Abstract
Epigenetic mechanisms play critical roles in stem cell biology by maintaining pluripotency of stem cells and promoting differentiation of more mature derivatives. If similar mechanisms are relevant for the cancer stem cell (CSC) model, then epigenetic modulation might enrich the CSC population, thereby facilitating CSC isolation and rigorous evaluation. To test this hypothesis, primary human cancer cells and liver cancer cell lines were treated with zebularine (ZEB), a potent DNA-methyltransferase1-inhibitor and putative CSCs were isolated by the Side Population (SP) approach. The CSC properties of ZEB-treated and untreated subpopulations were tested by standard in vitro and in vivo assays. Whole transcriptome profiling of isolated CSC was performed to generate CSC signatures. Clinical relevance of the CSC signatures was evaluated in diverse primary human cancers. Epigenetic modulation increased frequency of cells with CSC properties in the SP fraction isolated from human cancer cells as judged by self-renewal, superior tumor-initiating capacity in serial transplantations and direct cell tracking experiments. Integrative transcriptome analysis revealed common traits enriched for stemness-associated genes, although each individual CSC gene expression signature exhibited activation of different oncogenic pathways (e.g. EGFR, SRC and MYC). The common CSC signature was associated with malignant progression, enriched in poorly differentiated tumors and was highly predictive of prognosis in liver and other cancers patients.
CONCLUSION
Epigenetic modulation may provide a tool for prospective isolation and in-depth analysis of CSC. The liver CSC gene signatures are defined by a pernicious interaction of unique oncogene-specific and common stemness traits. These data should facilitate the identifications of therapeutic tools targeting both unique and common features of CSC.
Keywords: Cancer Stem Cells, Liver Cancer, Side-population, Epigenetics
It is increasingly recognized that many solid tumors contain a subset of cells which possess functional properties ascribed to normal stem cells, such as self-renewal, unlimited proliferative capacity and pluripotency, leading to a hierarchical model of cancer with a CSC population at the apex of tumor formation (1). The CSC hypothesis posits that CSCs are responsible not only for tumor initiation but also generation of metastasis and local recurrence after therapy (2).
The existence of CSCs (also referred to as tumor-initiating cells) has been shown in a variety of solid tumors including liver cancer (3, 4). However, CSCs have highly variable antigenic and functional properties even when derived from the same tumor type, thus highlighting heterogeneity as a cardinal problem in CSC biology. It is conceivable that CSC phenotype may be corrupted by distinct oncogenic events and influenced by various factors, including tissue microenvironment, resulting in an assortment of CSCs (5). Therefore, defining both unique and common CSC properties is essential for both understanding CSC biology and effective therapeutic translation.
Currently, most studies focusing on liver CSCs rely on cell surface markers, primarily single markers. This approach identified stem-like cancer cells with clonogenic and tumorigenic capacity strongly supporting the existence of CSCs in HCC (6–8). Nonetheless, antigenic approaches have several shortcomings including cross-reactivity, lack of specificity, and antibody-dependent toxicity (9, 10). Further, it has been recently shown that the primary tumor onco-genotypes can influence the marker phenotypes of CSCs raising questions regarding the utility of single markers in molecularly diverse malignancies (5, 11). Alternatively, the side population (SP) approach, which is based on the functional property of CSCs to exclude Hoechst-33342-dye via ABCG2-transporters, might have certain advantages for prospective isolation and characterization of CSC from liver and other cancers (4, 12).
Here, we aimed to identify and characterize core CSC properties by using the SP approach in combination with epigenetic modulation, a key mechanism in cancer development and progression (13, 14). Our recent results further indicate that high-risk hepatoblast-like (HB type) HCC characterized by the worst prognosis may be resistant to treatment with epigenetic-based treatment modalities (15). Therefore, we hypothesized that epigenetic modulation would not affect the core CSCs due to their intrinsic property of multidrug resistance while promoting the differentiated status of the non-CSC thereby forcing them out of the SP pool (16–18). The specific objectives were to (i) address the utility of epigenetic modulation for prospective isolation of core CSCs using short-term treatment of liver cancer cell lines with the potent DNMT1-inhibitor zebularine (ZEB) (ii) validate the findings in primary human cancers and (iii) provide an in-depth functional and molecular characterization of isolated CSCs using integrative genomics analysis. Our results demonstrate that ZEB treatment increased representation of highly tumorigenic cells with CSC properties within the SP fraction from cancer cell lines and primary cancer cells while reducing the tumorigenic capacity of the non-SP fraction. Transcriptome analyses of the CSC expression signatures revealed the diversity of activated oncogenic pathways and preservation of the core stemness-associated gene-sets.
