Abstract
Magnetic resonance imaging techniques have literally revolutionized neuroimaging with an unprecedented ability to explore tissue structure and function. Over the last three decades, the sensitivity and array of imaging techniques available have improved providing ever finer structural information and more sensitive functional techniques. Among these methods, diffusion imaging techniques have facilitated the generation of fiber-tract maps of the brain enabling an examination of issues related to brain structure and neural connectivity. Despite the potential utility of the techniques described, validation has not yet been achieved on biological samples.
Recently, using newly developed surface microcoils on small samples at high magnetic fields, we demonstrated the ability of MR microscopy to image individual neurons in mammalian brain tissue. In the present work, we combine MR microscopy with the highest resolution (15µm) fiber tracking yet reported and demonstrate the accuracy of the fiber tract maps with direct histological validation. Thus it becomes possible to delineate fiber structure in tissues at the cellular level. A semi-quantitative approach was used to estimate the cell overlap fraction (cOF) and fiber tract overlap fraction (tOF), with cOF’s of 94, 92 and 100%, and tOF’s of 84, 86 and 100%, in rat cervical, rat lumbar, and pig spinal cord tissue, respectively. These methods provide a way to directly validate fiber tracking techniques with histology so that contemporary tracking techniques may be compared and refined using the microstructural details of a biological template as a ground truth.
Introduction
Magnetic Resonance Imaging (MRI) has matured over the last three decades into a leading diagnostic imaging technique, and is now the modality of choice for many studies. This is especially true for neurological applications as MRI offers a non-invasive means of observing structures of the central nervous system encased in the skull and spinal column. Key to the success of MRI has been a growing array of techniques and capabilities offering differing structural, mechanical and functional information in living tissues. For example, different mechanisms (the relaxation times T1 and T2, diffusion) facilitate varied image contrast, flow may be detected to generate angiograms, rapid imaging techniques may visualize cardiac motion in real time, and over the last decade functional MRI (fMRI) has enabled the visualization of brain activity and is revolutionizing the fields of cognitive and functional neuroscience (1).
Over the last two decades, diffusion imaging techniques have also risen to prominence enabled by improvements in both hardware and technology. Images sensitized to the self-diffusion of water first showed clinical potential for detecting ischemic brain tissue (2). In this case, image contrast arises because water diffusion is restricted by the tissue’s constituents to varying degrees. Techniques were then developed for sensitizing the MR water diffusion signal in such a way as to determine the magnitude of the diffusion along different spatial axes, thus enabling the determination of anisotropic diffusion in tissues. When applied to nervous tissue, this technique, in combination with a tracking algorithm, is capable of producing synthetic fiber constructs intended to predict the spatial and orientational characteristics of white-matter tracts contained within the tissue under investigation (3, 4). These maps offer a wealth of opportunity for examining brain structure and connectivity, detecting and monitoring damage or diseases of the nervous system, and aiding in planning surgical interventions to minimize nerve damage (5).
To our knowledge, and as discussed in a recent review (6), direct histological validation of fiber tracking has not yet been achieved and is a pressing issue. Further, the inherently low SNR (signal-to-noise ratio) of MR has limited most diffusion tractography to relatively low spatial resolutions so that only the largest of fiber bundles can be observed while retaining predictive accuracy; however, previous studies achieved resolutions in the 100–150 micron range (7–11) by employing a small detector coil at high field strength, which together and dictate the resolution and imaging time achievable with MR. Thus, the highest resolutions are achieved at high magnetic fields on small samples, or alternatively small portions of larger samples, through the use of local radio frequency (RF) coils. Several groups have performed tractography studies at so-called ‘microscopic’ resolutions broadly accepted as less than 100 micrometers (12). To our knowledge, the highest resolutions achieved on fixed, isolated rat (13) and human (14) hippocampi were taken at in-plane resolutions of 50 and 60 micrometers, respectively. Isolated mouse hippocampus has been imaged at 40×80×80 micrometers (15). At these resolutions, DTI is sometimes referred to as diffusion tensor microscopy (DTM), a term first coined in 1999 (16, 17).
In a recent study, we used prototype surface microsurface coils on excised pieces of rat spinal cord at 14.1 Tesla to improve SNR and achieve spatial resolutions of 5–15 micrometers in diffusion weighted images (18). At this level of resolution, cell bodies of individual mammalian neurons were visualized for the first time. Additionally, the sample employed—a spinal cord cross-section approximately 3mm × 3mm × 25µm—could then be examined and imaged via light microscopy thus providing a histological template with which to correlate structures visualized in the MR images.
In the present study, we build upon previous microimaging experiments and apply diffusion tensor microscopy at higher resolutions (15µm) than previously reported, and with direct histological correlation. In this way, direct correlation of the MR-derived, synthetic fiber tracts and histology is possible, thus using histology as a structural ground truth for evaluation of tractography techniques. Additionally, at this level of resolution the organization of nerve fibers with respect to the positions of individual neurons becomes possible, opening up the feasibility of addressing cellular connectivity issues.
Methods
All imaging was performed on a 600MHz Bruker imaging spectrometer interfaced with surface microcoils developed by Bruker Instruments Inc. (19, 20). In these studies, four-turn, circular coils 500 micrometers in diameter were employed.
