Abstract
RecQ helicases are members of an evolutionary conserved family of DNA helicases. They are homologous to the RecQ helicase of E. coli, the founding member of the family. These enzymes include gene products of disease-causing genes in Bloom, Werner, and Rothmund-Thomson syndrome. To date, these proteins have been implicated in many aspects of DNA metabolism, including DNA replication, repair, and recombination. We reported here that RECQL5, a newer member of the human RecQ helicase family, physically interacts with SWI/SNF complex and RNAPII core complex within the context of a super complex. RECQL5 was detected in the RNAPII holoenzyme but not in purified RNAPII core complex. Together, these data link RECQL5 to the assembly of the RNAPII transcription machinery and suggest that this helicase may have a regulatory role in RNAPII transcription or an RNAPII-related process or processes.
Keywords: RecQ helicases, DNA helicases, RECQL5-SWI/SNF-RNAPII super complex, purification, E. Coli
Introduction
In eukaryotic cells, genomic DNA of eukaryotes is compacted within the nucleus in the form of chromatin in which individual DNA fibre wraps around histone octamers to form nucleosomes [1]. Thus, chromatin-remodelling factors play important roles during transcription, particularly those that are mediated by RNA polymerase II (RNAPII). First, chromatin-remodeling factors can enhance the recruitment of RNA polymerase to the promoter-proximal region immediately upstream of the transcription start site (TSS), a prerequisite for the establishment of transcriptional machinery, i.e. the formation of the so-called preinitiation complex (PIC) [2]. Therefore, the presence of nucleosomes at the vicinity of individual TSS represents a potential structural barrier for the establishment of PIC [3, 4]. Recent studies have revealed that most eukaryotic genes contain an evolutionary conserved nucleosome-free region (NFR) immediately upstream of their TSS's [3, 5–7]. This NFR provides a site for the initial recruitment of transcription regulators. However, other chromatin modulators, such as SWI/SNF chromatin-remodeling complex, are required in order to facilitate the assembly of a bulky PIC complex at a promoter [3, 8]. In addition, chromatin-remodeling factors also have important roles in other aspects of RNAPII transcription. For examples, they can function to modulate the process of transcription elongation and/or pre-mRNA processing [8]. The mechanism by which these chromatin modulators modulate RNAPII transcription in mammalian cells has not been fully understood to date.
RECQL5 is a member of the human RecQ DNA helicase family. DNA helicases are enzymes that catalyze the conversion of individual double stranded DNA molecules into their corresponding single stranded forms and therefore are involved in many important aspects of DNA transaction, including gene transcription, DNA replication, repair and recombination [9]. The human RecQ helicase family are members of the RecQ helicase super-family, which share a high degree of homology with the founding member, the RecQ helicase of E. coli [10]. To date, RecQ helicases have been shown to have important roles in DNA repair, recombination and DNA replication [11–13], consistent with their intrinsic DNA helicase activities. The functional importance of the human RecQ helicases are underscored by the recent discovery that mutations in three different RecQ helicase-encoding genes give rise to several human genetic diseases, including Bloom, Werner, and Rothmund-Thomson syndrome, respectively [14].
RECQL5 and RECQL represent two additional members of the mammalian RecQ helicase family. The RECQL5 gene was first cloned in 1998 based on its homology to other members of the RecQ helicase family [15]. It encodes multiple transcripts via alternative RNA processing [16]. However, to date, only the largest predicted protein product from these transcripts, i.e. REC-QL5beta, have been detected in a significant quantity in both mice and humans [16, 17], suggesting that it is the main isoform expressed in mammalian cells. In vitro biochemical studies showed that RECQL5 could unwind double strand DNA as other helicases. Interestingly, it also exhibits a unique single strand annealing activity [18], has high affinities to fork-liked structures [19], and contains a PCNA-interacting pocket (PIP) motif and could interact with PCNA both in vitro and in vivo [19]. In addition, RECQL5 interacts with RAD51 and the MRE11-RAD50-NBS1 complex [20, 21]. Functional studies in mice and human cultured cells have shown that Recql5/RECQL5 helicase has important roles in both DNA replication and homologous recombination [20, 22, 23]. Moreover, Recql5 knockout mice are prone to sporadic cancers [20], signifying the functional importance of this unique member of the mammalian RecQ helicase family in tumor suppression.
