SUMMARY
In response to viral infection, RIG-I–like RNA helicases bind to viral RNA and activate the mitochondrial protein MAVS, which in turn activates the transcription factors IRF3 and NF-κB to induce type-I interferons. We have previously shown that RIG-I binds to unanchored lysine-63 (K63) polyubiquitin chains and that this binding is important for MAVS activation; however, the mechanism underlying MAVS activation is not understood. Here we show that viral infection induces the formation of very large MAVS aggregates, which potently activate IRF3 in the cytosol. We find that a fraction of recombinant MAVS protein forms fibrils capable of activating IRF3. Remarkably, the MAVS fibrils behave like prions and effectively convert endogenous MAVS into functional aggregates. We also show that, in the presence of K63 ubiquitin chains, RIG-I catalyzes the conversion of MAVS on the mitochondrial membrane to prion-like aggregates. These results suggest that a prion-like conformational switch of MAVS activates and propagates the antiviral signaling cascade.
INTRODUCTION
Innate immunity is an evolutionarily conserved defense mechanism against microbial infections (Iwasaki and Medzhitov, 2010; Ronald and Beutler, 2010; Takeuchi and Akira, 2010). In higher organisms, an antiviral innate immune response is triggered by the recognition of viral nucleic acids by germline-encoded pathogen recognition receptors, including Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) (Barbalat et al., 2010; Rehwinkel and Reis e Sousa, 2010). Several TLRs, including TLR3, 7, 8 and 9, detect viral RNA and DNA in the endosome, whereas RLRs bind to viral RNA in the cytoplasm. Both TLR and RLR pathways activate signaling cascades that lead to the production of an arsenal of effector molecules that suppress viral replication and assembly. Prominent among the antiviral molecules are type-I interferons, including IFNα and IFNβ, which activate the JAK-STAT pathway to fight viral infection.
RLRs comprise RIG-I, MDA5 and LGP2, all of which contain an RNA helicase domain (Yoneyama and Fujita, 2009; Yoneyama et al., 2004). RIG-I also contains a C-terminal regulatory domain (RD) that binds to viral RNA harboring 5′-triphosphate (5′-pppRNA)(Hornung et al., 2006; Pichlmair et al., 2006). RIG-I and MDA5 detect distinct classes of RNA viruses (Kato et al., 2006). Both RIG-I and MDA5 contain two CARD domains in tandem at the N-terminus, whereas LGP2 lacks the CARD domains. The binding of viral RNA to the C-termini of RIG-I and MDA5 presumably induces a conformational change that exposes the N-terminal CARD domain, which interacts with the CARD domain of the mitochondrial adaptor protein MAVS (also known as IPS1, VISA or CARDIF) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS then activates the cytosolic kinases IKK and TBK1, which activate the transcription factors NF-κB and IRF3, respectively. NF-κB and IRF3 translocate into the nucleus, where they function cooperatively to induce type-I interferons and other antiviral molecules (McWhirter et al., 2005).
To understand the mechanism of signal transduction in the RIG-I pathway, we have recently established a cell-free system in which viral RNA triggers the activation of IRF3 and IKK in cytosolic extracts in the presence of mitochondria (Zeng et al., 2010). Using this system, we discovered that the CARD domains of RIG-I bind to unanchored K63 polyubiquitin chains, and that this binding is important for RIG-I activation. The binding of full-length RIG-I to ubiquitin chains depends on ATP and 5′-pppRNA, suggesting that RIG-I activation involves sequential binding of viral RNA and unanchored K63 polyubiquitin chains to RIG-I RD and CARDs, respectively. We have also shown that mitochondria isolated from virus-infected cells can activate IKK and TBK1 in the cytoplasm, and that this activity depends on MAVS on the mitochondrial membrane (Zeng et al., 2009). Interestingly, K63 polyubiquitination also plays an important role in TBK1 activation by MAVS.
The mechanism by which MAVS is activated by RIG-I and ubiquitin chains is still not understood. The nature of the active form of MAVS has also remained a mystery. In this report, we show that MAVS forms very large aggregates after viral infection, and that these aggregates are highly potent in activating IRF3 in the cytoplasm. Remarkably, these aggregates form self-perpetuating fiber-like polymers that can efficiently convert endogenous MAVS into functional aggregates. These properties closely resemble prions, which are infectious protein conformations found in pathological as well as physiological conditions (Chien et al., 2004; Halfmann and Lindquist, 2010; Tuite and Serio, 2010). Initially found to be the causative agent of fatal neurological diseases (Prusiner, 1998), including Scrapies in sheep and goats, bovine spongiform encephalopathy (BSE or “mad cow disease”) in cattle and Creutzfeldt-Jacob disease (CJD) in human, the prion PrP is the founding member of a growing list of proteins that can form self-perpetuating aggregates, several of which have evolved to serve physiological functions. For example, several prions in fungi regulate phenotypic switches that may confer selectable advantages (Halfmann and Lindquist, 2010). Thus, the prion-based conformational switch can be a robust epigenetic mechanism that regulates protein functions and cellular phenotypes. Properties of prions include fibrous aggregates, resistance to detergent and protease, and most importantly, the ability to infect the endogenous protein and convert the native conformation into fibrous aggregates. Strikingly, MAVS possesses all of these prion-like properties. The formation of MAVS aggregates leads to a gain of function, and the conformational switch is highly efficient and tightly regulated by viral infection. Also quite remarkably, in vitro incubation of RIG-I and mitochondria in the presence of K63 polyubiquitin chains efficiently converts endogenous MAVS into functional aggregates.
RESULTS
Viral Infection Induces the Formation of Large MAVS Signaling Complexes
To understand how MAVS is activated by viral infection, we used differential centrifugation to isolate crude mitochondria (P5) from HEK293T cells, which were infected with Sendai virus (+SeV) or not infected (−SeV). The mitochondrial proteins were extracted in a buffer containing the nonionic detergent n-dodecyl beta-D-maltoside (DDM; 1%), and then fractionated by sucrose gradient ultracentrifugation. Aliquots of the fractions were analyzed by immunoblotting with a MAVS antibody, whereas other aliquots were incubated with 35S-IRF3 and HEK293T cytosolic extracts in the presence of ATP. The dimerization of IRF3, which is caused by its phosphorylation by TBK1 and represents the hallmark of its activation, was measured by native gel electrophoresis (Panne et al., 2007; Yoneyama et al., 2002). As shown in Figure 1A, viral infection led to the formation of a very large complex containing MAVS, which activated IRF3 in the cytosol. This complex was much larger than 26S proteasome, and sedimented towards the bottom of the centrifuge tube containing 50–60% sucrose. We have previously shown that our MAVS antibody, which was raised against residues 131–291 of MAVS, detected two major bands on SDS-PAGE (Seth et al., 2005). The upper band represents full-length MAVS, whereas the lower band is a truncated form of MAVS, which lacks the N-terminus but retains the C-terminal transmembrane domain. Interestingly, only the full-length MAVS formed a large complex capable of activating IRF3. Furthermore, almost all full-length MAVS shifted to the large complex in response to viral infection.