MATERIALS AND METHODS
Cell Lines and Zebularine Treatment
Detailed description hepatoma cell lines are provided in Supporting Information. Cells were treated with 100 µM ZEB (Drug Synthesis and Chemistry Branch, Division of Cancer Treatment and Diagnosis, NCI) for 3 days. Optimal dose (100 µM) was selected based on published data and efficiency in reducing SP fraction without overt cell toxicity.
Primary Tumor Cell Lines
Primary tumors were obtained from patients undergoing surgery at the NCI Surgery Branch and from the UPMC, Pittsburgh in accordance with ethical guidelines. Tumors were processed directly or after one round of xenotransplantation as described in the Supporting Information.
SP Analysis by Flow Cytometry
SP analysis was performed as described (19). Cells were incubated at 37°C for 90 min with 15 µg/ml of Hoechst-33342 (Invitrogen), either alone or in presence of 50 µmol/L of Fumitremorgin C (FMT) (Sigma). Only viable cells were FACS sorted (based on 7-AAD exclusion) followed by confirmation of purity and viability and used only when both parameters were >90%.
Animals and Tumor Xenografts
All procedures were performed with approval of and in accordance with the guidelines of the National Institutes of Health animal care committee. Hepatoma cells (1×106) and FACS-sorted SP and non-SP cells (102–104) (mixed 1:1 in DMEM/Matrigel) (BD Bioscience) were transplanted subcutaneously (s.c.) into nude/athymic or non-obese diabetic/severe combined imunodeficient (NOD/SCID) mice. Control injections with DMEM and Matrigel did not produce tumors. Limiting dilution analysis was performed as described (http://bioinf.wehi.edu.au/software/limdil/index.html) and tumor-initiating cell frequency was calculated for each transplanted fraction (20). Kaplan-Meier analysis of tumor incidences was performed using Gehan-Breslow-Wilcoxon and Mantel-Cox Test.
Colony and Sphere Formation
Colony formation was assessed by agar-based assays. 103 SP and non-SP cells were re-suspended in 75 µl of DMEM containing 0.3% agar and 10% FBS and added on top of pre-solidified 0.6% agar in 96-well plates. Sphere formation was monitored for 14 days and average number of spheres (per 5 view fields) was calculated for each fraction in three independent replicates.
Microarray Analysis
200 ng RNA from 3–4 independent FACS-sorting experiments were linearly amplified as recommended by the manufacturer (Ambion, Austin, TX). For in vitro transcription, reactions were incubated for 16 h at 37°C. Hybridization, washing, detection (Cy3-streptavidin, Amersham Biosciences, GE Healthcare), and scanning were performed on illumina® iScan system (Illumina®) following protocols supplied by the manufacturer. Biotinylated cRNA (750 ng/sample) was hybridized on Sentrix beadchips human Ref-8v3 (~ 24,000 RefSeq transcripts) for 18 h at 58°C while rocking (5 rpm). Image analysis and data extraction were performed using illumina® GenomeScan Software. Detailed descriptions of performed analyses are provided in Supporting Information.
Databases
Oncomine Cancer Microarray database (http://www.oncomine.org) was used to conduct a meta-analysis for the predictive value of the classifier signature in 40 different cancer types as described (21).
RESULTS
Zebularine Reduces the SP Size while Increasing Representation of Cells with CSC Properties within SP Fraction
In agreement with previously published data (4), we found that the SP fraction is enriched in tumor-initiating cells (Supporting Table 1A). Among 10 cancer cell lines, only those with relatively high SP frequency (0.8–1.4%) developed tumors within 5 wk after s.c. transplantation into nude/athymic mice. These results were validated by limiting dilution analysis (LDA) of cells with high (Huh7, WRL68, PLC/PRF/5) or low (Hep3B, Huh1) SP frequency in NOD/SCID mice (Supporting Table 1B). Regardless of origin (15), a 3-day exposure to zebularine caused a consistent albeit varying reduction in SP frequency (Fig. 1A and B) which reversed to the levels found in parental cells lines one week after discontinuation of zebularine treatment (data not shown), suggesting a transient nature of the ZEB effect on the size of the SP population.
Fig. 1.
Treatment with Zebularine Reduces Frequency while Increasing Clonogenicty of SP Cells. (A) Effect of Zebularine on SP frequency. Data presented as mean percentage ± SD of 3 independent experiments. (B) Live-cell FACS profiles for Huh7 cells untreated and treated with 100 µM zebularine for 3 days. SP cells were identified by Hoechst 33342 staining and the use of blue and red filters. Cells were incubated with Hoechst 33342 in presence (top) and absence (bottom) of Fumitremorgin C to identify the SP gate with cell percentages noted. (C and D) Bar graph showing the number of spheres formed in soft agar by untreated (C) or zebularine-treated cells (D). Data are presented as mean ± SD of triplicate experiments and expressed relative to corresponding non-SP cells. (* P<0.05; ** P<0.01, *** P<0.001). (E) Representative images of spheres formed in soft agar by SP and non-SP cells sorted by FACS form untreated and zebularine-treated PLC/PRF/5 cells. (X40).