Rat and pig spinal cords were exposed by spinal laminectomy and tissue from both cervical and lumbar enlargements removed by gross dissection. Transverse slices of perfusion-fixed (rat) or immersion-fixed (pig) cord (4% formaldehyde) were sectioned in increments ranging from 25µm to 300µm using a vibratome and washed overnight in phosphate buffered saline (PBS) as described previously (18). Portions of the ventral horn were excised and a single tissue slice for each imaging study was selected containing alpha-motor neurons as well as the boundary between gray and white matter, and placed directly over the coil face with the aid of a dissecting microscope. Due to the fact that the sample well depth (500µm) exceeded the depth of the tissue slice (25µm to 100µm), a tissue retention device was employed to prevent shifting and ensure that the structures of interest stayed within the coil’s excitation profile. The separate pieces of this device which consisted of a nylon mesh insert and nylon retention ring were soaked in PBS overnight prior to use in an attempt to eliminate air bubbles which otherwise tended to become trapped on their surface. After securing the sample in place and adding additional PBS, the tissue well was sealed with adhesive polymerase chain reaction (PCR) film. Heavy diffusion weighting—such as that employed in the scans of the current study—eliminates free-water signal originating from PBS surrounding the tissue slice. As such, the effective slice thickness reported for the imaging is the physical thickness of the tissue slice itself. This method allows for accurate correlation with histology as demonstrated previously (18).
HARDI diffusion tensor datasets (21 directions uniformly distributed on the sphere) of rat cord (TR = 500ms, TE = 36ms, Δ= 17ms, δ= 6ms, resolution = 15.6µm in-plane, effective slice thickness = 25µm, b = 3750 s1mm−2, the number of averages at b=0 was 60, the number of averages at b = 3750 was 30, scan time = 49h) and pig cord (effective slice thickness = 100µm, number of averages at b(0) = 20, number of averages at b(1800) = 10, scan time = 16h 25min) were also collected. MR scan parameters not listed for the pig dataset were unchanged from those listed in the rat experiments.
Following MR microscopy (MRM) analysis, tissue sections were immersed in Nissl stain (0.5% cresyl violet acetate, 0.3% glacial acetic acid, 99.2% ddH2O) for two to four minutes. This stain was chosen because of the need to identify cell body positions in the histology for the purpose of performing image registration. After staining, slices were placed in a destaining bath (0.3% glacial acetic acid, 99.7% ddH2O) for approximately one minute before being placed in PBS (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, and 1.8mM KH2PO4: pH 7.4 at an osmotic strength of 300mOsm). Slices were then wet-mounted to precleaned, glass microscopy slides using Histomount (National Diagnostics, HS-103) mounting medium. Lastly, histological images were attained at 200X magnification using a digital camera (QImaging, Retiga 4000R Fast 1394 Color) attached to a microscope (Axioplan 2, Zeiss) and processed with software (QCapture Pro 6.0) available from QImaging.
Following a noise regularization step, the diagonalized diffusion tensor and primary eigenvector describing the preferred axis of diffusion was determined with Matlab (The MathWorks Inc.) using non-linear least squares regression. The apparent diffusion coefficient (ADC) was determined as one third of the trace of the diffusion tensor and the full b-value matrix was employed. Diffusion tensor tractography was performed using the DTI tools software package (DTI Tools, Freiburg University Hospital, Germany) which employs the FACT algorithm (21). Trackings were performed across the entire FOV in order to show all predicted structure present in the data as opposed to methods which track only selected fibers. All presented MR/histology overlays were produced using image registration that involved combinations of rotation, translation and uniform scaling only (e.g. 'similarity' transformation in Matlab R2008a, image processing toolbox). The positions of alpha-motor neurons in the diffusion weighted images (see figure 1B and C) and/or the mean diffusivity (ADC) map were used to align the histology and the structural information extracted from the MR DTI data, i.e. in-plane primary diffusion directions (in-plane coordinates of the primary eigenvector) and tractography results. In this manner, the co-registration was based on directly identifiable landmarks present in both MR data and the histology. Therefore only those cells were picked out that are identifiable in both imaging techniques. A semi-quantitative measure of the registration was then generated by calculating how many of the cell positions that align compared to the total number of cells used in the registration. In this study, this is referred to as the cell overlap fraction (cOF) based on an overlap score used by Anbeek et al. (22) and Dyrby et al. (23). Similarly, the quality of the tractography was assessed by counting, in the histology, the number of visible fibers inside the coil FOV. Fiber counting was limited to white matter regions where the fibers are well defined on histology. The fibers were grouped into minor and major tracts, and a collected overlap fraction was calculated i.e. number of tracts successfully traced by the algorithm divided by total number of visible tracts. The grouping allows us to introduce a penalty system: If the tractography does not identify a major tract it counts as −1 in the sum of identified tracts, a non-identified minor tract counts as zero. We refer to this measure as the tract overlap fraction, or tOF.