Figure 1. Viral Infection Induces the Formation of Large MAVS Particles That Activate IRF3.
(A) Crude mitochondria isolated from HEK293T cells infected with Sendai virus for 14 hours (+SeV) or uninfected (−SeV) were solubilized in a buffer containing 1% DDM, then subjected to sucrose gradient ultracentrifugation. Aliquots of the fractions were immunoblotted with a MAVS antibody, or incubated with 35S-IRF3 and cytosolic extracts in the presence of ATP at 30°C for 60 min, followed by native gel electrophoresis and autoradiography. Arrows indicate the positions of proteins used as molecular size markers, including 20S and 26S proteasome. ΔMAVS denotes a truncated MAVS lacking the N-terminus. (B) YFP-MAVS was expressed in Mavs−/− MEF cells by retroviral transduction, and the cells expressing low levels of YFP-MAVS were sorted by FACS. These cells were infected with Sendai virus for 13 hours, stained with Mitotracker, and then visualized by confocal fluorescent microscopy. The images are representative of >50 % of the cells under examination. (C) Crude mitochondrial extracts were prepared from HEK293T cells infected with Sendai virus for the indicated time, then aliquots of the extracts were analyzed by SDD-AGE, SDS-PAGE, or IRF3 dimerization assays. (D) Crude mitochondrial extracts were treated with or without β-mercaptoethanol (BME at 0.35, 3.5 and 35 mM), followed by SDD-AGE. (E) HEK293T cells were treated with geldanamycin or 17-AAG at the indicated concentrations for 1 hour before Sendai virus infection. 9 hours later, the activation of endogenous IRF3 in the cytosolic extracts were analyzed by native gel electrophoresis. Mitochondria (P5) were prepared and analyzed by SDD-AGE or SDS-PAGE using a MAVS antibody. An aliquot of P5 was incubated with cytosolic extracts, 35S-IRF3 and ATP followed by native gel electrophoresis to measure MAVS activity.
To visualize MAVS protein in cells, we expressed YFP-tagged MAVS in Mavs-deficient murine embryonic fibroblasts (MEFs) by retroviral transduction. Confocal fluorescent microscopy revealed that the staining pattern of YFP-MAVS overlapped with that of the mitochondrial marker Mitotracker in the absence of virus infection (Fig. 1B). Strikingly, after infection with Sendai virus, YFP-MAVS appeared to form clusters that partially overlapped with Mitotracker, suggesting that MAVS forms aggregates in response to viral infection.
The large size of the active MAVS complex, together with our previous observation that MAVS in virus-infected cells is more resistant to detergent extraction (Seth et al., 2005), led us to test whether MAVS forms detergent-resistant aggregates. We employed a method called semi-denaturing detergent agarose gel electrophoresis (SDD-AGE), which was previously used for the detection of prion-like structures (Alberti et al., 2009). In SDD-AGE, the crude mitochondria (P5) from cells infected with Sendai virus for different lengths of time were resuspended in a sample buffer containing 2% SDS, and then separated on 1.5% agarose gel by electrophoresis in a running buffer containing 0.1% SDS (Fig. 1C). Strikingly, a smear of SDS-resistant high molecular weight MAVS aggregates appeared after 9 hours of viral infection, much like prions (Alberti et al., 2009). These aggregates were not detected in cells depleted of MAVS by RNAi, which blocked IRF3 activation by Sendai virus (Supplementary Figure S1A). The kinetics of MAVS aggregate formation correlated with IRF3 activation by mitochondria from the virus-infected cells (Fig. 1C). These results indicate that MAVS forms very large and highly active signaling complexes following viral infection.
In Figure 1C, we noted that our MAVS antibody could barely detect MAVS on SDD-AGE during the early time course of viral infection, but was able to detect MAVS in the same samples when they were separated by the standard SDS polyacrylamide gel electrophoresis (SDS-PAGE). A major difference between SDD-AGE and SDS-PAGE is the presence of a reducing agent (β-mercaptoethanol or BME) in the latter but not in the former sample buffer. Interestingly, when crude mitochondria were resuspended in sample buffers containing different concentrations of BME followed by SDD-AGE, the smear of high molecular weight MAVS aggregates disappeared (Fig. 1D). These results suggest that the SDS-resistant MAVS aggregates may contain disulfide bonds and that the functional aggregates are preferentially detected by our MAVS antibody.
To determine if reduction of the MAVS aggregates alters their activity and/or aggregation, we resuspended mitochondria from Sendai virus infected cells in a buffer containing 1% DDM and 10 mM DTT, and then fractionated the mitochondrial extracts by sucrose gradient ultracentrifugation. MAVS still sedimented as very large particles after the DTT treatment, and these particles were fully capable of activating IRF3 in the cytosol (Supplementary Figure S1B). Control experiments showed that the DTT-treated particles in high-density sucrose fractions no longer formed detectable MAVS aggregates on SDD-AGE (Supplementary Figure S1C). Thus, DTT treatment prevented the detection of MAVS aggregates using the SDD-AGE assay, but did not cause the breakdown of the MAVS aggregates, which could be isolated by ultracentrifugation. These MAVS aggregates were still active in causing IRF3 dimerization (Figure S1B). However, DTT treatment of cells blocked MAVS aggregation as well as IRF3 activation by Sendai virus (Figure S1D). This effect was more evident when DTT was applied during the early time course of virus infection than during the late time course (Supplementary Figure S1E). Thus, it is possible that disulfide bond formation facilitates MAVS aggregation, but the maintenance of the MAVS aggregates and its activity does not require the disulfide bonds. In any case, SDD-AGE without a reducing agent provides a sensitive assay for the detection of SDS-resistant, functional MAVS aggregates induced by virus infection.
Previous studies have identified several chemicals that inhibit IRF3 phosphorylation by RNA viruses and poly[I:C] (Iwamura et al., 2001). Among these is the Hsp90 inhibitor geldanamycin, which inhibits IRF3 phosphorylation through an unknown mechanism. We found that geldanamycin and its analogue (17-AAG) at concentrations that inhibited IRF3 activation also blocked MAVS aggregation induce by Sendai virus (Fig. 1D). Further, mitochondria isolated from cells treated with the drugs failed to activate IRF3 when incubated with cytosolic extracts. In contrast, cytosolic extracts from geldanamycin-treated cells could still support IRF3 activation when incubated with mitochondria from virus-infected cells (Supplementary Figure S1F). Interestingly, the cytosolic extracts from Sendai virus infected cells were refractory to activation by mitochondria from virus-infected cells (lane 2, Figure S1F), suggesting that some signaling proteins in the cytosol were desensitized following their activation. Taken together, these results suggest that geldanamycin and 17-AAG inhibit IRF3 activation by preventing MAVS aggregation on the mitochondria.