We then used a variety of standard in vitro and in vivo assays to examine whether ZEB increased the frequency of CSCs. In the absence of ZEB treatment, sphere-forming ability of SP cells was higher (Huh7, WRL68 and KMCH) or comparable (WITT and PLC) with that of non-SP cells as estimated by soft-agar- and Matrigel-based spheroid assays (Fig. 1C and not shown). Consistent in all the cell lines, ZEB treatment increased the frequency of SP-derived tumor-spheres relative to non-SP (Fig. 1D,E). Similar effects were observed using fluorescence-based colony-forming assays (data not shown). Thus, epigenetic modulation amplified sphere-forming and clonogenic potential of SP cells suggesting relative enrichment of CSCs within the SP fraction. In support of this, qRT-PCR analysis revealed upregulation of CSCs (ABCG2, CD133, GPC3 and c-KIT) and “stemness” (OCT4, NANOG, SOX2) associated genes in ZEB-treated SP cells as compared to non-SP cells albeit to a different degree (Supporting Fig. 1).
Limiting dilution analysis confirmed a higher frequency of tumor-initiating cells within SP fractions from Huh7, WRL68, and KMCH as compared to non-SP cells at 8 wk after s.c. transplantation into NOD/SCID mice, although by 10 wk the differences became less pronounced in untreated cells (Table 1). ZEB remarkably increased the number of tumor-initiating cells in SP fractions (7 fold, P=6.12×10−5; Table 1). As few as 100 ZEB-treated SP cells produced tumors while 1,000 non-SP cells gave rise to less (KMCH, WRL68) or no tumors (Huh7) (Supporting Table 2). Selected animals injected with non-SP cells from Huh7 and KMCH were followed over 20 wk after transplantation without tumor growth. LDA performed at 10 wk revealed a 14.8-fold increase in tumor-initiating capacity of SP cells compared to non-SP (P=5.45×10−6). Histologically, tumors derived from SP and non-SP cells were similar and recapitulated features of the parental tumors regardless of ZEB treatment (Supporting Fig. 2).
Table 1.
Zebularine Increases Tumorigenic Potential of SP Cells
| Treatment | Weeks | Cell fraction | No. injected cells |
TIF | CI 95% | p-value | ||
|---|---|---|---|---|---|---|---|---|
| 100 | 1000 | 10000 | ||||||
| 1/348– | ||||||||
| Untreated | 8w | SP | 6/12 | 7/12 | 12/12 | 1/657 | 1/1243 | |
| 1/1428– | ||||||||
| non-SP | 0/12 | 6/12 | 11/12 | 1/2804 | 1/5509 | 0.001 | ||
| 1/103– | ||||||||
| ZEB | 8w | SP | 4/12 | 12/12 | 12/12 | 1/217 | 1/457 | |
| 1/1156– | ||||||||
| non-SP | 0/12 | 4/12 | 12/12 | 1/2341 | 1/4741 | <0.0001 | ||
| 1/270– | ||||||||
| Untreated | 10w | SP | 7/12 | 8/12 | 12/12 | 1/507 | 1/952 | |
| 1/266– | ||||||||
| non-SP | 3/12 | 10/12 | 12/12 | 1/499 | 1/936 | n.s | ||
| 1/36– | ||||||||
| ZEB | 10w | SP | 9/12 | 12/12 | 12/12 | 1/73 | 1/147 | |
| 1/535– | ||||||||
| non-SP | 0/12 | 8/12 | 12/12 | 1/1073 | 1/2150 | <0.0001 | ||
Limiting dilution analysis (LDA) of FACS sorted SP and non-SP cells isolated from Huh7, WRL68, KMCH liver cancer cell lines and inoculated into both flanks of NOD/SCID mice (n=4 for each line). Tumor-initiating frequency (TIF) was calculated at 8 wk and 10 wk as described in Material and Methods and considered statistically significant at P <0.05.
In addition, ZEB-treated SP cells were analyzed for self-renewal potential. Cells were re-isolated from Huh7 and KMCH xenograft-tumors established from 100 SP-ZEB cells, propagated in short term cultures, treated with ZEB for 3 days and FACS-sorted for SP and non-SP fractions before re-transplanting into secondary recipients. Consistent in both cell lines, SP cells not only sustained tumorigenic potential in serial transplantations but progressively increased in frequency (Fig. 2A,B). KMCH secondary tumors developed with a shorter latency. Conversely, the corresponding non-SP cells showed either dramatic increase in tumor latency and decrease in tumor incidence (KMCH) or no tumor growth at all (Huh7) (Fig. 2C,D). Together the data show that ZEB significantly enhanced the tumorigenic potential of SP cells while reducing it in non-SP cells.
