Abstract
AIM: To analyze the association of three IL28B single nucleotide polymorphisms with response to therapy in Chilean patients infected with hepatitis C virus (HCV).
METHODS: We studied two groups of patients with chronic HCV infection (genotype 1), under standard combined treatment with pegylated interferon plus ribavirin. One group consisted of 50 patients with sustained virological response, whereas the second group consisted of 49 null responders. In order to analyze the IL28B single nucleotide polymorphisms rs12979860, rs12980275 and rs8099917, samples were used for polymerase chain reaction amplification, and the genotyping was performed by restriction fragment length polymorphism.
RESULTS: The IL28B rs12979860 CC, rs12980275 AA and rs8099917 TT genotypes were much more frequently found in patients with sustained virological response compared to null responders (38%, 44% and 50% vs 2%, 8.2% and 8.2%, respectively). These differences were highly significant in all three cases (P < 0.0001).
CONCLUSION: The three IL28B polymorphisms studied are strongly associated with sustained virological response to therapy in Chilean patients with chronic HCV (genotype 1).
Keywords: IL28B, Hepatitis C virus, Chile, Pegylated interferon, Ribavirin
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) is a global health problem that affects more than 170 million people worldwide, with 3-4 million new cases each year[1]. Most (70%-80%) HCV infections persist, and about 30% of individuals with a persistent infection develop chronic liver diseases, including cirrhosis, and hepatocellular carcinoma[2].
The most effective current standard therapy for chronic HCV infection consists of subcutaneous injections of long-acting pegylated interferon-α (PEG-IFN) plus oral treatment with ribavirin (RIB). This therapy, however, yields a sustained virological response (SVR) in only 40%-50% of patients who are infected with HCV genotype 1, the most common viral genotype[3]. In Chile, HCV genotype 1 is also the most prevalent[4]. Thus, since a significant number of patients will fail to respond, or will experience significant side-effects, the identification of host and viral determinants predicting virologic response is of major interest.
Recently, three independent research groups have reported the results of separate genome-wide association studies (GWAS), supporting the association of SVR in HCV genotype 1 with single nucleotide polymorphisms (SNPs) near the gene region IL28B encoding interferon lambda 3. In the first study, performed with European-American, African-American, and Hispanic individuals, the rs12979860 SNP was most strongly associated with SVR, which is located 3 kilobases upstream of the IL28B gene. The minor allele (T) was associated with a lower rate of SVR (26% in those with genotype TT and 79% in those with genotype CC)[5]. In the second study, carried out with 293 Australian patients, a significant association between the SNP rs8099917 and SVR was found. This was further validated by an independent cohort of 555 European individuals. From 392 patients who achieved SVR, 247 (63%) were homozygotes for the allele T, which was significantly higher than genotype GG (SVR of 3.8%)[6]. Similar findings were also reported in a Japanese study. Results of a GWAS showed a significant association between treatment response with two SNPs (rs12980275 and rs8099917), both located in the IL28B gene region, with the latter being the same SNP found by Australian researchers. In this case, for the SNP rs8099917, the G allele was associated with a significantly lower SVR (0% for genotype GG and 78% for genotype TT). For the SNP rs12980275, homozygotes for the allele A had a SVR rate of 85%, which was significantly higher than genotype GG[7].
The aim of this study was to investigate the association between these three IL28B polymorphisms and the virological response in treatment-naïve Chilean patients infected with HCV genotype 1, which is the most prevalent viral isolate within Latin-American populations.
MATERIALS AND METHODS
Patient samples
The present study involved serum samples collected (January 2002-July 2010) at the Clinical Hospital University of Chile (Santiago, Chile) from 99 Chilean patients with chronic HCV infection. Patients who received at least 80% of the recommended dose of PEG-IFNα2a and RIB were considered assessable for response to treatment. We included 50 patients that achieved SVR (defined as an undetectable HCV RNA in serum more than 24 wk after treatment termination), and 49 null responder patients (NR) (defined as those who did not achieve an early virological response, < 2 log10 decrease in viral load at week 12 of treatment). Among these patients, no co-infections with human immunodeficiency virus or hepatitis B virus were included. Both groups of patients were similar in terms of basal viral load, age, gender, and clinical degree of the liver disease. This study was approved by the ethics committee of the Clinical Hospital University of Chile (protocol number 394/10).