Intriguingly, several recent studies have revealed a direct physical interaction between RECQL5 and RPB1, the largest subunit of the RNA polymerase II (RNAPII) core complex [24–27]. Moreover, a recent study has shown that RECQL5 affects both initiation and elongation of RNAPII-mediated in vitro transcription from naked DNA templates [27]. Here, we report the immunoaffinity purification of a novel RECQL5-containing complex of a very high molecular mass using newly produced anti-RECQL5 polyclonal antibodies. Mass spectrometry analysis revealed that this complex comprises primarily the components of the RNAPII core complex and the SWI/SNF chromatin-remodeling complex. RECQL5 is present in RNAPII holoenzyme. These findings in conjunction with those from previous studies reveal novel temporal and structural information regarding the interaction between RECQL5 and RNAPII and suggest that RECQL5 may have a role in RNAPII transcription during the initial assembly of the PIC and/or at the elongation phase of RNAPII transcription.
Materials and methods
Antibodies and Other Reagents
Anti-RPB1 antibodies were purchased from commercial vendors (8WG16, H5, H14 from Convance; N20 from Santa Cruz). Antibodies for BRG1, BAF170, BAF155, and SNF5 were kindly provided by Dr. Weidong Wang's group at the National Institute of Aging, USA. Rabbit polyclonal anti-RECQL5 antibodies were produced by Pocono Rabbit Farm and Laboratory Inc (PA) using a recombinant polypeptide corresponding to amino acid 661 to 880 of human REC-QL5beta. The antigen was produced in E. coli. The antibodies were purified by a two-step affinity column chromatography (a CNBR-GST column followed by a CNBR-HQ5C antigen column) procedure as described [28]. All of the other reagents, unless specified otherwise, were purchased from Sigma (Sigma, MO).
Plasmid Constructs
pGEX-2TK-HQ5C, the vector that was used to generate the antigen for producing anti-RECQL5, was constructed as follows. First, a pair of oligos: 5'-GATCTGCAGAGCTCGGAGCAG-3', and 5'-GATCCTGCTCCGAGCTCTGCA-3’ was ligated into BamHI-cut pGEX-2TK vector (Amersham, NJ) converting the BamHI site into a BamHI-SacI sequence to obtain pGEX-2TK-BS. The sequence corresponding to amino acid 661 to 880 of human RECQL5beta was first amplified by PCR with the appropriate primers: 5'-CTAGGAGCTCAAAGGCTCCTGCCCGTTCCAG-3’ and 5'-CGTAGGATCCTTATACGACGGAGGGCTTGG CTG-3'. This PCR product was then digested with BamHI plus Sacl to a BamHI- Sac I fragment and cloned into BamHI-SacI doubly digested PGEX-2TK-BS to derive pGEX-2TK-HQ5C, which is expected to express a GST-HQ5C fusion protein when transformed into the E coli strain BL21. pFasrBac-RECQL5beta, the vector that was used to produce the recombinant human RECQL5beta protein in insect cells was constructed as the following: First, the coding region of RECQL5beta cDNA was amplified as two fragments by PCR using two different pair of primers. The first pair of primers (5'-CAAGCTTGGCTAAGATGAGCAGCCACCATA-3’ and 5'-GGGATCCTCCTGCTAGAAGCCTCTTTC) generate the 5’ portion of the coding region as a Hindlll and BamHI fragment, whereas the second pair of primers (5'-AGGATCCCCAGGCTGACT GTGAAGG-3’ and 5'-GTCTAGATCTCTGGGGGCCA CACAGGCCATG-3') amplify the 3’ portion as a BamHI-Xbal fragment in which a sequence coding for three copies of the FLAG epitope peptide was included in frame at the C terminal end of the RECQL5beta open reading frame. These two fragments were then cloned into Hindlll-Xbal cut pCDNA3.1mychisA (Invitrogen) to obtain pCDNA3.1RECQL5-3flag. Subsequently, the RECQL5beta-3flag cassette of pCDNA3. lRECQL5-3flag was retrieved as a Notl-Xbal fragment and cloned into Notl-Xbal cut pFast-Bac vector (Invitrogen) to derive pFastBac-RECQL5beta-3flag. Introduction of this plasmid into DHl0BacTM cells generated the REC-QL5beta-3flag Bacmid that was then used to produce recombinant flag-RECQL5 protein from insect cells.
Production of Recombinant RECQL5 proteins
The protocol for this experiment has been described[29]. Briefly, Sf9 insect cells were infected with the recombinant Bacmid pFastBac-RECQL5beta for 4 consecutive passages. The final protein induction was done in a 250 ml suspension culture after 48 h of infection. Then the harvested cell pellet was lysed in 20 ml BLB buffer (20 mM Tris-HCl, pH7.9, 500 mM NaCl, ImM EDTA, 0.1% NP40 and 10% glycerol supplied with protease inhibitor cocktail, Roche). After clarification and centrifugation, 200 μl of M2 agarose beads (Sigma, MO) was used to capture the recombinant protein by incubation for 12 hours at 4°C, the beads-protein complex was washed three times in BC300, 2 times in BC100. Finally, the beads were loaded onto a 1.5 ml microcentrifuge spin column (Bio-Rad, CA). BC100 with the 200 μg/ml of 3XFIag peptide (Sigma, MO) was added to elute the protein by continuous rocking for 1 hour at 4°C. The elution was repeated four times. The eluted protein was concentrated by Amicon Ultra Centrifugal Filter (Millipore, MA).