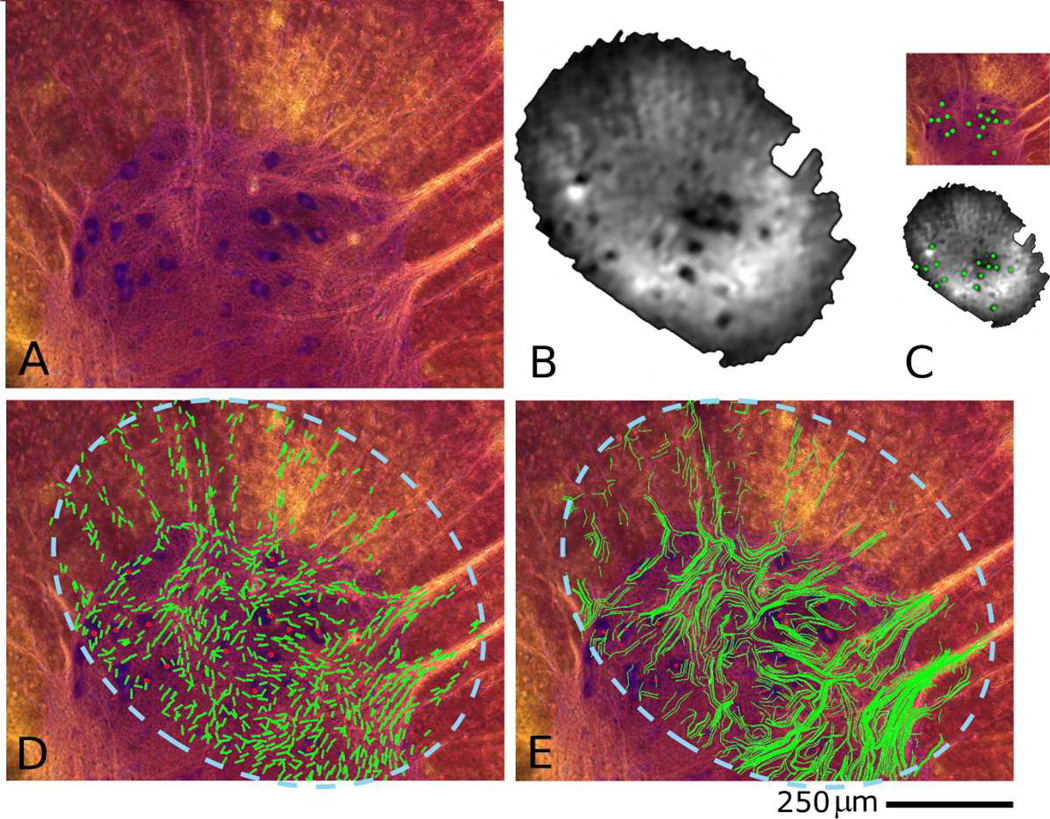
Figure 1.
Nissl stained histology section (A) and diffusion weighted image (B) of a transverse spinal cord section from the cervical enlargement of a rat. Corresponding cell clusters can be identified as marked in (C) with green dots. A 21-direction HARDI data set was acquired and processed to generate a diffusion orientation map as shown in (D) and overlaid on the histology. The dotted blue line illustrates the FOV of the RF coil represented by (B). (E) The resultant diffusion tensor tractography data (green tracts) calculated using the FACT algorithm also overlaying the histology. The data in (D) and (E) were co-registered using the spatial coordinates of cell bodies (red dots). Scale bar applies for histology images (A, D and E) only.
Results
Figure 1 shows an example data set illustrating the cell localization to align the histology and MRI. The diffusion weighted image highlights the cell bodies which can then be seen to correspond to the positions of the cells in the histology. On the same sample, Figure 1 then shows the diffusion tensor data set with in-plane components of the diffusion eigenvectors as well as the calculated tractography. Primary eigenvectors whose in-plane length was less than 60% of their total length were removed from the orientation map for the purpose of visual clarity. Note especially the alignment of the diffusion-predicted tracts over the white-matter tracts visible in the histology, and the correlation between the histologically and MR determined cell positions. The coregistration of these data sets was based on 16 cell body positions identified in both MR images and histology. Inspection of the registration gives a cellular overlap fraction (cOF) of 15/16 = 94%. Our selection of visible white-matter tracts and their grouping for assessing the fraction identified by the tracking algorithm is illustrated in Figure 2. Inspection of the tractography overlay in Figure 1 then shows that the tOF measure for this registration was 2/2 major tracts and 14/17 minor tracts or 16/19 = 84% in total.