The Active MAVS Complex Is Composed of MAVS Polymers
To facilitate purification of the active MAVS complex, we generated a HEK293T cell line stably expressing Flag-MAVS. Analysis of the mitochondrial extracts from this cell line by sucrose gradient ultracentrifugation revealed that a fraction of Flag-MAVS formed a large complex capable of activating IRF3 dimerization even in the absence of viral infection, suggesting that overexpression caused a small fraction of Flag-MAVS to form the active complex (Supplementary Figure S2A, upper). Sendai virus infection caused the vast majority of MAVS to form the active complex (Figure S2A, lower). However, despite much effort, we were unable to immunoprecipitate the active MAVS complex with antibodies against Flag or MAVS under native conditions. We therefore attempted to carry out immunoprecipitation under a partially denaturing condition that could maintain the activity of the MAVS complex. We found that when the MAVS complex was solubilized in 2.5M guanidine-HCl and then dialyzed in a buffer containing 0.5M guanidine-HCl, it could be immunoprecipitated with the Flag antibody and the dialysis restored its ability to activate IRF3 (Supplementary Figure S2B). Based on these experiments, we devised a protocol to purify the functional Flag-MAVS particles from Sendai virus-infected cells (Fig. 2A and 2B). As a control, we also purified Flag-MAVS from uninfected cells. In both cases, silver staining of the purified particles revealed a predominant band that corresponded to Flag-MAVS itself (Fig. 2B), which was verified by mass spectrometry and immunoblotting (data not shown). Importantly, only Flag-MAVS purified from the virus-infected cells formed aggregates and was capable of activating IRF3 when incubated with cytosolic extracts (Fig. 2C). These results suggest that the active MAVS particles consist predominantly of the MAVS protein itself, which likely forms polymers.
Figure 2. Purification of Active MAVS Particles.
(A) A protocol for purification of active Flag-MAVS particles. (B) Silver staining of Flag-MAVS purified from HEK293T cells infected with Sendai virus or not infected. (C) IRF3 dimerization assay using purified Flag-MAVS shown in (B). MAVS was also analyzed by immunoblotting following SDD-AGE and SDS-PAGE.
Recombinant MAVS Protein Forms Fibrous Polymers That Activate IRF3
To test directly whether MAVS alone could form functional polymers, we attempted to express and purify recombinant MAVS protein in E. coli. Since full-length MAVS containing the C-terminal transmembrane domain (TM) was largely insoluble when expressed in E. coli, we expressed and purified TM-deleted MAVS (MAVSΔTM) from HEK293T cells, and then tested its ability to activate IRF3 in the cytosol (Supplementary Figure S3A & S3B). Interestingly, although the TM domain is absolutely required for MAVS to activate IRF3 and induce IFN in intact cells (Seth et al., 2005), in vitro incubation of MAVSΔTM with cytosolic extracts led to IRF3 dimerization (Fig. S3B, lane 1). This result suggests that the activity of MAVSΔTM is blocked in intact cells by an unknown mechanism, but unleashed in the in vitro assay (see below). We took advantage of this assay to test a panel of MAVS deletion mutants and found that the proline-rich region (residues 103–153) and the C-terminus (residues 460–540) were dispensable for IRF3 activation, whereas the CARD domain was essential (Supplementary Figure S3B). Based on these results, we expressed in E. coli a variant of MAVS lacking TM and proline-rich region as a fusion protein with Sumo, a ubiquitin-like protein known to facilitate expression of fusion partners in soluble forms. We purified this protein, termed Sumo-MAVS, to apparent homogeneity and found that it potently activated IRF3 in the cytosolic extracts (Supplementary Fig. S3C).
Interestingly, when Sumo-MAVS was analyzed by gel filtration on Superdex-200, a fraction of the protein eluted in the void volume, and these high molecular weight forms activated IRF3 when they were incubated with cytosolic extracts (Peak I; Fig. 3A). In contrast, the low molecular weight forms of Sumo-MAVS had no activity (Peak II). Negative-stain electron microscopy of the protein particles showed that Sumo-MAVS in Peak I formed large fiber-like polymers, whereas the protein in Peak II formed much smaller particles with globular shapes (Fig. 3B). When Peak II was stored at 4°C for one or two days, it gradually converted to Peak I, indicating that the low molecular weight forms of Sumo-MAVS spontaneously formed the fibrous polymers (Supplementary Fig. S3D). Removal of the Sumo tag caused the majority of MAVS to elute in Peak I, which was also capable of activating IRF3 (data not shown). We also expressed and purified mouse MAVS lacking the TM domain as a His6-tagged protein. The mouse MAVS protein also formed long fibers and were capable of activating IRF3 in cytosolic extracts (Fig. 3C). The average diameter of the mouse MAVS fibers was smaller than that of the human Sumo-MAVS fibers, presumably because the presence of Sumo rendered the fiber thicker. These results suggest that the ability of MAVS to form fibrous polymers is evolutionally conserved, and is independent of the purification tags.
Figure 3. Recombinant MAVS Forms Fiber-Like Polymers that Convert Endogenous MAVS into Functional Aggregates.
(A) Sumo-MAVS lacking the proline-rich and TM domains was expressed and purified from E. coli as described in Experimental Procedures, then further fractionated by gel filtration on Superdex-200. Each fraction was analyzed by Coomassie blue staining and IRF3 dimerization assay. (B) Two peaks of the Sumo-MAVS protein as shown in (A) were imaged by electron microscopy using negative staining. (C) Mouse MAVS lacking the TM domain was expressed as a His6-tagged protein in E. coli and affinity purified. The purified protein was analyzed by IRF3 dimerization assay and electron microscopy. (D) Peak I and II of Sumo-MAVS as shown in (A) was diluted 5 fold serially as indicated, and then incubated with mitochondria from HEK293T cells (total volume: 10 μl). The mitochondria were solubilized in a buffer containing 1% DDM, and then analyzed by SDD-AGE and SDS-PAGE with a MAVS antibody. (E) The mitochondria incubated with Sumo-MAVS as described in (D) were subsequently incubated with 35S-IRF3 and cytosolic extracts to measure IRF3 dimerization.