Fig. 2.
SP Cells Possess Self-Renewal Capacity in Serial Transplantations. (A) Changes in SP frequency after serial transplantations of SP-ZEB cells from Huh7 and KMCH. Data presented as mean percentage ± SD of three independent experiments. Cells were isolated from tumors derived from 100 SP-ZEB cells, re-treated with ZEB, and FACS sorted for SP and non-SP cells before serial re-transplantation. (B) Representative FACS profiles for Huh7 cells. SP cells were gated based on inhibition with Fumitremorgin C. (C) Graphs show kinetics of tumor development after serial transplantations of ZEB-treated SP and non-SP cells isolated from Huh7 (upper panels) and KMCH (lower panels) cell lines. 1000 cells of each fraction were transplanted into flanks of NOD/SCID mice (n=4) and tumor formation was monitored by weekly palpation. P <0.05 was considered statistically significant.
To provide evidence that the cancer stem cell-enriching effect of zebularine was due to inhibition of DNMT1, we determined the protein levels of DNMT1, as well as DNMT3a and DNMT3b both in SP and non-SP cells. For this purpose we used primary liver cancer cells which display a relatively large SP population to obtain sufficient numbers of cells for western blot analysis (Fig. 4). Consistent with the results of our recent study (15) we found that zebularine treatment caused a complete inhibition of DNMT1 expression both in the SP and non-SP cells (Supporting Fig.3). In accordance with the literature (15, 22), zebularine treatment did not affect the protein levels of DNMT3a and DNMT3b confirming its specificity (Supporting Fig.3).
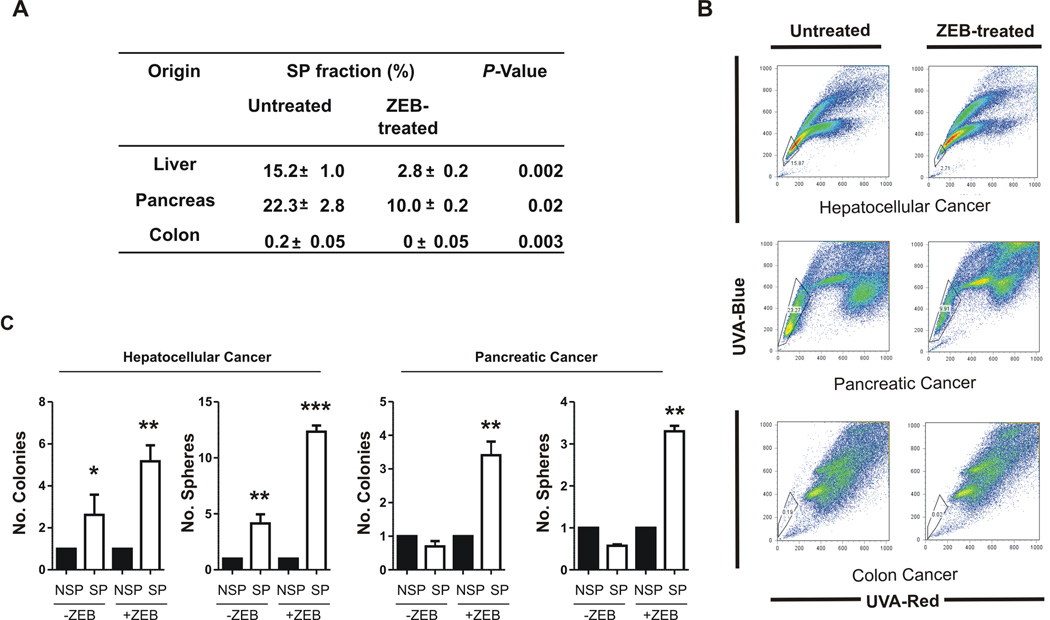
Fig. 4.
ZEB effect on primary human cancer cells. (A) Effect of Zebularine on SP frequency of primary human cancers cells from gastrointestinal and hepatobilary cancers. Data presented as mean percentage ± SD. All experiments performed in triplicates. (B) Representative SP profiles of untreated and zebularine- treated cells from liver, pancreatic and colon cancer (top) gated based on inhibition with Fumitremorgin C (bottom). (C) Bar graph showing the number of colonies and spheres formed by untreated or zebularine-treated sorted liver and pancreatic cancer cells cultured on monolayer or Matrigel. The data presented as mean ± SD of triplicate experiment. (* P<0.05; ** P<0.01, *** P<0.001)
Sphere Formation and Tumorigenicity in Direct Growth Competition
To directly compare tumorigenic potential of SP and non-SP cells exposed to ZEB we used lineage-tracking experiments in vivo and in vitro (Fig. 3). Huh7 cells transduced with lentiviral vectors expressing green (GFP) or red (mCherry) fluorescent proteins were sorted for SP (green) and non-SP (red) cells, mixed in 1:1 ratio and cultured at low-cell density to allow clonal expansion (using plain or Matrigel-coated dishes) or transplanted into NOD/SCID mice. The majority of colonies and spheres were derived from GFP-expressing SP cells after 2 wk and 3 wk of culture (Figure 3A,B). Experiments with reverse labeling of SP and non-SP cells produced comparable results (not shown). Frequency of sphere forming units in mixed cultures was consistently higher than that observed in individual cultures implying a role for microenvironment in propagation of tumor growth.