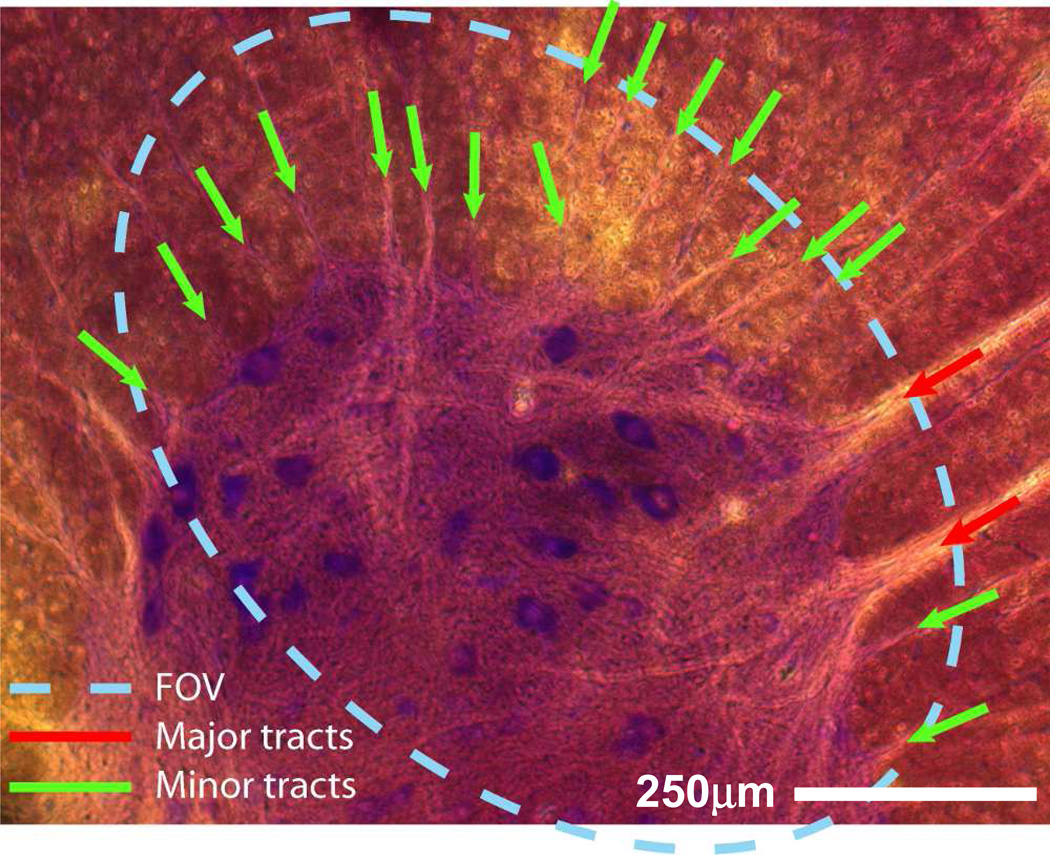
Figure 2.
In order to measure the quality of the tractography using the histology as a ground truth, we have identified white-matter tracts in the coil’s field-of-view (FOV) that we would expect the tracking algorithm to predict accurately. We have limited this selection to the fibers in the white matter which are clearly defined in the histology. The bundles are divided into major and minor tracts as shown in the figure: there are 2 major tracts and 17 minor tracts inside the FOV in this case.
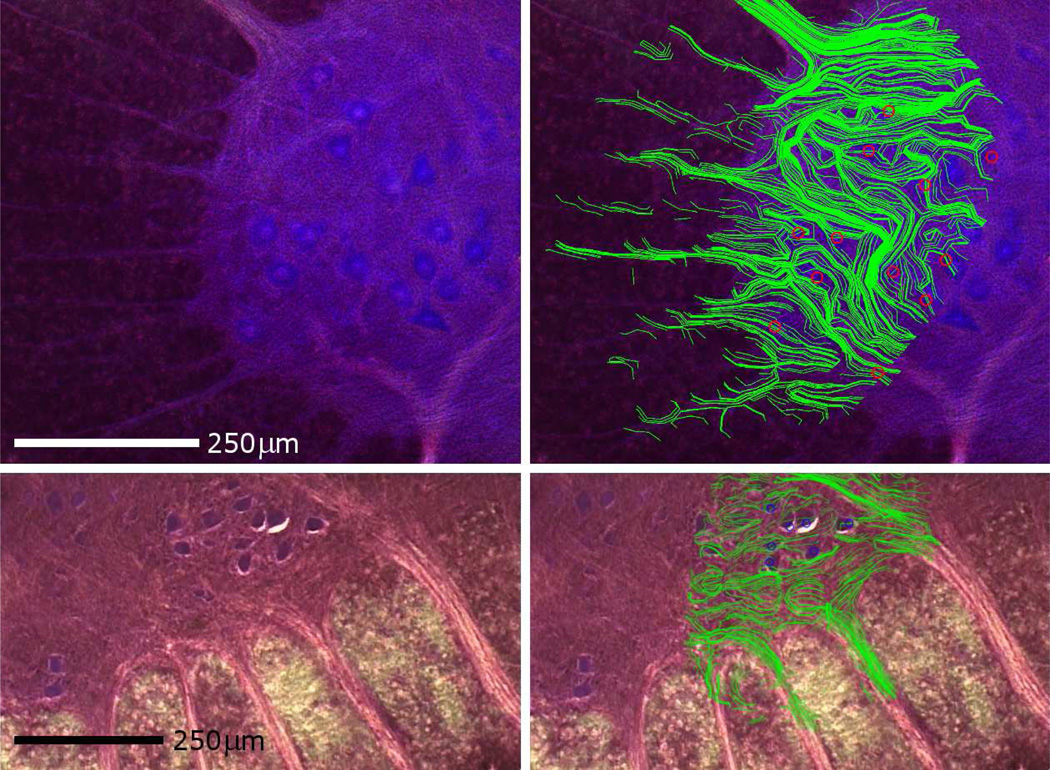
In figure 3 the top row offers a similar example performed in rat spinal cord tissue from the lumbar enlargement again illustrating a clear correlation between histology and tractography. For this registration cOF = 11/12 = 92%, and tOF = 12/14 = 86% based on 1 major tracts and 13 minor tracts. In the same figure, the bottom row shows the same techniques applied to a 100µm thick slice of pig spinal cord. The registration quality was excellent for this sample with a cOF and tOF of 100%.