MAVS Fibrils Have A Prion-Like Activity That Converts Endogenous MAVS Into Functional Aggregates
A hallmark of prions is their ability to convert endogenous proteins from their native conformations into prion-like fibrils. To test if the MAVS fibrils have the prion-like activity, we incubated the Peak I and Peak II fractions of Sumo-MAVS with mitochondria from HEK293T cells at room temperature for 30 minutes, and then analyzed endogenous MAVS in the mitochondrial extracts by SDD-AGE (Fig. 3D). Significantly, MAVS formed large aggregates after the mitochondria were incubated with Peak I, but not Peak II. Even highly diluted Peak I (~80 ng/ml Sumo-MAVS; Fig. 3D, lane 3), which was not detectable by the MAVS antibody, caused detectable aggregation of endogenous MAVS, suggesting a catalytic mechanism of this conformational conversion, which is reminiscent of prion-like infection. The mitochondria also gained the ability to activate IRF3 after incubation with Peak I, and the activity was detectable with a concentration of Peak I as low as 16 ng/ml (Fig. 3E, lane 6). In contrast, Peak II was unable to activate the mitochondria even at high concentrations. High concentrations of Peak I alone modestly activated IRF3, but this activity was significantly enhanced in the presence of mitochondria (e.g, compare lanes 3 & 4 in Fig. 3E).
The CARD Domain of MAVS Forms Protease-Resistant Fibrils With A Prion-Like Activity
Most prions form fiber-like structures that are resistant to protease digestion (Tuite and Serio, 2010). To determine if the MAVS fibrils are resistant to proteolysis, we fractionated Sumo-MAVS on Superdex-200 and digested Peak I and Peak II with proteinase K (PK). Two prominent PK-resistant fragments appeared when Peak I was digested for 2 hours, whereas such fragments were much less in the Peak II sample (Fig. 4A). The Peak II sample contained a faint band that was relatively resistant to PK and this band was identified as Hsp70 by mass spectrometry (Fig. 4A, lanes 7–10; data not shown). Fractionation of the PK-digested Sumo-MAVS on Superdex-200 led to the separation of two peaks, the first peak eluting in the void volume, similar to Peak I of undigested Sumo-MAVS (Fig. 4B). The second peak from the gel filtration column contained predominantly Hsp70, as determined by mass spectrometry (data not shown). Peak I contained a doublet with molecular weights of ~ 30 kDa. Both bands, designated as PK-MAVS, were excised for mass spectrometry, which identified multiple peptides of SUMO and the N-terminus of MAVS, but none after residue 218 of MAVS (Supplementary Figure S4A). These results suggest that PK-MAVS contains a fragment from Sumo and the N-terminus of MAVS including the entire CARD domain (Figure S4B). Negative-stain electron microscopy of PK-MAVS revealed that it formed long fibers with an average diameter of 12.6 +/− 0.69 nm and an overall shape similar to that of the prion PrP (Fig. 4C; Supplementary Figure S4C).
Figure 4. MAVS CARD Domain Forms Protease-Resistant Prion-Like Fibers That Convert Endogenous MAVS Into Functional Aggregates.
(A) Sumo-MAVS in Peaks I and II from Superdex-200 was digested with proteinase K (PK) for the indicated time at 22°C, then analyzed by Coomassie blue staining. (B) Sumo-MAVS was treated with or without proteinase K for 1 hour and then fractionated on Superdex-200 (2.4 ml column). The fractions were analyzed by Coomassie blue staining. Mass spectrometry showed that the protease-resistant fragments in Peak I contained the N-terminus of MAVS including the intact CARD domain (PK-MAVS; Figure S4A), and that the protease-resistant protein in Peak II was Hsp70. (C) The fraction containing PK-MAVS shown in B (lane 9) and the prion PrP were imaged by electron microscopy using negative staining. (D) PK-MAVS and PrP at the indicated amounts were incubated with the mitochondria from HEK293T cells at 22°C for 30 min (total volume: 10 μl). The mitochondria were subsequently incubated with 35S-IRF3 and cytosolic extracts to measure IRF3 dimerization. Mitochondrial extracts were also analyzed by SDD-AGE and SDS-PAGE using a MAVS antibody, which reacts very weakly with PK-MAVS due to the removal of the epitopes. (E) The CARD domain of MAVS is required for its conversion into aggregated forms by PK-MAVS. Full-length MAVS and MAVSΔCARD were expressed in Mavs−/− MEF cells by retroviral transduction. Mitochondria (P5) from these cells were incubated with or without PK-MAVS for 30 minutes before mitochondrial proteins were separated by sucrose gradient ultracentrifugation. (F) The MAVS CARD domain was expressed in HEK293T cells as a Flag fusion protein and affinity purified. The purified protein was analyzed by silver staining, IRF3 dimerization assay and electron microscopy.
The PK-MAVS fragment was incubated with mitochondria, which were subsequently analyzed for their ability to activate IRF3 and form aggregates (Fig. 4D). Strikingly, following incubation with even highly diluted PK-MAVS (40–200 ng/ml; lanes 4 & 5), the mitochondria gained the ability to activate IRF3. Moreover, endogenous MAVS formed large aggregates as revealed by SDD-AGE. In contrast, neither PrP fibers nor ubiquitin (as a globular protein control) caused MAVS aggregation or IRF3 activation even at much higher concentrations (Fig. 4D, lanes 6–10). PK-MAVS alone did not activate IRF3 even at high concentrations (Fig. 4D, lane 2), indicating that degradation of the C-terminus, which contains binding sites for cytosolic signaling proteins such as TRAF2, TRAF3 and TRAF6 (Seth et al., 2005), abrogated its ability to activate IRF3. Thus, the PK-MAVS fibrils must act through endogenous MAVS to activate IRF3 in the cytoplasm. In support of this notion, mitochondria from cells depleted of MAVS by RNAi were unable to support IRF3 activation by PK-MAVS (Supplementary Figure S4D). Reconstitution of MAVS-deficient MEF cells with full-length MAVS, but not a mutant lacking the CARD domain (MAVS-ΔCARD), supported IRF3 activation by PK-MAVS (Figure S4E). Furthermore, sucrose gradient ultracentrifugation revealed that full-length MAVS, but not MAVS-ΔCARD, formed high molecular weight particles after the mitochondria were in contact with PK-MAVS (Fig. 4E), indicating that the CARD domain of MAVS on the mitochondrial surface is required for its conversion to the active form by PK-MAVS. These results suggest that MAVS activation occurred through a prion-like conformational switch, which was triggered and templated by the PK-MAVS fibrils, likely through interaction between the CARD domains of the infectious agent (PK-MAVS) and that of endogenous MAVS. We estimated that approximately 1 ng of PK-MAVS caused the conversion of 16 ng of endogenous MAVS into functional aggregates within 30 minutes, again suggesting a prion-like catalytic mechanism.
Since PK-MAVS contains the CARD domain as well as other sequences (Supplementary Fig. S4A), we tested whether the CARD domain alone is sufficient to form functional fibrils. We expressed Flag-MAVS CARD-only (residues 1–100) in HEK293T cells and purified it to apparent homogeneity (Figure 4F). This protein alone did not activate IRF3, but its incubation with the mitochondria led to IRF3 activation. Electron microscopy showed that the CARD domain formed long fibers with an average diameter of 8.39 +/− 1.1 nm. This diameter was smaller than that of PK-MAVS fibers, likely because it did not contain the extra N-terminal and C-terminal extension sequences found in PK-MAVS.
