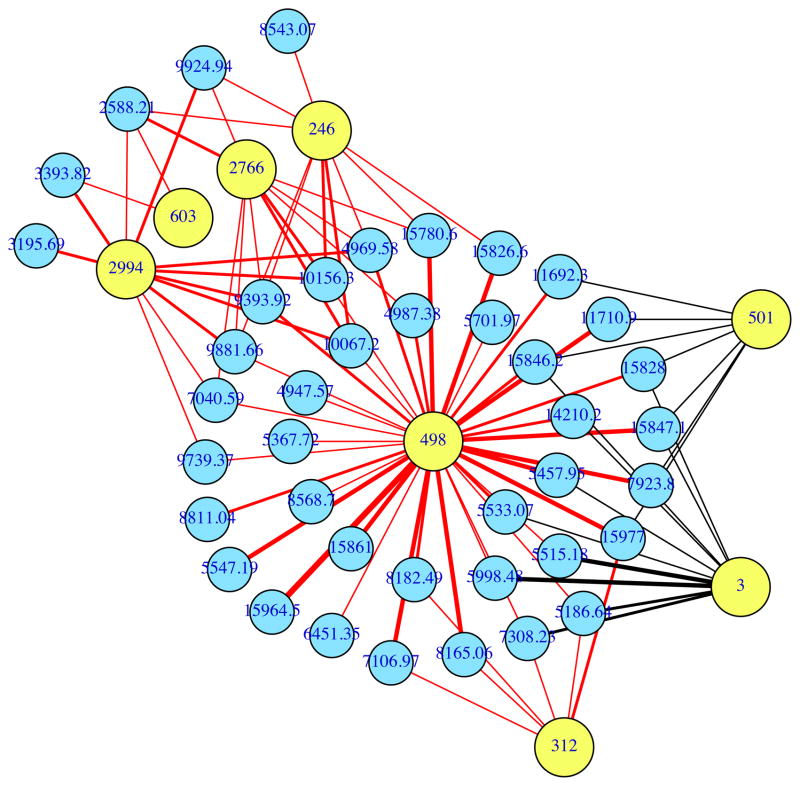
Indeed, the level of the innate inflammatory protein, complement C3a, was correlated with such changes in microbial composition (star) (Figure 4). Levels of 5 index bacterial phylotypes were negatively correlated with local C3a, and, as expected, all of these phylotypes were also depleted in UC or CD (Figure S2). In addition, some taxa identified in this analysis, such as Faecalibacterium 2994, which has high sequence identity (97%) to Faecalibacterium prausnitzii, have demonstrated protective anti-inflammatory roles in a mouse model of colitis, and are depleted in IBD.29 However, two phylotypes (Ruminococcus 03 and Clostridium 501) exhibited positive associations with C3a, suggesting that these organisms may have contributed to inducing this inflammatory environment (Figure 4). In addition to C3a, Ruminococcus 03 and Clostridium 501 also exhibited positive associations with haptoglobin (circles), which is notable since both of these host products contribute to epithelial permeability, another hallmark of IBD-associated inflammation.30,31 Finally, these two phylotypes also exhibited a positive association with serum amyloid A (triangles), a product of epithelial and resident mucosal hemopoeitic cells, which contributes to the initiation of local innate inflammatory response. Recent work examining several members of the Clostridiales, a taxon that includes these two phylotypes, has uncovered examples of both pro- or anti-inflammatory pathways.32,33 One of these Clostridiales, termed segmented filamentous bacteria (SFBs), was able to trigger both the generation of TH17 cells33,34 and intestinal inflammation,35 in part by inducing intestinal cells to produce serum amyloid A.33
Thus, the correlation analysis performed in this study not only identified disease-associated bacterial and protein biomarkers, but it also provided an experimental framework to uncover potential in situ host-microbe interactions. For instance, our study and others have shown that IBD is associated with decreases in specific bacterial phylotypes in the mucosal habitat.29,36,37 The question is why does this occur? By examining the relationships between the microorganisms and a measure of host response (in this study, the metaproteome), it becomes evident that host-defense processes could be driving these population shifts. Concerning our putative proinflammatory phylotypes (Ruminococcus 03 and Clostridium 501), while the RCCA identified these two bacteria as key variables in this study, and Pearson’s correlation demonstrates their statistical significance (P = 0.004 with haptoglobulin in the sigmoid colon for both phylotypes), tests relating their amounts to disease type would not have classified them as important variables: P values = 0.368 and 0.133 for Ruminococcus 03 and Clostridium 501, respectively, using ANOVA and sigmoid colon samples. These examples provide compelling evidence for the utility of examining relationships between host and microbial variables, because they enable key organisms and host responses to be both detected and linked. The idea of focusing on the linkages between variables is gaining strength as several groups have independently converged on this concept in the fields of host-microbe interactions (this study) and network analysis.38,39 Inspired by these network studies, we reanalyzed the strongest relationships identified by the RCCA (from Figure 4) using a network analysis; an example of potential specific host-microbial interactions uncovered and visualized by this strategy is illustrated in Figure 5.