Genotyping of SNPs rs12979860, rs12980275, rs8099917, and RFLP analyses
Genomic DNA was prepared from peripheral blood lymphocytes. The rs12979860, rs12980275, and rs8099917 SNPs genotyping was carried out by polymerase chain reaction (PCR), and restriction fragment length polymorphism (RFLP). For rs12979860, oligonucleotide primers were: 5’- AGG GCC CCT AAC CTC TGC ACA GTC T -3’ (sense), and 5’- GCT GAG GGA CCG CTA CGT AAG TCA CC -3’ (antisense). For rs12980275, primer sequences were: 5’- GAG AGC AAG AGG AGG GAA GGA A -3’ (sense), and 5’- GTG TGC CAT TAG CCA GTC AGA T -3’ (antisense). For rs8099917, oligonucleotide primers were: 5’- TTC ACC ATC CTC CTC TCA TCC CTC AT -3’ (sense) and 5’- TCC TAA ATT GAC GGG CCA TCT GTT TC -3’ (antisense). PCR reaction conditions (30 μL) were: initial denaturation at 94 °C for 10 min, followed by 40 cycles of: denaturation at 94 °C for 1 min, annealing at 58 °C for 40 s, and extension at 72 °C for 1 min. The PCR product for rs12979860, rs12980275 and rs8099917 was of 403, 441 and 401 base pairs, respectively.
In order to perform RFLP assay for the rs12979860 genotype, 20 μL of amplicons were digested with 5U of BstU I restriction endonuclease (New England Biolabs, MA, United States) at 60 °C for 2 h. BstU I digestion of allele CC yields fragments of 184, 105, 89 and 25 base pairs, whereas DNA containing the allele TT polymorphism yields fragments of 184, 130 and 89 base pairs. For the RFLP assay for the rs12980275 genotype, 20 μL of amplicons were digested with 5U of Bsl I restriction endonuclease (New England Biolabs, MA, United States) at 55 °C for 2 h. Bsl I digestion of allele AA yields fragments of 121 and 320 base pairs, whereas DNA containing the allele GG polymorphism yields fragments of 121, 30 and 290 base pairs. For the RFLP assay for the rs8099917 genotype, 20 μL of amplicons were digested with 1U of Mae III restriction endonuclease (Roche Molecular Systems, Branchburg, NJ, United States) at 55 °C for 2 h. Mae III digestion of allele TT yields fragments of 105, 110 and 186 base pairs, whereas DNA containing the allele GG polymorphism yields fragments of 105, 110, 39 and 147 base pairs. Restriction digestion products for each were separated on agarose gels stained with ethidium bromide for visualization on a UV trans-illuminator.
Statistical analysis
Genotypic frequencies were obtained by direct counting, and statistical analysis was performed by the χ2 test [calculated on 2 × 2 contingency tables, assuming a recessive model (CC vs CT + TT for rs12979860; AA vs AG + GG for rs12980275; TT vs GT + GG for rs8099917)]. P values less than 0.05 were considered statistically significant.
RESULTS
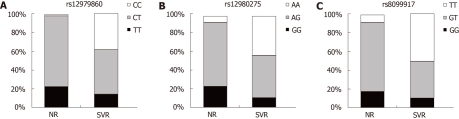
In the current study, results from all three recently known IL28B polymorphisms influencing the therapy response against HCV, rs12979860, rs12980275 and rs8099917, were available for all Chilean patients with SVR and NR, as shown in Figure 1. For the rs12979860 genotype (Figure 1A), the homozygous CC was found in 19 of 50 patients with SVR, vs 1 of 49 in NR patients (P < 0.0001). The proportion of patients with the rs12979860 CC, CT and TT genotypes was 38%, 48% and 14%, respectively, in those with SVR. In NR patients, this proportion was 2%, 76% and 22%, respectively. For the rs12980275 genotype, the homozygous AA was found in 22 of 50 cases with SVR, vs 4 of 49 in patients NR (P < 0.0001). The proportion of patients with the rs12980275 AA, AG and GG genotypes was 44%, 46% and 10% in those with SVR. In NR patients, this proportion was 8.2%, 69.4% and 22.4%, respectively, as indicated in Figure 1B. For the rs8099917 genotype, as shown in Figure 1C, the homozygous TT was found in 25 of 50 patients with SVR, vs 4 of 49 in patients NR (P < 0.0001). The proportion of patients with rs8099917 TT, GT and GG genotypes was 50%, 40% and 10% in those with SVR. In patients NR, this proportion was 8.2%, 75.5% and 16.3%, respectively.