Fig. 3.
Cell Tracking Experiments Demonstrate Superior Self-Renewal Ability of SP Cells versus non-SP Cells. (A) Experimental design. Huh7 cells stably transduced with either green fluorescent protein (GFP) or red fluorescent protein (mCherry) were FACS-sorted for SP (green) and non-SP (red) cells to allow cell tracking in vitro and in vivo. (B) Graph bars showing the number of colonies formed by 1000 SP (green) and nonSP (red) cells 14 days after plating. Direct fluorescence image of colonies formed by SP and non-SP cells (X40) shown on the right. Data represent means of 3 replicates ± SD. P <0.05 calculated by paired t-test were considered statistically significant. (C) Bar graphs showing the number of spheres formed in Matrigel by 1000 SP and non-SP cells 21 days after plating. Direct fluorescence (left) and bright (right) field images showing a sphere formed by a green SP cell. Scale bars, 100µm. Data represent means of 6 replicates ± SD. P-values were calculated by paired t-test and P <0.05 were considered statistically significant. (D) Representative ex vivo confocal image of the whole tumor at 8 wk after s.c. transplantation of 1000 SP and non-SP (1:1) cells into NOD/SCID mice (n=4). The vast majority of tumor composed from cells with green fluorescence phenotype indicating that SP cells were responsible for tumor growth. Arrows point to minor areas formed by non-SP-red cells. Scale bar, 200 µm. Inset, macroscopic image of tumor at 6 wk after transplantation.
More dramatic differences in tumor-initiating potency between ZEB-treated SP and non-SP cells were observed when a relative contribution of each fraction was evaluated in xenograft tumors initiated by a 1:1 mixture of SP (GFP) and non-SP (mCherry) cells. Ex-vivo whole confocal imaging demonstrated that the vast majority of tumor cells expressed GFP, indicating their SP origin (Fig. 3C,D).
Effect of Zebularine on SP Cells from Primary Cancer Cells
Finally, the effect of ZEB treatment was validated in freshly isolated tumor cells from different human gastro-intestinal and hepatobiliary cancers (Fig. 4). Consistent with our findings in cell lines, ZEB reduced SP size in primary tumor cells which was paralleled by increased spheroid- and colony-forming ability (Fig. 4A–C). We also found upregulation of CSC and pluripotency associated genes albeit to various degrees in cancers of different origin (Supporting Fig. 1). Pancreatic cancer SP cells displayed dramatic increase in the expression of CSC and pluripotency markers as compared to non-SP cells (Supporting Fig. 1). Liver cancer SP cells displayed a strong upregulation of NANOG (23-fold) and OCT4 (3-fold) whereas the expression of selected CSC markers was comparable (EpCAM, cKIT) or undetectable (CD133, SOX2, GPC3) (Supporting Fig. 1). Notably, ZEB treatment of colon cancer cells caused complete elimination of SP population, suggesting differential sensitivity of SP cells to ZEB effect. These results show that epigenetic modulation in combination with SP approach provides an important tool to enrich for cells possessing CSC properties.
Molecular Characteristics of Putative Liver CSCs
To make use of high frequency of cells with CSC properties in the ZEB-treated SP fraction, we performed global gene expression analysis and compared transcriptomes of ZEB-treated SP and non-SP cells. The number of differentially expressed genes, as determined by Bootstrap t-test, varied from 2041 genes in KMCH to 2731 genes in Huh7 and 4133 genes in WRL68, underlining CSCs’ heterogeneity in liver cancer cell lines (Supporting Fig. 4A). Pathway enrichment analysis revealed that each individual cell line was characterized by activation of unique oncogenic pathways with known associations to HCC, such as EGFR (Huh7), MYC (WRL68) and SRC (KMCH) (Supporting Fig. 4B). Furthermore, all cell lines demonstrated ubiquitous activation of NF-kB signaling, suggesting that CSCs exhibit a generalized increase in stress resistance and survival.