Cell Culture and Nuclear Extract preparation
Nuclear extracts were prepared from HeLa S3 cells as described [30]. Briefly, HeLa S3 cells were grown in suspension culture and expanded to 60 liters. Cells were then harvested at log phase by centrifugation. Cell pellets were re-suspended in five volumes of Low Salt Buffer A (20 mM TRIS (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 μM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol (DTT)) and incubated for 10 minutes on ice. The cell suspension was then centrifuged at 420 g for 5 minutes. The cell pellet was re-suspended again in two volumes of the same low salt buffer and processed using a Dounce homogenizer with a type B pestle. The crude nuclear pellet was obtained by centrifugation at 10,000 g for 10 minutes and re-suspended in 0.5 volume of Low Salt Buffer B (20 mM TRIS (pH 7.9), 1.5 mM MgCl2, 25% glycerol, 10 mM NaCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT plus cocktail inhibitors). An equal volume of High Salt Buffer (20 mM TRIS (pH 7.9), 1.5 mM MgCl2, 25% glycerol, 1.2 M NaCI, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT plus cocktail inhibitors) was then added drop by drop and incubated with rotation at 4°C for 30 minutes. After the incubation, nuclear extract was harvested by centrifugation at 12,000 g for 15 minutes followed by an overnight dialysis in Storage Buffer (20 TRIS (pH 7.9), 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 uM PMSF, and 0.5 mM DTT) to obtain the final preparation of nuclear extract. This nuclear extract was snap frozen and stored at-80°C.
Pll ion-exchange chromatography
The procedure has been described previously [30]. Briefly, for each experiment, 60 ml of nuclear extract was loaded onto a 60-ml P11 phosphocellulose column (Whatman) at a flow rate of 1 column volume (CV)/hour. The column was washed with 3 CVs of BC100 or BC buffer (20% glycerol, 20 mM Tris-HCl, pH 7.9, at 4°C, 0.2 mM EDTA, 0.5 mM PMSF, 1.0 mM DTT) with 100 mM KCl and then step-eluted with 2.5 CVs each of BC300, BC500, BC850, BC1200 to obtain P.3, P.5, P.85, and P1.2 fractions, respectively. These individual fractions, when necessary, were dialyzed overnight against 4 liters of BC100 for 90 minutes, and then centrifuged at 14,000 rpm for 15 minutes to remove insoluble debris.
Antibody affinity purification and mass spectrometry analysis
The experiment was performed as described [28]. Briefly, anti-RECQL5 antibodies were immorbilized onto agarose beads. The binding efficiency of the antibodies was estamiated by PAGE gel electrophoresis followed by Commasie staining. Then, 20 μl of beads (with approximately 4 ug antibodies) was incubated with 2 ml of P.5 fraction from the P11 fractionation experiment at 4°C for 6 hours. The beads were then washed three times with BC300 plus two times with BC100. Bound polypeptides were eluted in 0.1 M glycine (pH 2.5) twice and the elutant was neutralized by 1/10 volume of 1 M Tris-HCl (PH 8.0). In the meantime, a control set of experiments was performed using preim-mune IgG-conjugated beads. An aliquot of the elutants was run on a 12% gel and visualized by silver staining (ICN Biomedicals, Aurora, OH). A sample with the best enrichment was run on a regular PAGE gel, stained with GelcodeTM Blue Stain Reagents (Pierce, IL). The lane with the IP product was excised. Then, three portions (the part above the 250 kDa marker, the part below the 22 kDa marker, and a segment of about 6 mm corresponding to the IgG band, respectively) were removed. Finally, the rest of sample was sliced into segments of 2-3 mm, depending on their relative position to visible bands (if there was any nearby) and analyzed using a LC-MS/MS system (ProtTech, PA).
In vitro interaction between RECQL5 and the RNAPII core complex
For in vitro co-incubation between purified recombinant RECQL5 and RNAPII core complex, 200 ng of purified RNA pol II core complex was mixed with 500 ng of RECQL5 protein and incubated for 2 hours at 4°C. Following incubation, 1 μg of RECQL5 antibodies and 20 μl of protein A beads were added to capture the immuno-complex for an additional hour. The beads were washed three times with BC300 and 2 times with BC100 plus 0.1% Nonidet P 40. Bound polypeptides were then eluted in 40 μl of 2X sample buffer. Similar experiments were carried out using purified RNAPII-holo enzyme [29], rather than purified RNAPII core complex.