Figure 1.
Distribution of IL28B single nucleotide polymorphisms by response to combined therapy with pegylated interferon-α plus oral treatment with ribavirin, in Chilean patients with chronic hepatitis C (genotype 1). A: rs12979860 genotype; B: rs12980275 genotype; C: rs8099917 genotype. SVR: Sustained virological response (n = 50); NR: Null responders (n = 49).
DISCUSSION
Throughout the results shown herein, we have confirmed that the three recently identified genetic polymorphisms in the interferon λ3 gene region are strongly associated with the response to treatment with PEG-IFN/RIB in Chilean patients infected with HCV genotype 1. Moreover, our current study also represents the first analysis of these SNPs from Latin-American regions, where the genotype 1 of HCV is the most prevalent.
The significant genetic results on common IL28B polymorphisms with respect to treatment response in individuals with chronic hepatitis C infection may open the possibility of a personalized medicine for the treatment of this progressive disease. Further studies are now required to determine whether patients infected with genotype 1 of HCV, and bearing a favorable SNP, will benefit or not from a shorter treatment duration with the current therapy scheme. This might reduce the cost and side effects associated with longer term treatment[8]. The way in which SNP responder genotypes influence the outcomes of anti-viral strategies including those based upon protease and polymerase inhibition, requires immediate investigation. Understanding the clinical implications of these findings will be a major research goal for the immediate future.
COMMENTS
Background
The current standard therapy for chronic hepatitis C virus (HCV) infection genotype 1 consists of pegylated interferon alfa plus ribavirin for a period of 48 wk. This regimen, however, yields a sustained virological response in only 40%-50% of patients. Because a significant number of patients will fail to respond or will have significant side effects, it is of major interest for both patient care and economic approach to predict non response.
Research frontiers
Recently, several independent research groups have reported results of genome-wide association studies, supporting the association of sustained virological response in HCV genotype 1 with single nucleotide polymorphisms (rs12979860, rs12980275 and rs8099917) near the gene region IL28B, encoding interferon lambda 3.
Innovations and breakthroughs
This study represents the first analysis of these well-know IL28B polymorphisms in patients with chronic hepatitis C infection from Latin-American regions, where genotype 1 is the most commonly found. The report shows that the three IL28B polymorphisms are associated with the sustained virological response in Chilean patients treated with standard therapy.
Applications
This study may contribute to better treatment strategies of hepatitis C. Genotyping of these IL28B polymorphisms will aid clinical decisions, improve current standard of care, and potentially lead to the integration of other agents in the future, providing an opportunity for clinicians to individualize treatment regimens for hepatitis C patients.
Peer review
The authors investigated the association between genetic IL28B polymorphisms, which encode interferon lambda 3, with the response to the treatment against hepatitis C with standard combined therapy: pegylated interferon-α plus ribavirin. They described that the rs12979860 CC, rs12980275 AA and rs8099917 TT genotypes were much more frequently found in patients with sustained virological response compared to null responder patients. This study may contribute to better treatment strategies for hepatitis C.
Footnotes
Supported by The grant OAIC 394/10 (to M.V.) from Hospital Clínico Universidad de Chile
Peer reviewer: Damiao Carlos Moraes Santos, DCM, PhD, Department of Quality Control (DEQUE), Bio-Manguinhos - FIOCRUZ, Avenida Brazil, 4365 Manguinhos, 21045-900 Rio de Janeiro, Brazil
S- Editor Sun H L- Editor Rutherford A E- Editor Li JY
References
- 1.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 3.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, Nyberg LM, Lee WM, Ghalib RH, Schiff ER, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz G, Velasco M, Thiers V, Hurtado C, Brahm J, Larrondo-Lillo M, Guglielmetti A, Smok G, Brechot C, Lamas E. Prevalence and genotypes of hepatitis C virus in blood donors and in patients with chronic liver disease and hepatocarcinoma in a Chilean population. Rev Med Chil. 1998;126:1035–1042. [PubMed] [Google Scholar]
- 5.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 6.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 8.Clark PJ, Thompson AJ, McHutchison JG. IL28B genomic-based treatment paradigms for patients with chronic hepatitis C infection: the future of personalized HCV therapies. Am J Gastroenterol. 2011;106:38–45. doi: 10.1038/ajg.2010.370. [DOI] [PubMed] [Google Scholar]