To more specifically identify genes related to liver CSCs, we looked for commonly dysregulated genes across all three cell lines. We found 1259 differentially expressed genes between SP-ZEB and non-SP-ZEB cells with 617 genes displaying more than 1.5-fold expression changes (Fig. 5, Supporting Table 4). This 617-gene set very efficiently separated SP-ZEB and non-SP-ZEB and therefore referred to as common SP-ZEB signature (Fig. 5A). Gene set enrichment analysis (GSEA), demonstrated that the SP-ZEB gene signature overlapped with two previously published gene sets associated with adult stem cells and hepatoblasts (HB) (Fig. 5B) (23, 24). Conversely, non-SP-ZEB cells showed a negative correlation suggesting a more differentiated phenotype (23). Again, the common signature was characterized by activation of NF-kB as well as IL6 and WNT/β-catenin signaling pathways known to be involved in cancer (Supporting Fig. 5A,B,D). Notably, we found that genes centered around the core HNF4α network, critical for hepatocyte differentiation, were consistently downregulated (Fig. 5C; Supporting Fig. 5C). Ingenuity Pathway analyses revealed that in addition to genes dysregulated in various cancers (AXIN2, EDN1, EP400, RICTOR, ADAMTS1, HOXA13, AGGF1, CCND1) and/or during invasion and metastasis (SNAI2, TIMP4, MMP25, RHOB, CCL20, CTAG2, CGN, CX3CL1, LGR4), the common SP-ZEB signature also contained stem/progenitor and liver development associated genes (CK19, FOXA2, CLDN2, SOX9, SOX4, DMBT1, MED12, AMD1). GSEA further identified abundance of gene sets involved in cytoskeleton architecture, vascular development, and JNK signaling in SP cells (Supporting Table 3). Selected targets of the generated signature (SOX4 and p-NF-kB) were validated using human tissue microarray of 95 HCC and 10 normal livers (25, 26). Significant enrichment of both markers was found during malignant progression (p-NF-kB: P<0.001; SOX4: P<0.05) (Supporting Fig. 5E,F).
Fig. 5.
Common Gene expression Signatures of Putative Liver CSCs. (A) Unsupervised hierachial cluster analysis of SP and non-SP fractions from Huh7, WRL68 and KMCH cell lines after zebularine treatment. A common gene expression signature of 617 differentially expressed genes from three SP-ZEB fractions identified by Bootstrap t-test (P-values ≤ 0.05 and fold-change ≥1.5). Note clear separation of SP (red) and non-SP (green) cells into 2 clusters. (B) GSEA based on two previously published gene sets: Adult tissue stem cell genes(23) and upregulated genes of the HB-signature.(24) For the adult tissue stem module available homologue genes were used. Results demonstrate enrichment of both gene sets in SP-ZEB cells. Enrichment score (ES) reflects degree of over-representation for each group at the peak of the entire set. Statistical significance calculated by nominal P-value of the ES by using an empirical phenotype-based permutation test. False positives are calculated by the false discovery rate (FDR).(C) Top 5 functional networks for the SP-ZEB-signature.
CSC Gene Signature Is Independent of Zebularine Effect
To differentiate between the contribution of Fraction- (SP, non-SP) and Treatment- (ZEB-treated, untreated) dependent effects on CSC gene signature, we employed a Two-Way-Anova statistical approach which takes into consideration all potential causative factors. Comparison of molecular profiles derived from untreated and treated SP and non-SP populations showed that the majority of the common CSC signature (932/1259 genes, 74%) overlapped with the SP fraction-dependent gene set and thus was defined by CSC properties but not ZEB exposure. We confirmed this conclusion by a further comparison of the CSC signature with zebularine methylation signature which was generated for Huh7, WRL68 and KMCH cells using Illumina Infinium HumanMethylation27 microarray involving 27,578 CpG sites. Only 28 genes overlapped between the 617-gene CSC signature and ZEB methylation signature (990 genes) indicating that the described CSC signature was reflective of intrinsic CSC properties. Finally, screening the promoters of 118 genes, which best classified HCC patients according to clinical outcome (Fig. 6B) for the presence of 5′-CpG islands using the EMBOSS CpGplot/report (GC content >50%, ratio of CpG-to-GpC >0.6, and minimum length 200 bp (27) revealed that only 52.5% of genes contained promoter CpG islands as compared to 60% expected average within the human genome (28).
Fig. 6.
SP-ZEB Signature Predicts HCC Patients Prognosis. (A) Hierarchical cluster analysis of 53 human HCCs based on the SP-ZEB signature (579 of 617 genes remained when filtered for 80% presence of genes). Bars under cluster tree represent integrated cell fractions and overlap with previously generated HCC subclasses (subtype A, B, HB and HC). (B) Identification of gene classifiers predictive for patient survival. A class random variance model was used to identify classifier genes (α≤0.001) and validation of correct classification was performed by seven different algorithm using 1000 random permutations. (C) Validation of the SP-ZEB gene signature in poor prognosis patients (subtype A) by GSEA.(D) Kaplan-Meier plots of overall survival and (E) recurrence of the HCC (Mantel-Cox test). Recurrence data were available only for 20 patients. (F) Association of the classifier signature with clinical outcome of cancer patients from different types of cancer. Integrative meta-analysis of genomic data from 40 primary tumors using the Oncomine Microarray database. Data presented as mean odds ratio ± SD with P-values < 0.001 and odds ratios >2 set as a threshold.