Our finding that the CARD domain of MAVS is capable of activating endogenous MAVS on the mitochondrial membrane in vitro is in contrast with our previous reports that the mitochondrial localization of MAVS is essential for its function in vivo (Li et al., 2005; Seth et al., 2005). Consistent with our previous reports, transfection of Flag-MAVS CARD-only into a HEK293T IFNβ-luciferase reporter cell line failed to induce the luciferase reporter or IRF3 dimerization (data not shown). When the MAVS CARD domain was fused to the TM domain, this fusion protein, termed mini-MAVS, strongly induced IFNβ and caused IRF3 dimerization (Fig. S4F). Interestingly, depletion of endogenous MAVS by RNAi abrogated IFNβ induction by mini-MAVS (Fig. S4F, lanes 5 & 6), indicating that mini-MAVS must act through endogenous MAVS to induce IFNβ. Thus, it appeared that endogenous MAVS on the mitochondria were prevented from being activated by the cytosolic MAVS CARD domain in intact cells through an unknown mechanism. Intriguingly, when the MAVS CARD domain is appended to the TM domain, it is highly potent in activating endogenous MAVS and IRF3, suggesting that the mitochondrial localization facilitates MAVS aggregation in cells.
MAVS Aggregates Recruit TRAF2 and TRAF6
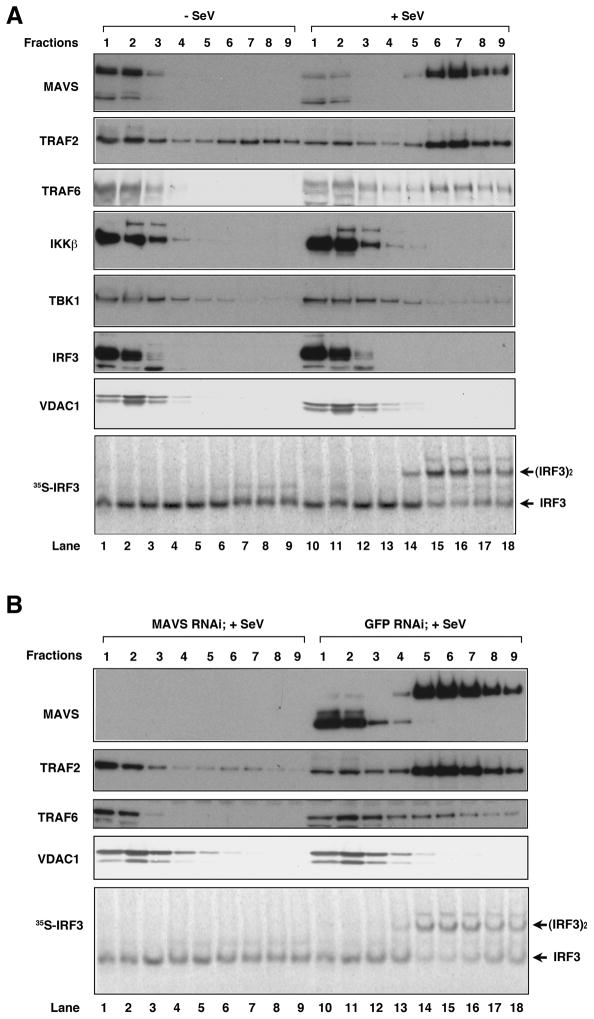
Our observation that mini-MAVS requires endogenous MAVS to induce IFNβ suggests that the sequence between the CARD and TM domains of MAVS, which contain binding sites for TRAFs and other cytosolic signaling proteins, may mediate the recruitment of these proteins to MAVS aggregates. To test this possibility, we examined several signaling proteins known to be involved in NF-κB and IRF3 activation by immunoblotting following sucrose gradient ultracentrifugation of mitochondrial extracts (Figure 5). Interestingly, TRAF2 and TRAF6, but not IKKβ, TBK1 or IRF3, were found to sediment in the high molecular weight fractions together with MAVS in response to Sendai virus infection (Fig. 5A). The shifting of TRAF2 and TRAF6 to the high molecular weight fractions was abrogated in cells depleted of MAVS by RNAi (Fig. 5B). VDAC1, a mitochondrial outer membrane protein, did not co-migrate with MAVS after virus infection, suggesting that virus-induced formation of the MAVS complex does not lead to non-specific aggregation of resident mitochondrial proteins. Further work is needed to understand how the recruitment of TRAF2, TRAF6 and potentially other signaling proteins to MAVS aggregates lead to the activation of NF-κB and IRF3.
Figure 5. TRAF2 and TRAF6 are recruited to MAVS aggregates in response to virus infection.
(A) Crude mitochondria (P5) were isolated from cells infected with Sendai virus or not infected, solubilized in 1% DDM and then separated by sucrose gradient ultracentrifugation. Fractions were analyzed by immunoblotting with the indicated antibodies. Aliquots of the fractions were incubated with cytosolic extracts, 35S-IRF3 and ATP followed by native gel electrophoresis. (B) Similar to (A), except that cells were transfected with siRNA against MAVS or GFP (control) before virus infection.
RIG-I and K63 Polyubiquitin Promote MAVS Aggregation on the Mitochondrial Membrane
We have previously shown that RIG-I binds to K63 polyubiquitin chains through the N-terminal tandem CARD domains and that this binding is essential for IRF3 activation and interferon induction (Zeng et al., 2010). To determine if RIG-I can promote MAVS aggregation in vitro, we incubated full-length RIG-I protein with the mitochondria in the presence or absence of 5′-pppRNA and ubiquitin chains. Strikingly, after RIG-I was incubated with 5′-pppRNA, ATP and K63-Ub4, it caused very rapid formation of MAVS aggregates on the mitochondrial membrane (Fig. 6A). This activity required RNA and K63-Ub4, and was not induced by K48-Ub4 or mono-Ub. Overexpression of the RIG-I N-terminus can activate IRF3 and induce IFN-β independently of viral RNA (Yoneyama et al., 2004). Purified GST-RIG-I(N) also caused robust MAVS aggregation when it was incubated with the mitochondria and K63-Ub4, but not K48-Ub4 or mono-Ub (Fig. 6B). This activity did not require ATP and was unaffected by EDTA, which chelates magnesium (Fig. 6B, lanes 10 & 11). The MAVS aggregates were not observed in cells treated with MAVS siRNA, confirming the identity of these aggregates (Supplementary Figure S5A). Similar to RIG-I(N), overexpression of MDA5(N) in HEK293T cells led to aggregation of endogenous MAVS and dimerization of IRF3, and mutations of two conserved residues within the first CARD domain of MDA5 abrogated its ability to induce IRF3 dimerization and MAVS aggregation (Figure S5B). Titration experiments showed that ~60 nM K63-Ub4 (less for RIG-I[N]:K63-Ub4 complex) was able to convert ~130 nM MAVS into the aggregate forms within 30 minutes (Fig. 6C, lane 8). Kinetic experiments showed that MAVS aggregation was evident after 2 minutes of exposure of the mitochondria to the RIG-I(N):K63-Ub4 complex (Fig. 6D). SDD-AGE analysis showed that the SDS-resistant MAVS aggregates induced by RIG-I(N) and K63-Ub4 were sensitive to DTT treatment (Fig. 6E; top); however, DTT treatment did not affect in vitro activation of MAVS by RIG-I(N) and K63-Ub4 (Fig. 6E, bottom). Furthermore, the DTT-reduced MAVS still sedimented as high molecular weight particles after sucrose gradient ultracentrifugation (Fig. 6F). Thus, the MAVS aggregates induced in vitro by RIG-I and ubiquitin chains behaved similarly to those in cells triggered by viral infection.