Figure 3.
Top row: Nissl-stained false color histology of a fixed, transverse section from the lumbar enlargement of a rat spinal cord (left) then overlaid with fiber tract maps (right) generated as described in Figure 1. The locations of alpha-motor neurons (blue) match well (cOF = 11/12) with cell positions determined form the MR images (red circles). The tractography map shows clear delineation of axonal tracts as evidenced by the correlative overlay in the top right panel (tOF = 12/14, based on 1 major and 13 minor tracts). Bottom row: Histology (left) with overlaid fiber tracks (right) from a section of pig spinal cord tissue (300µm thick). cOF = 1 for this data set. Structures within the pig spinal cord are morphologically similar to those found in human tissue and thus represent a closer tissue approximation than can be attained from rat samples. The fiber tracks correlate well between the histology and fiber tract map with a tOF = 1 based on four major bundles. The distortion in tractography on the lower left is due to an air bubble next to the tissue sample during the MR microscopy.
Discussion
There are now a plethora of diffusion imaging techniques and tracking algorithms for visualizing fiber tracts with various advantages and disadvantages. Foremost in these issues are the data acquisition time (which can become lengthy and restrictive for in vivo studies), spatial resolution, and the ability to distinguish crossing fibers. With this large range of techniques and issues, validation and comparisons of the various techniques is a pressing issue and quoting Hubbard and Parker (6); “The validation of tractography is fundamental to the implementation of the technique as a useful biomedical tool.”
Summarizing Hubbard and Parkers book chapter (6), several approaches which rely on foreknowledge of the constructs under investigation have been used in the attempt to link tensor measurements employed in tractographic analysis with fiber morphology. These include software phantoms (computer simulations) (23–25), physical phantoms (i.e. arrays of microtubes (26), polymer fibers (27), or other biological structures composed of fibers such as muscle tissue (28, 29), spinal cord (30), optic tracts (31), or peripheral nerves), or in vivo and in vitro animal and human studies. For the in vivo and in vitro studies, tracts are validated against surgical dissection, invasive tracers (non-MR) and MR visible tracers (23), prior anatomical knowledge (32, 33), or circumstantially; i.e. using evidence from alternative techniques such as fMRI and PET, and by inferences from lesion studies (34). Further, there have been studies linking the fractional anisotropy measured in DTI to histologically derived measures of fiber orientation in humans (35) and animal models (36–38).
In this work, we demonstrate for the first time the ability of MR techniques to generate tractography maps at microscopic resolutions with direct histological correlation on isolated fixed tissue. We accomplished this by using the locations of individual neuronal cell bodies visible in both datasets to co-register MR and light microscopy images. As demonstrated above, our method produces robust correlation between MR data and histology using simple linear registration of the images, which is very promising if the method is to be used for comparing the precision of various tractography methods. We note that as in many previous studies, our diffusion data is from a single slice of tissue and thus spatially two-dimensional. However there is sufficient in-plane continuity of the white-matter tracts in these samples that fibers may be followed over extensive lengths as evident in the images, and thus tractography approaches may be validated. Three dimensional tractography will require multislice MRI on thicker tissue slices, true three dimensional imaging techniques, or similar separate data acquisitions on adjacent tissue slices that can then be combined. Since our long term goal is to perform high-resolution DTM on live tissue, requiring relatively thin slices of tissue (>500µm) so that the tissue is well perfused, the latter option appears most viable. To improve efficiency in that regard, several tissue slices may be imaged simultaneously with a multislice perfusion chamber as demonstrated in a previous study (39).
Although the rat studies exhibit a high level of quantitative correlation between MR and histology data, the pig study herein shows a perfect correlation. This is attributable to the overall larger size of the tissue structures in the pig sample, which are more easily visualized at these spatial resolutions. Similar to the rat tissue, cell bodies in the pig spinal cord can be clearly visualized showing that cell contrast is achievable in multiple mammalian tissue models. Also, the size and shape of tissue components in the pig spinal cord, such as alpha-motor neuron perikarya and axon bundles, are far more similar to those found in human tissue than the equivalent and much smaller components seen in the rat. Such morphological similarity, we believe, offers a more suitable tissue model for predicting what we might expect to see when performing our analysis on a human spinal cord slice. Since direct validation is not feasible in in vivo human studies, tractography techniques can be validated by comparing their predictions to actual tissue microstructure via comparison to histology. Such methods can be used to compare the accuracy of different tractography techniques in an excised tissue model because they are the same tractography techniques which are routinely applied in clinical imaging. Additionally, having a variety of animal models available provides more options for future studies concerning validation with pathological changes, such as traumatic brain injury, degenerative diseases, lesions etc.