FIGURE 5.
Network analysis of the relationships between the amounts of selected bacterial phylotypes and proteins in MLI samples from the second cohort of IBD and healthy subjects. The variables examined were the 8 bacteria and 43 proteins depicted in Figure 4. Phylotypes and proteins are depicted as yellow and blue circles, respectively. Only correlations > 0.4 or < −0.4 are shown. Negative and positive associations are depicted as red and black lines, respectively. The thickness of each line indicates the strength of the correlation. The interaction map was constructed using the social networking package (SNA) in R (http://cran.rproject.org/web/packages/sna/index.html).
Our correlation analysis also corroborates a current concept in IBD etiology, positing that shifts in the populations of common intestinal microbiota are contributing factors. An examination of the 2 strongest pairwise correlations showed that these bacteria-protein relationships likely represent biologic interactions that are shared among all subjects (Figure S3), as even though many subjects cluster by disease type (CD and UC versus HC), there is also crossover among groups. These results also indicate that some host-microbe interactions involve binary states. We speculate that phylotypes that are highly sensitive to inflammatory proteins represent one instance where binary relationships will exist. Deciphering the intricacies of such relationships, and how other factors such as host genetics influence them, will certainly lead to a greater understanding of the underlying biology in IBD.
Lastly, we compared four different methods for identifying putatively important host and microbe variables – a crucial but daunting task given the need to identify a small number of key variables from enormously complex datasets. To facilitate the comparisons, we identified 8 bacteria and 43 proteins from each method (Tables S3 and S4). For the two means-based methods, ANOVA and nearest shrunken centroids, we identified the variables whose amounts best sorted the subjects by disease type. For the RCCA, we identified the variables that were the strongest contributors to the interrelationships defined by the first two canonical variates. For the fourth method, we performed an analysis that included identifying the bacterial phylotypes that exhibited the largest numbers of strong relationships (LNSR) with the proteins (see Supporting Information for details).
Focusing on the proteins first, the two correlation methods identified higher percentages (93%, LNSR; 88%, RCCA) of proteins previously associated with IBD than the means-based methods (77%, ANOVA; 73%, nearest shrunken centroids) (Table S3, see IBD column). However, given the lack of a comprehensive model of IBD pathogenesis, the meaning of these results remains uncertain. What is clear is that, compared to the other methods, the LNSR correlation method uncovered a key observation that supports its underlying rationale. Using the LNSR method, 53% of the proteins with a database match were immune system molecules, compared to only 9%, 12% and 14% for the other methods (Table S3, blue bold text). These molecules are induced by an integrated host immune response through contact with microorganisms and their products, and as such are indicators of intimate host-microbe interactions occurring in the MLI habitat. In contrast, the most predominant proteins identified by the traditional approach (e.g., transthyretin, hemoglobin and serum amyloid) were high abundance proteins commonly and non-specifically associated with many settings of tissue injury. We anticipate that the LNSR method may yield new and important clues regarding upstream host-microbe interactions associated with IBD, and that these new capabilities will complement existing strategies and enhance investigations of causal relationships in IBD and other multifactorial etiologies. A similar comparison analysis was also performed on the bacterial phylotypes (Table S4).