Figure 6. RIG-I and K63-Ub4 induce MAVS aggregation and activation on the mitochondrial membrane.
(A) Full-length RIG-I was incubated with 5′-pppRNA and/or ubiquitin chains as indicated at 22°C for 10 min. The mixtures were then incubated with mitochondria from HEK293T cells for 30 min before mitochondrial extracts were analyzed by SDD-AGE and SDS-PAGE. 0.25 μg of ubiquitin chains or mono-Ub was analyzed by silver staining (right panel). (B) GST-RIG-I(N) was incubated with different ubiquitin chains at 22°C for 10 min in the presence or absence of ATP or EDTA as indicated. The mixture was then incubated with mitochondria from HEK293T cells for the indicated time, followed by analysis of the mitochondrial extracts using SDD-AGE and SDS-PAGE. (C) Similar to (B) except that varying amounts of GST-RIG-I(N) and K63-Ub4 were used, and their incubation time with mitochondria were kept at 30 min. (D) GST-RIG-I(N) was incubated with K63-Ub4 and then with mitochondria for the indicated time, followed by analysis of the mitochondrial extracts using SDD-AGE and SDS-PAGE. (E) GST-RIG-I(N) was incubated with K63-Ub4 before incubation with mitochondria in the presence or absence of DTT. The mitochondrial extracts were analyzed by SDD-AGE, SDS-PAGE and IRF3 dimerization assays. Mitochondria from Sendai virus-infected cells were used as a positive control. (F) Mitochondrial extracts from (E) were fractionated by sucrose gradient ultracentrifugation followed by SDS-PAGE and immunoblotting with a MAVS antibody.
DISCUSSION
We have previously shown that MAVS becomes more resistant to extraction with detergent from the mitochondrial membrane after viral infection (Seth et al., 2005). Recent microscopy studies show that MAVS redistributes in the mitochondria to form speckle-like aggregates in cells in response to viral infection (Onoguchi et al., 2010). In this report, we show that viral infection induces the formation of very large MAVS aggregates on the mitochondrial membrane. Importantly, we provide direct biochemical evidence that these aggregates are highly potent in activating IRF3 in cytosolic extracts. Furthermore, the aggregation of MAVS could be robustly induced in vitro by incubation of mitochondria with RIG-I and K63 ubiquitin chains. Most remarkably, our new data reveal that the CARD domains of MAVS form protease-resistant prion-like fibrils, which effectively convert endogenous MAVS on the mitochondria into functional aggregates. Based on these results and other published data, we propose a model of MAVS activation that involves the following steps (Figure 7): 1) RIG-I binds to viral RNA through the C-terminal RD domain and the helicase domain; 2) RIG-I hydrolyzes ATP, undergoes a conformational change and forms a dimer that exposes the N-terminal CARD domains; 3) the CARD domains recruit TRIM25 and other ubiquitination enzymes to synthesize unanchored K63 polyubiquitin chains, which bind to the CARD domains; 4) the ubiquitin-bound CARD domains of RIG-I interact with the CARD domain of MAVS, which is anchored to the mitochondrial outer membrane through its C-terminal TM domain; 5) the CARD domain of MAVS rapidly forms prion-like aggregates, which convert other MAVS molecules into aggregates in a highly processive manner; 6) the large MAVS aggregates interact with cytosolic signaling proteins, such as TRAFs, resulting in the activation of IKK and TBK1.
Figure 7. A model of MAVS activation involving a prion-like conformational switch induced by RIG-I.
Following sequential binding of RIG-I to viral RNA (5′-ppp RNA) and K63 ubiquitin chains, the CARD domains of RIG-I interact with the CARD domain of MAVS. This interaction induces a conformational change of the MAVS CARD (depicted by the change in color and shape of the CARD), which in turn converts other MAVS on the mitochondrial outer membrane into prion-like aggregates. These aggregates activate cytosolic signaling cascades to turn on NF-κB and IRF3, leading to induction of type-I interferons and other antiviral molecules. MAVS on the mitochondrial membrane of uninfected cells may be a multimer but is depicted as a monomer for simplicity. Also not depicted is the possibility that MAVS on the surface of two adjacent mitochondria could aggregate through the N-terminal CARD domains, thereby propagating the antiviral signal between mitochondria.
Prions are self-propagating protein aggregates best known for causing fatal neurodegenerative diseases (Prusiner, 1998). However, accumulating evidence through studies in fungi and other organisms suggests that prion-catalyzed conformational switches can regulate phenotypes in a way that is not detrimental, and in some cases beneficial, to a cell or organism (Halfmann and Lindquist, 2010; Tuite and Serio, 2010). A recent example of beneficial prions is provided by the invertebrate Aplysia translation regulator CPEB, which forms self-sustaining polymers that contribute to long-term facilitation in sensory neurons (Si et al., 2010; Si et al., 2003). Our finding that MAVS forms highly active, self-perpetuating fiber-like polymers provides another example of beneficial prions, in this case regulating mammalian antiviral immune defense.
MAVS shares many hallmarks of a prion, including: a) the ability to infect the endogenous protein and convert it into the aggregate forms; b) the formation of fiber-like polymers; c) resistance to protease digestion; d) resistance to detergent solubilization. Surprisingly, although endogenous MAVS aggregates from virus-stimulated cells were resistant to 2% SDS as analyzed by SDD-AGE, these aggregates were sensitive to treatment with reducing agents such as DTT, suggesting disulfide bond formation within functional MAVS aggregates. Interestingly, disulfide bond formation has also been found in some prions such as PrP (Stanker et al., 2010). However, even after DTT treatment, MAVS still sediments as very large and active particles after sucrose gradient ultracentrifugation, suggesting that disulfide bond formation is not essential to maintain the aggregation and activity of MAVS.