Prognostic Value of the SP-ZEB Signatures for Survival of Cancer Patients
To test the clinical significance of the SP-ZEB signature, we integrated individual SP-signatures with our previously published gene expression dataset from 139 human HCC (24). Kaplan-Meier analysis showed that each SP signature independently classified HCC patients according to survival (Supporting Fig. 4D). All three SP signatures were enriched in the poorly differentiated HCC-subtype A including tumors previously defined by hepatic stem cell-like traits and worse clinical outcome (hepatoblst, HB subtype) (Supporting Fig. 4C) (24, 29). These findings were confirmed by integrative analysis of the common SP-ZEB signature using gene expression data from 53 HCC patients generated on illumina® beadchips (Fig. 6A,C). To narrow down the common SP-ZEB signature to genes most significantly associated with the identified clusters, we generated a 118-gene classifier using Leave-One-Out cross-validation and confirmed its predictive value by 7 different prediction models (Fig. 6B). The 118-gene set successfully differentiated HCC patients according to overall survival (P<0.006) and disease recurrence (P<0.02) (Fig. 6D,E). Notably, removal of genes involved in proliferation and cell cycle did not impact the ability of the signature to classify liver cancer patients according to clinical outcome (P=0.01) (30, 31). Furthermore, a meta-analysis performed on gene expression data from 40 different primary tumor types demonstrated that the 118-gene classifier also predicted survival of patients with other tumors (e.g. lung, breast, kidney) and successfully classified lung adeno-carcinoma according to clinical outcome (Fig. 6F, Supporting Fig. 6A,B) suggesting prognostic utility of the SP-ZEB signature for cancers other than HCC (21, 32). Complete list of these genes is provided in Supporting Information Table 5. Of note, ZEB treatment of lung adenocarcinoma cell line A549 led to a significant reduction in SP size (from 7±1% to 3±1%; P=0.008) further indicating the potential of ZEB for isolation and characterization of CSC (Supporting Fig. 6C).
DISCUSSION
In this study we demonstrate that epigenetic modulation of liver cancer cells may facilitate functional isolation of CSC cells possessing self-renewal and tumor-initiating capacity. Transcriptome analyses of highly enriched CSC populations isolated from different liver cancer cell lines revealed that CSCs maintain a common “stemness” gene expression signature while exhibiting activation of unique oncogenic pathways. Clinically, the common CSC signature was enriched in liver cancer with poor differentiated status and was highly predictive of tumor recurrence and overall survival of HCC patients supporting the translational value of this approach.
The common CSC gene signature was independent of potential treatment (ZEB) effects, as demonstrated by Two-Way-ANOVA statistical analysis, computational prediction of promoter CpG islands and comparison with ZEB-response methylation signature. These observations support the idea that treatment with zebularine did not affect the core CSCs while promoting the differentiation status of the non-CSC thereby forcing them out of the SP pool (16, 17). In agreement with these findings, we have recently demonstrated that high-risk hepatoblast-like (HB type) HCC characterized by the progenitor cell signature may be resistant to treatment with zebularine. Importantly, all examined cell lines showed an enrichment of cells with CSC properties within SP fraction albeit to a different degree despite the differential sensitivity to ZEB treatment (15). The latter is consistent with the intrinsic resistance of CSC to therapy, including epigenetic therapy, and underlies the necessity of CSC targeting to advance the therapeutic strategies against liver cancer.
Epigenetic modulation of liver (and other) cancer cell populations preferentially increased the frequency of tumor-initiating cells within the SP fraction. This conclusion is based on a greater colony forming capacity of ZEB-treated SP cells in vitro and parallel loss of clonogenic potential in corresponding non-SP cells indicating a better separation and a higher purity of the isolated fractions. Likewise, limiting dilution as well as serial transplantation experiments demonstrated a progressive increase in self-renewal of SP cells while corresponding non-SPs were gradually losing tumorigenic potential (Table 1., Fig. 3,4). Direct cell tracking experiments further emphasized greater tumor-initiating ability of ZEB-treated SP cells over non-SP cells. This effect was reproduced in primary human cancer cells of hepatobiliary and gastro-intestinal origin whereby validating the findings from established cell lines.
Following ZEB treatment we also found enrichment of selected CSCs related genes, such as CD133, c-KIT, GPC-3 and ABCG2 in addition to genes involved in maintenance of stemness in embryonic stem cells (OCT4, NANOG and SOX2) (Supporting Fig. 1). Epithelial tumors harboring gene expression profiles enriched for embryonic-stem-cell-like traits are more aggressive and have worse prognosis, supporting the notion that these tumors possess CSC signatures (33).