It should be noted that the fixation procedure itself has well described effects on the MR properties of tissue which are dependent on the type of fixative used, the fixation procedure (immersion or perfusion), and the post-mortem interval before fixation (40, 41). Similarly, perfusion fixation results in decreased mean diffusivity of neural tissues as compared to living tissue (42), and also results in a decreased fractional anisotropy (FA) value that was attributed to the fixatives’ effects on membranous structures within the tissues under investigation. Because these effects are inherent to studies conducted on fixed tissue samples, it is very important to keep in mind these alterations when reporting quantitative data and comparing studies. However, in the current study, which focuses on qualitative measures relating to the microstructural organization of white matter, the fact that our DTI data offers such robust levels of tract prediction when subjected to tractographic analysis is indicative that necessary levels of tissue microstructure have been preserved with our fixation methods. Furthermore, because fractional anisotropy parameters of white matter structures are expected to increase in living tissue constructs, improvements in the predictive power of these techniques are expected when applied during future live-slice studies.
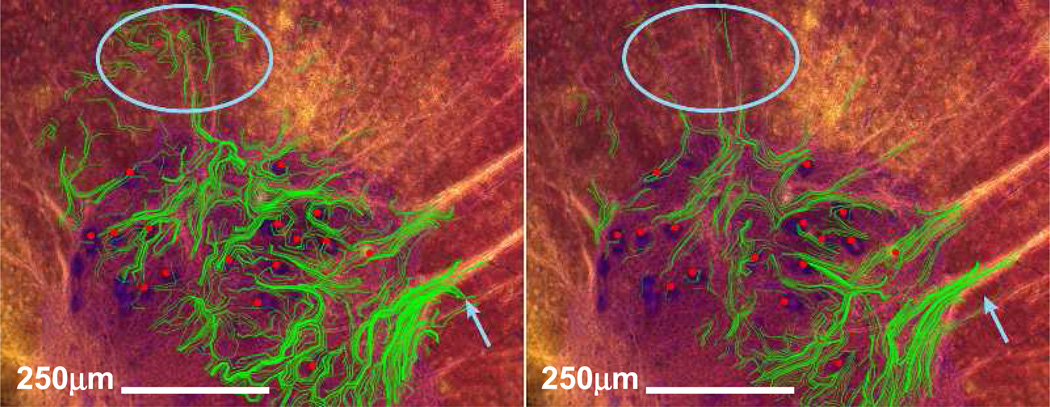
The methods presented in the current study could enable not only validation of tractography techniques, but also offer a practical way of comparing the effectiveness and accuracy of different DTT imaging techniques, data processing, and analysis. The ability to perform such techniques in biological tissue would satisfy a crucial requirement in the development of MR tractography. As an example, Figure 4 shows one of our data sets processed using all 21 weighting directions compared with an analysis of a 6 direction subset of the same data set. These 6 directions were chosen from the 21 direction data set to produce a uniform spatial sampling. As expected, the 6 direction data set qualitatively appears less robust than the 21 direction data set, with many potential fiber tracts not following the underlying histology, for example, as indicated in two particular areas in blue, but prevalent through the figure. Although employing the 21 direction HARDI data set in tractographic analysis results in increased accuracy when predicting spatial properties of white-matter tracts, the 6 direction data set can be collected in less than one third of the time. In this manner, the relationship between desired accuracy and time limitations inherent to MR imaging can be explored.
Figure 4.
Fiber track maps (green lines) overlying cell positions (red dots) generated using a 6 direction DTI design (left) or using a 21 direction DTI design (right). The 6 direction subset was selected from the 21 direction set so that a uniform spatial sampling was attained. Note the more robust fitting in the 21-direction data set, for example within the blue circle and at the blue arrow.
The predictive power exhibited by the 6 direction data suggests the feasibility of performing tractography and functional imaging on living, excised tissue at cellular-level resolutions which would allow us to interrogate the link between structural imaging, neural connectivity, and function in both normal tissues and pathological tissues undergoing therapeutic intervention. Although the scan times in this study are quite long, the use of smaller coils, higher magnetic fields and more efficient imaging techniques are expected to improve this issue dramatically as well as facilitate higher spatial resolutions. Additionally, the use of small perfusion chambers could enable the examination of live tissue slices in the MR systems allowing for controlled studies on isolated tissues. For example, we recently demonstrated diffusion signal changes in brain slices arising from chemically induced functional activation (43). We are thus presently developing a microperfusion chamber compatible with the surface microcoils and anticipate that microscopic tractography and cell mapping with direct histological validation will be feasible using both acute and organotypic brain-slice preparations in the near future.
Acknowledgements
Funding provided by the NIH (P41 RR16105 and RO1 NS36992), the NSF through the National High Magnetic Field Laboratory, the KTI (Switzerland, CTI project 6364.1 KTS-NM), the Danish National Research Foundation (95093538-2458, project 100297), and the Augustinus Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JF and BH performed the majority of the imaging and data analysis. MK assisted with histological techniques. MF and DS assisted with the microcoil development and application. PV advised on MRI, provided funding and collaborated through the exchange and support of BH. SJB assisted with initial imaging studies, provided the MRM expertise, funding, and co-wrote the paper with the authors editing the manuscript.
Author Information
None of the authors have any competing financial interests.