It remains to be determined whether MAVS forms one or a few very large aggregates, or the aggregates are broken down to smaller fragments, which then form new “seeds” to multiply the aggregates. It would also be interesting to investigate how cells resolve these mitochondrial aggregates after an immune response is called into motion. Although there is evidence that MAVS is degraded by the ubiquitin-proteasome pathway (You et al., 2009), other mechanisms such as mitophagy or chaperone-mediated refolding are potentially involved in clearing the MAVS aggregates. Interestingly, we found that geldanamycin and its analog 17-AAG, which was previously known to inhibit IRF3 activation by RNA viruses, block MAVS aggregation. The dose response of the drugs shows an excellent correlation between MAVS aggregation and IRF3 dimerization, suggesting that MAVS aggregation is required for its function. It remains to be determined whether the effect of geldanamycin is due to its inhibition of Hsp90. It is possible that Hsp90 facilitates ordered assembly of the functional MAVS fibers by preventing non-specific aggregation.
Many prions form amyloids consisting predominantly of β-sheets that may be detected with dyes such as Congo red (Chien et al., 2004; Sawaya et al., 2007). However, we have been unable to observe staining of MAVS aggregates with Congo red (data not shown). Like the CARD domains of other proteins, MAVS CARD forms a six-helix bundle (Potter et al., 2008). Likewise, other priongenic proteins, such as the native form of PrP (PrPc), form alpha-helical folds before they are converted to the aggregate forms (Chien et al., 2004). Further studies are required to determine the atomic structure of the MAVS fibers and to understand how the fiber structure gains the competence to initiate downstream signaling. Importantly, MAVS fibers, but not PrP fibers, are able to induce endogenous MAVS aggregation, indicating specificity in this conformation-based mechanism of cell signaling.
CARD and CARD-like domains are present in a large variety of proteins, especially those involved in immune defense and cell death (Park et al., 2007). CARD domains are well known to mediate protein-protein interactions, and the CARD domains of RIG-I and MAVS likely mediate the interaction between these proteins. Surprisingly, our studies reveal that the CARD domains of RIG-I and MAVS have additional unique functions. The tandem CARD domains of RIG-I, but not the MAVS CARD, bind specifically to K63 polyubiquitin chains (Zeng et al., 2010). On the other hand, the CARD domain of MAVS, but not those of RIG-I, can form prion-like aggregates. The primary sequences of the CARD domains of RIG-I, MDA5 and MAVS are distantly related to conventional CARD domains found in other proteins. Interestingly, while the CARD domain of MAVS shares very limited sequence homology with those of RIG-I and MDA5, the CARD domains of MAVS from different species have high degrees of sequence homology, and both mouse and human MAVS can form prion-like functional fibers (Fig. 3). Thus, the MAVS CARD domain may have evolved to acquire the propensity to form prion-like aggregates, which obviously benefit the host organisms by mounting rigorous antiviral immune defense.
In cells, the CARD domain of MAVS must be appended to the mitochondrial targeting domain (TM) in order to induce IRF3 activation (Seth et al., 2005). In fact, overexpression of mini-MAVS that contains only the CARD and TM domains is sufficient to activate IRF3 and induce IFNβ in cells (Seth et al., 2005); Figure S4F). The importance of the mitochondrial localization of MAVS is underscored by the fact that hepatitis C virus employs the viral protease NS3/4A to cleave MAVS off the mitochondrial membrane, thereby suppressing type-I interferon production (Li et al., 2005; Meylan et al., 2005). Surprisingly, we found that recombinant MAVS lacking the TM domain (MAVSΔTM) could activate IRF3 when it is incubated with cytosolic extracts. Even a fragment containing only the CARD domain of MAVS is sufficient to form aggregates in vitro. The CARD domain aggregates can also activate IRF3 in the cytosol, but in this case the activity requires intact mitochondria containing endogenous MAVS (Fig. 4D–F). These biochemical results are consistent with our new finding that the induction of IFNβ by mini-MAVS in cells requires endogenous MAVS (Supplementary Fig. S4F). Taken together, our results suggest that the CARD domain of MAVS mediates aggregation, whereas the intervening sequence between CARD and TM domains is important for recruiting cytosolic signaling proteins to activate IKK and TBK1.
While the vast majority of MAVS is located on the mitochondrial membrane, a very small fraction of MAVS is located on the peroxisomal membrane (Dixit et al., 2010). When MAVS was engineered to express predominantly on peroxisomal membrane, it failed to induce type-I interferons, but could still induce some antiviral genes such as viperin to inhibit viral infection through an interferon-independent mechanism. Our crude mitochondrial preparation likely contains peroxisomes, raising the interesting possibility that a small fraction of MAVS that is located on the peroxisomal membrane may also form aggregates to induce viperin and other antiviral molecules.
Although overexpression of MAVS in cells is sufficient to cause its aggregation and induce type-I interferons, the aggregation and activation of endogenous MAVS is tightly regulated by viral infection. We found that viral infection causes almost complete conversion of endogenous full length MAVS into the aggregate forms. Such a highly efficient aggregation of MAVS can be reproduced in vitro by a simple incubation of mitochondria, RIG-I CARD domains and K63-Ub4. Moreover, endogenous MAVS rapidly aggregates upon exposure of the mitochondria to the fibers consisting of MAVS CARD domain. These results suggest an amplification cascade in which the RIG-I:Ub chain complex causes some MAVS molecules to form aggregates, which then function as prion-like “seeds” to convert other MAVS molecules to form aggregates. Indeed, we found that sub-stoichiometric amounts of K63-Ub4 and the MAVS CARD fibrils could cause almost complete conversion of endogenous MAVS into functional aggregates within 30 minutes in vitro, suggesting that the RIG-I:Ub chain complex and MAVS fibrils function like catalysts. This is consistent with our previous estimate that less than 20 molecules of viral RNA and K63 ubiquitin chains in a cell are sufficient to cause detectable IRF3 activation (Zeng et al., 2010). Thus, the RIG-I pathway appears to be highly sensitive to viral infection. Our finding of the prion-like conformational switch of MAVS provides a mechanism underlying this ultrasensitive and robust antiviral response.
EXPERIMENTAL PROCEDURES
Purification of Functional Flag-MAVS Particles from Virus-infected Cells
HEK293T cells stably expressing Flag-MAVS (human) were infected with Sendai virus for 14 hours, then lysed in Buffer A by repeated douncing. After differential centrifugation as described above, mitochondria were further purified by sucrose density ultracentrifugation. Briefly, mitochondria resuspended in Buffer B (20mM HEPES, pH7.4, 10% Glycerol, 0.5mM EGTA, 0.25 M D-mannitol) were loaded on top of a centrifuge tube containing 1 ml of 50% sucrose in phosphate buffered saline (PBS) on the bottom layer and 1 ml of 40% sucrose in PBS on the top layer. After centrifugation at 100,000 × g for 30 minutes, mitochondria enriched at the interface of two layers were collected and solubilized with PBS containing 1% DDM. The mitochondrial lysate was loaded onto a sucrose gradient (30%–60%) and centrifuged at 170,000 × g for 2 hours. Nine fractions with equal volume were taken from the top to bottom of the tube. Fractions containing MAVS (corresponding to about 50–60% sucrose) were pooled and then guanidine-HCl was added to 2.5M. The mixture was dialyzed against PBS containing 0.5M Guanidine-HCl and 0.2% DDM overnight. Flag-MAVS was purified from the dialyzed mixture using anti-Flag (M2) agarose beads and eluted with the Flag-peptide (200 μg/ml). All procedures were performed at 4°C.