Global transcriptome analysis revealed that each individual CSC signature was characterized by a dominant oncogenic network, such as MYC, EGFR and SRC, known to be associated with phenotypically different cancer subtypes (34–36) supporting recent findings that primary tumor genotype is an important determinant of CSCs (5). Additionally, the liver CSCs shared a common gene expression signature indicating that a variety of oncogenic pathways can exploit similar gene networks associated with stemness and self-renewal (WNT/β-Catenin, IL6), development (SOX4, SOX9, MED12, AMD1), and hepatic progenitor/stem cells (CK19, DMBT1). In agreement with the concept that CSCs are generally responsible for seeding of local and distant metastasis (17), we found disruption of mTOR and JNK pathways and genes important for vasculogenesis and cytoskeleton organization including activated RHOA/B kinases. Another key common feature of CSCs was activation of NF-kB signaling known to increase stress resistance and survival (26, 37, 38). Thus, specific targeting of common CSC traits can complement currently used multiple pathways inhibitors (e.g. sorafenib) and advance discovery of novel individualized therapies (39).
The capacity of CSC signature to classify HCC patients according to prognosis further underlines the clinical importance of these findings. Integration of individual SP and common SP-ZEB signatures with human HCC showed significant associations with less-differentiated tumors and enrichment of gene-expression signatures of the progenitor cell sybtype HB. Furthermore, the 118-gene classifier signature demonstrated high predictive power for tumors other than HCC.
In conclusion, the common stemness-enriched CSC gene signature exhibits a pernicious interaction with a variety of known oncogenic pathways and correlates with poor clinical status and bad prognosis in liver and other cancers. Furthermore, epigenetic modulation of cancer cells is a useful tool to increase the relative representation of highly tumorigenic cells with CSC characteristics within the SP fraction without notable changes in their properties and improves the identification of therapeutic targets specifically directed towards CSCs.
Supplementary Material
Acknowledgements
Financial Support
This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
We thank Barbara Taylor and Gordon Wiegand, NCI, for help with FACS sorting; Susan Garfield and Langston Lim, NCI for confocal microscopy assistance; Gregory Gores, Mayo Clinic, and David S. Schrump, NCI Surgery Branch, for providing cell lines.
Abbreviations
- CSC
cancer stem cells
- ZEB
Zebularine
- SP
Side-Population
- HCC
Hepatocellular Carcinoma
- DNMT-1
DNA- methyltransferase1
Footnotes
Disclosures
The authors have nothing to disclose.
Transcript profiling: GEO accession number: GSE24520
Supporting Information: The Supporting Information includes supporting Materials and Methods, 6 tables and 6 figures.
Contributor Information
Jens U. Marquardt, Email: Jens_marquardt@nih.gov.
Chiara Raggi, Email: Chiara_raggi@nih.gov.
Jesper B. Andersen, Email: Jesper_andersen@nih.gov.
Daekwan Seo, Email: Deakwan_seo@nih.gov.
Itzhak Avital, Email: Avitali@mail.nih.gov.
David Geller, Email: gellerda@upmc.edu.
Yun-Han Lee, Email: Yunhan.lee@nih.gov.
Mitsuteru Kitade, Email: Kitadem@mail.nih.gov.
Agnes Holczbauer, Email: Holzcbauera@mail.nih.gov.
Elizabeth A. Conner, Email: Liz_conner@nih.gov.
Valentina M. Factor, Email: Valentina_factor@nih.gov.
Reference List
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 5.Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, et al. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010;7:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Potgens AJ, Schmitz U, Kaufmann P, Frank HG. Monoclonal antibody CD133-2 (AC141) against hematopoietic stem cell antigen CD133 shows crossreactivity with cytokeratin 18. J Histochem Cytochem. 2002;50:1131–1134. doi: 10.1177/002215540205000814. [DOI] [PubMed] [Google Scholar]
- 10.Taussig DC, Miraki-Moud F, njos-Afonso F, Pearce DJ, Allen K, Ridler C, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112:568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan JP, Minna JD. Tumor oncogenotypes and lung cancer stem cell identity. Cell Stem Cell. 2010;7:2–4. doi: 10.1016/j.stem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, et al. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JB, Factor VM, Marquardt JU, Raggi C, Lee YH, Seo D, et al. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001338. 54ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 18.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26:2813–2820. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 23.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 25.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 27.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 28.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 30.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Conner EA, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 37.Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253–260. doi: 10.1385/SCR:1:3:253. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Yan HX, Chen L, Li Q, He YQ, Yu LX, et al. Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287–4295. doi: 10.1158/0008-5472.CAN-07-6691. [DOI] [PubMed] [Google Scholar]
- 39.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.