References
- 1.Van Bruggen N, Roberts TPL, editors. Biomedical Imaging in Experimental Neuroscience (Methods and New Frontiers in Neuroscience) CRC; 2002. [Google Scholar]
- 2.Moseley ME, Kucharczyk J, Mintorovitch J, Cohen Y, Kurhanewicz J, Derugin N, Asgari H, Norman D. Diffusion-weighted MR imaging of acute stroke: correlation with T2-weighted and magnetic susceptibility-enhanced MR imaging in cats. Am. J. Neuroradiol. 1990;11(3):423–429. [PMC free article] [PubMed] [Google Scholar]
- 3.Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori S. Introduction to Diffusion Tensor Imaging. Elsevier Science. 2007 [Google Scholar]
- 5.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev. Neurosci. 2003;4(6):469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard PL, Parker GJM. Validation of Tractography. Chapter 16 in: McNab, J.A., Jbabdi, S., Deoni, S.C., Douaud, G., Behrens, T.E., Miller, K.L. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage. 2009;46(3):775–785. doi: 10.1016/j.neuroimage.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Boretius S, Würfel J, Zipp F, Frahm J, Michaelis T. High-field diffusion tensor imaging of mouse brain in vivo using single-shot STEAM MRI. J. Neurosci. Methods. 2007;161(1):112–117. doi: 10.1016/j.jneumeth.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Roebroeck A, Galuske R, Formisano E, Chiry O, Bratzke H, Ronen I, Kim DS, Goebel R. High-resolution diffusion tensor imaging and tractography of the human optic chiasm at 9.4 T. Neuroimage. 2008;39(1):157–168. doi: 10.1016/j.neuroimage.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Van Zijl PC, Laterra J, Salhotra A, Lal B, Mori S, Zhou J. Unique patterns of diffusion directionality in rat brain tumors revealed by high-resolution diffusion tensor MRI. Magn. Reson. Med. 2007;58(3):454–462. doi: 10.1002/mrm.21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chahboune H, Ment LR, Stewart WB, Ma X, Rothman DL, Hyder F. Neurodevelopment of C57B/L6 mouse brain assessed by in vivo diffusion tensor imaging. NMR Biomed. 2007;20(3):375–382. doi: 10.1002/nbm.1130. [DOI] [PubMed] [Google Scholar]
- 11.D'Arceuil HE, Westmoreland S, de Crespigny AJ. An approach to high resolution diffusion tensor imaging in fixed primate brain. Neuroimage. 2007;35(2):553–565. doi: 10.1016/j.neuroimage.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 12.Benveniste H, Blackband SJ. Translational neuroscience and MR microscopy. Lancet Neurology. 2005;5(6):536–544. doi: 10.1016/S1474-4422(06)70472-0. [DOI] [PubMed] [Google Scholar]
- 13.Shepherd TM, Ozarslan E, King MA, Mareci TH, Blackband SJ. Structural insights from high-resolution diffusion tensor imaging and tractography of the isolated rat hippocampus. Neuroimage. 2006;32(4):1499–1509. doi: 10.1016/j.neuroimage.2006.04.210. [DOI] [PubMed] [Google Scholar]
- 14.Shepherd TM, Ozarslan E, Yachnis AT, King MA, Blackband SJ. Diffusion tensor microscopy indicates the cytoarchitectural basis for diffusion anisotropy in the human hippocampus. Am. J. Neuroradiol. 2007;28(5):958–964. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Van Zijl PC, Mori S. Three-dimensional diffusion tensor magnetic resonance microimaging of adult mouse brain and hippocampus. Neuroimage. 2002;15(4):892–901. doi: 10.1006/nimg.2001.1012. [DOI] [PubMed] [Google Scholar]
- 16.Hsu EW, Setton LA. Diffusion tensor microscopy of the intervertebral disc anulus fibrosus. Magn. Reson. Med. 1999;41(5):992–999. doi: 10.1002/(sici)1522-2594(199905)41:5<992::aid-mrm19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 17.Inglis BA, Neubauer D, Yang L, Plant D, Mareci TH, Muir D. Diffusion tensor MR imaging and comparative histology of glioma engrafted in the rat spinal cord. Am. J.Neuroradiol. 1999;20(4):713–716. [PMC free article] [PubMed] [Google Scholar]
- 18.Flint JJ, Lee CH, Hansen B, Fey M, Schmidig D, Bui JD, King MA, Vestergaard-Poulsen P, Blackband SJ. Magnetic resonance microscopy of mammalian neurons. Neuroimage. 2009;46(4):1037–1040. doi: 10.1016/j.neuroimage.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massic C, Boreo G, Vincent F, Abenhaim J, Besse P-A, Popovic RS. High-Q factor RF planar microcoils for micro scale NMR spectroscopy. Sensors and Actuators A: Physical. 2002;97:280–288. [Google Scholar]
- 20.Weiger M, Schmidig D, Denoth S, Massin C, Vincent F, Schnekel M, Fey M. NMR-microscopy with isotropic resolution of 3.0 µm using dedicated hardware and optimized methods. Concepts in Magnetic Resonance Part B. 33B(2):84–93. [Google Scholar]