Purification of Flag-MAVS from uninfected cells was carried out as above except that after isolation of mitochondria by sucrose gradient ultracentrifugation, the mitochondrial lysate was loaded on top of 40% sucrose cushion and centrifuged at 170,000 × g for 2 hours. The fraction above the sucrose cushion was incubated with anti-Flag (M2) agarose beads, followed by elution of Flag-MAVS with the Flag peptide.
Expression, Purification, and Protease Digestion of Recombinant MAVS Proteins
The bacterial expression vector pET-28a-His6-Sumo-MAVS or pET14b-mMAVS was transformed into BL-21(pLys). Protein expression was induced with 0.2 mM IPTG at 18°C for 4 hours. After sonication in Buffer C (10mM Tris-HCl, pH8.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 0.5 mM DTT, 5% glycerol, 0.5mM PMSF, 0.5mM AEBSF, 10mM imidazole), cell lysates were centrifuged at 50,000 × g for 30 minutes. His6-Sumo-MAVS and His6-mMAVS in the supernatant were purified using Ni-NTA beads, loaded onto a Hitrap-Q column and then eluted with Buffer D (10mM Tris pH7.5, 5% glycerol, 2mM DTT, 1mM EDTA, 0.5mM PMSF, 0.5mM AEBSF) containing a gradient of NaCl from 0.1 M to 0.5M. The fractions containing His6-Sumo-MAVS, which was eluted with approximately 300 mM NaCl, were pooled and loaded onto a Superdex-200 gel filtration column equilibrated with Buffer E (10mM Tris-HCl, pH 8.0, 150mM NaCl, 1mM DTT, 1mM EDTA, 0.5mM PMSF, 0.5mM AEBSF). FPLC (GE Healthcare) and a 24 ml Superdex-200 were used for large scale purification, whereas a SMART or ETTAN purification system (GE Healthcare) and a 2.4 ml Superdex-200 column were used for small scale purification.
Purified His6-Sumo-MAVS was digested with proteinase K (mass ratio 50:1) at room temperature (~22°C) for the indicated time. To isolate the protease-resistant fragments, the reaction mixture was fractionated on Superdex-200 using the ETTAN system.
Flag-MAVS containing only the CARD domain was expressed in HEK293T cells by transient transfection of pcDNA3-Flag-MAVS-CARD. 48 hours after transfection, cells were lysed in Buffer F (20mM Tris-HCl, pH 8.0, 150mM NaCl, 10% glycerol, 0.1% Triton X-100, 1mM DTT and protease inhibitors). After centrifugation at 10, 000 × g for 10 min, Flag-MAVS CARD was absorbed on anti-Flag agarose beads and eluted with the Flag peptide. The eluate was further fractioned on Superdex-200 using the ETTAN system.
In vitro IRF3 Activation Assay
Crude mitochondria (P5) and cytosolic extracts (S5) were prepared by differential centrifugation. Briefly, HEK293T cells were resuspended in Buffer A (10mM Tris-HCl, pH7.5, 10mM KCl, 1.5mM MgCl2, 0.25M D-mannitol, and Roche EDTA-free protease inhibitor cocktail) and then lysed by repeated douncing. After removing the cell debris by centrifugation at 1000 × g for 5 minutes, the supernatants were centrifuged at 10,000 × g for 10 minutes at 4°C to separate P5 and S5. 35S-IRF3 was synthesized by in vitro translation in rabbit reticulocyte lysates (TNT; Promega) using pcDNA3-Flag-IRF3 as the template. P5 and S5 were incubated with 35S-IRF3 and ATP, then IRF3 dimerization was analyzed by native gel electrophoresis as described (Zeng et al., 2009).
Semi-Denaturing Detergent Agarose Gel Electrophoresis (SDD-AGE)
SDD-AGE was performed according to a published protocol with minor modifications (Alberti et al., 2009). Briefly, Crude mitochondria (P5) were resuspended in 1 × sample buffer (0.5x TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue), and loaded onto a vertical 1.5% agarose gel (Bio-Rad). After electrophoresis in the running buffer (1 × TBE and 0.1% SDS) for 35 minutes with a constant voltage of 100V at 4°C, the proteins were transferred to Immobilon membrane (Millipore) for immunoblotting.
Induction of Endogenous MAVS Aggregation and Activation in vitro
Crude mitochondria (P5) were isolated from HEK293T cells, and incubated with indicated amounts of RIG-I, ubiquitin chains, or recombinant MAVS proteins at 22°C for different lengths of time. The reaction mixtures were centrifuged at 10, 000 × g for 10 minutes, then the pellets were analyzed by SDD-AGE and IRF3 dimerization assays. For ultracentrifugation analyses, the pellets were resuspended in PBS containing 1%DDM, treated with or without 10 mM DTT for 30 minutes, and then loaded on top of a sucrose gradient (20%–60%) and centrifuged at 170,000 × g for 2 hours.
Supplementary Material
Acknowledgments
We thank Xiaomo Jiang for the MDA5 plasmids. This work was supported by grants from NIH (RO1-GM63692) and the Welch Foundation (I-1389). H.Z. was supported by a Welch Foundation grant (I-1684) and partially by a EUREKA grant from NIH (R01-GM88745 to Q.-X.J.). B. S is supported by an NIH pre-doctoral training grant (GM007062). Z. J. C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and five figures and can be found with this article online at http://www.cell.com.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic Acid Recognition by the Innate Immune System. Annu Rev Immunol. 2010 doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, Okabe Y, Namiki H, Fujita T. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Tenoever BR, Maniatis T. Connecting mitochondria and innate immunity. Cell. 2005;122:645–647. doi: 10.1016/j.cell.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Julkunen I, Namiki H, Yoneyama M, Fujita T. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282:22816–22822. doi: 10.1074/jbc.M703019200. [DOI] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol. 2008;8:11. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Stanker LH, Serban AV, Cleveland E, Hnasko R, Lemus A, Safar J, DeArmond SJ, Prusiner SB. Conformation-dependent high-affinity monoclonal antibodies to prion proteins. J Immunol. 2010;185:729–737. doi: 10.4049/jimmunol.0902930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fujita T. Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res. 2002;22:73–76. doi: 10.1089/107999002753452674. [DOI] [PubMed] [Google Scholar]
- You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.