The results from this study also highlight several additional and potentially useful experimental design features. The first involves the importance of examining specific and narrowly defined taxa. For example, two closely related (96% identity of near full-length 16S rRNA genes) Ruminococcus phylotypes (312 and 323) were analyzed in this study. One of the phylotypes (Ruminococcus 312) demonstrated the ability to differentiate subjects by disease type while the other did not (Ruminococcus 323). Our investigations were able to detect these differences because we designed the qPCR assays to amplify sequences with a narrow range of identities (98.2–100%). The second consideration is the importance of validating population results using methods such as qPCR, as most PCR-based methods that are currently used to examine microbial composition, including high throughput sequence analyses, produce skewed results.40,41 Addressing both of the first two considerations, utilization of the PRISE software facilitates the development of sequence-selective qPCR assays.42 Finally, our studies also demonstrated that the phylotypes with the largest number of protein relationships were not the most abundant ones. Indeed, Akkermansia 498 was the least abundant index phylotype examined in this study (data not shown), and yet it exhibited both the largest number of protein relationships and the ability to differentiate subjects by disease type.
Supplementary Material
Acknowledgments
The research is supported in part by NIH grants 5R01AI078885 and P01DK46763, and Crohn’s and Colitis Foundation of America grant 1567 and Microbiome in IBD award. We are grateful to patients and physicians at UCLA and Cedars-Sinai Medical Center whose participation made this study possible. We thank the editor and reviewers for their important input regarding data analysis.
References
- 1.Mayer L. Evolving paradigms in the pathogenesis of IBD. J Gastroenterol. 2010;45:9–16. doi: 10.1007/s00535-009-0138-3. [DOI] [PubMed] [Google Scholar]
- 2.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 4.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 6.Hisamatsu T, Suzuki M, Reinecker HC, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 7.Kim TH, Payne U, Zhang X, et al. Altered host:pathogen interactions conferred by the Blau syndrome mutation of NOD2. Rheumatol Int. 2007;27:257–262. doi: 10.1007/s00296-006-0250-0. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 9.Rosenstiel P, Sina C, End C, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- 10.Zelinkova Z, van Beelen AJ, de Kort F, et al. Muramyl dipeptide-induced differential gene expression in NOD2 mutant and wild-type Crohn’s disease patient-derived dendritic cells. Inflamm Bowel Dis. 2008;14:186–194. doi: 10.1002/ibd.20308. [DOI] [PubMed] [Google Scholar]
- 11.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–112. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 13.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescott NJ, Fisher SA, Franke A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 15.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid D, Dengjel J, Schoor O, et al. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med. 2006;84:194–202. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- 17.Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 18.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 19.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 20.Sartor RB. Microbial influences in inflammatory bowel disease: role in pathogenesis and clinical implications. In: Sartor RB, Sandborn WJ, editors. Kirsner’s Inflammatory Bowel Diseases. Philadelphia, PA: Elsevier; 2004. pp. 138–162. [Google Scholar]
- 21.Kim SC, Tonkonogy SL, Albright CA, et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Kim SC, Tonkonogy SL, Karrasch T, et al. Dual-association of gnotobiotic IL-10-/-mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González I, Déjean S, Martin PGP, et al. CCA: An R Package to Extend Canonical Correlation Analysis. J Stat Softw. 2008;23:1–14. [Google Scholar]
- 25.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durbán A, Abellán JJ, Jiménez-Hernández N, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol. 2011;61:123–133. doi: 10.1007/s00248-010-9738-y. [DOI] [PubMed] [Google Scholar]
- 27.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presley LL, Wei B, Braun J, et al. Bacteria associated with immunoregulatory cells in mice. Appl Environ Microbiol. 2010;76:936–41. doi: 10.1128/AEM.01561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conyers G, Milks L, Conklyn M, et al. A factor in serum lowers resistance and opens tight junctions of MDCK cells. Am J Physiol. 1990;259:C577–585. doi: 10.1152/ajpcell.1990.259.4.C577. [DOI] [PubMed] [Google Scholar]
- 31.Tripathi A, Lammers KM, Goldblum S, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci USA. 2009;106:16799–16804. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Stepankova R, Powrie F, Kofronova O, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 36.Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 37.Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 38.Ahn YY, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 39.Evans TS, Lambiotte R. Line graphs, link partitions, and overlapping communities. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:016105. doi: 10.1103/PhysRevE.80.016105. [DOI] [PubMed] [Google Scholar]
- 40.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Q, Ruegger P, Bent E, et al. PRISE (PRImer SElector): software for designing sequence-selective PCR primers. J Microbiol Methods. 2008;72:263–267. doi: 10.1016/j.mimet.2007.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.