- 21.Mori S, Crain BJ, Chacko VP, Van Zijl PCM. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Anbeek P, Vincken KL, Osch vMJP, Bisschops RHC, Grond vdJ. Probabilistic segmentation of white matter lesions in MR imaging. NeuroImage. 2004;21:1037–1044. doi: 10.1016/j.neuroimage.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Dyrby TB, Søgaard LV, Parker GJ, Alexander DC, Lind NM, Baaré WFC, Hay-Schmidt A, Eriksen N, Pakkenberg B, Paulson OB, Jelsing J. Validation of in vitro tractography. Neuroimage. 2007;37:1267–1277. doi: 10.1016/j.neuroimage.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proc. Nat. Acad. Sci. USA. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DK, Simmons A, Williams SCR, Horsfield MA. Noninvasive assessment of axonal fiber connectivity in the human brain via diffusion tensor MRI. Magn. Reson. Med. 42:37–41. doi: 10.1002/(sici)1522-2594(199907)42:1<37::aid-mrm7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 26.Lin CP, Wedeen VJ, Chen JH, Yao C, Tseng WYI. Validation of diffusion spectrum magnetic resonance imaging with manganese- enhanced rat optic tracts and ex vivo phantoms. Neuroimage. 19:482–495. doi: 10.1016/s1053-8119(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 27.Perrin M, Poupon C, Rieul B, Leroux P, Constantinesco A, Mangin JF, LeBihan D. Validation of q-ball imaging with a diffusion fibre-crossing phantom on a clinical scanner. Phil. Trans. R. Soc. B. –Biological Sciences. 2005;360:881–891. doi: 10.1098/rstb.2005.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Doorn A, Bovendeerd PHM, Nicolay K, Drost MR, Janssen JD. Determination of muscle fibre orientation using diffusion weighted MRI. European J. Morphol. 1996;34:5–10. doi: 10.1076/ejom.34.1.5.13156. [DOI] [PubMed] [Google Scholar]
- 29.Van Donkelaar CC, Kretzers LJG, Bovendeerd PHM, Lataster LMA, Nicolay K, Janssen JD, Drost MR. Diffusion tensor imaging in biomechanical studies of skeletal muscle function. J. Anat. 1999;194(Pt 1):79–88. doi: 10.1046/j.1469-7580.1999.19410079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell JSW, Savadjiev P, Siddiqi K, Pike GB. Validation and regularization in diffusion MRI tractography. 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro. 2006:351–354. [Google Scholar]
- 31.Lin CP, Tseng WYI, Cheng HC, Chen JH. Validation of diffusion tensor magnetic resonance axonal fiber imaging with registered manganese-enhanced optic tracts. Neuroimage. 2001;14:1035–1047. doi: 10.1006/nimg.2001.0882. [DOI] [PubMed] [Google Scholar]
- 32.Stieltjes B, Kaufmann WE, Van Zijl PCM, Frederickson K, Pearlson GD, Solaiyappan M, Mori S. Diffusion tensor imaging and axonal tracking in the human brain stem. NeuroImage. 2001;14:723–735. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- 33.Catini M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fascicule in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 34.Leemans A, Sijbers J, Verhoye M, Vander Linden A, Van Dyck D. Mathematical framework for simulating diffusion tensor MR neural fiber bundles. Magn. Reson. Med. 2005;53:944–953. doi: 10.1002/mrm.20418. [DOI] [PubMed] [Google Scholar]
- 35.Simonyan K, Tovar-Moll F, Ostuni J, Hallett M, Kalasinsky VF, Lewin-Smith MR, Rushing EJ, Vortmeyer AO, Ludlow CL. Focal white matter changes in spasmodic dysphonia: a combined diffusion tensor imaging and neuropathological study. Brain. 2008;131(Pt 2):447–459. doi: 10.1093/brain/awm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Loy DN, Liang HF, Trinkaus K, Schmidt RE, Song SK. Noninvasive diffusion tensor imaging of evolving white matter pathology in a mouse model of acute spinal cord injury. Magn Reson Med. 2007;58(2):253–260. doi: 10.1002/mrm.21316. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Budde MD, Liang HF, Klein RS, Russell JH, Cross AH, Song SK. Detecting axon damage in spinal cord from a mouse model of multiple sclerosis. Neurobiol Dis. 2006;21(3):626–632. doi: 10.1016/j.nbd.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Leergaard TB, White NS, de Crespigny A, Bolstad I, D'Arceuil H, Bjaalie JG, Dale AM. Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One. 2010;5(1):e8595.3. doi: 10.1371/journal.pone.0008595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shepherd TM, Blackband SJ, Wirth ED., III. Simultaneous diffusion MRI measurements from multiple perfused rat hippocampal slices. Magn. Reson. Med. 2002;48(3):565–569. doi: 10.1002/mrm.10241. [DOI] [PubMed] [Google Scholar]
- 40.Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn. Reson. Med. 2009;62(1):26–34. doi: 10.1002/mrm.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepherd TM, Flint JJ, Thelwall PE, Stanisz GJ, Mareci TH, Yachnis AT, Blackband SJ. Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue—Implications for MRI studies of human autopsy samples. Neuroimage. 2009;44(3):820–826. doi: 10.1016/j.neuroimage.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mardi S, Hasan KM, Narayana PA. Diffusion tensor imaging of in vivo and excised rat spinal cord at 7T with an icosahedral encoding scheme. Magn. Reson. Med. 2005;53:118–125. doi: 10.1002/mrm.20304. [DOI] [PubMed] [Google Scholar]
- 43.Flint JJ, Hansen B, Vestergaard-Poulsen P, Blackband SJ. Diffusion weighted magnetic resonance imaging of neuronal activity in the hippocampal slice model. Neuroimage. 2009;46(2):411–418. doi: 10.1016/j.neuroimage.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]