Size exclusion column chromatography
P.5 or the immunoprecipitated product was fractionated by size exclusion column chromatography as described [31]. Briefly, for each experiment, a Superose 6 (10/300 GL) column (GE Healthcare) was equilibrated with modified BC200 (20 mM Tris-HCl, pH 7.9, 1 mM EDTA, 200 mM KCI, 5% glycerol and calibrated with a defined Gel Filtration Standard (BIO-RAD). Then, 200 μl of either P.5 fraction from the P11 fractionation experiment or the product of the immunoaffinity purification experiment was applied to the column. Fractions (500 μl) were collected and analyzed by SDS-PAGE followed by Western blotting.
Immunoprecipitation
The immunoprecipitation (IP) experiments were performed as described [32]. Briefly, for each experiment, the corresponding immunoglobin, i.e. IgG or IgM was used as negative control. Nuclear extract, whole cell lystate, chromatinbound protein portion or P.5 was first clarified by centrifugation and 2-5 μg of primary antibody was added to the clarified sample and the reaction was incubated for 6-12 hours in the cold room, protein A or protein G beads were added to capture the IgG and its associated antigens for additional 2 hours. The bead complex was washed sequentially three times each with BC300 and 2 times with BC100 plus 0.1% Nonidet P-40. The IgG-bound polypeptides were then eluted with 2X sample buffer.
Results
Identification of RECQL5-interacting polypeptides
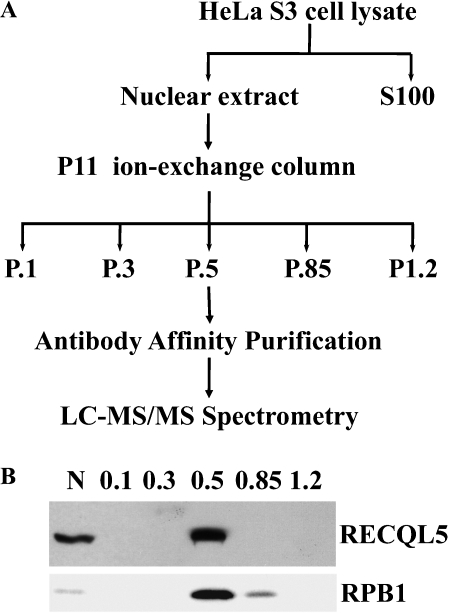
As part of our ongoing effort to elucidate the molecular basis through which RECQL5 functions in humans, we have undertaken a biochemical study to identify RECQL5-interacting polypeptides. First, a rabbit anti-RECQL5 polyclonal antiserum was raised against a recombinant polypeptide corresponding to the last 220 amino acids of the human RECQL5beta protein (referred to as RECQL5 hereon). The presumptive anti-RECQL5 antibodies were then purified by affinity chromatography against the antigen. Western blot with these antibodies showed that they could be used to detect a single band with the expected size for human RECQL5 in total lysates from HeLa cells (Figure 1B, and data not shown). Preliminary studies also indicated that these antibodies could be used to pull down RECQL5 (data not shown). Thus, these affinitypurified anti-RECQL5 antibodies were used to isolate RECQL5-containing complexes by immuno-affinity purification. To increase specificity, an ion exchange chromatography step was included as an additional measure to enrich RECQL5-containing complex(es) before the antibody-affinity purification step (Figure 1B). This resulted in a significant enrichment of the RECQL5-containing complexes in the fraction eluted with 0.5 mM KCl, the P.5 fraction (Figure 1B). Immunoprecipitation (IP) using this P.5 fraction resulted in a significant enrichment of a limited number of protein bands as revealed by silver staining of the SDS-PAGE gel of the IP products (Figure 2A). Importantly, mass spectrometry analysis of the IP product identified primarily components of RNAPII and SWI/SNF chromatin-remodeling complex (Fig 2B). The identified RNAPII components include RPB1, RPB2, RPB3, and RPB5, which represent essentially all components of RNAPII core complex within the 22-250 kDa range. The identified SWI/SNF proteins include BRG1, BAF170. BAF155, BAF60b, BAF57, BAF47/SNF5 and β-actin, which appear to represent all but BAF53 of a BGRl-containing SWI/SNF BAF complex within the range of molecular weight between 22 to 250 kDa [33, 34]. In addition, five other proteins (INT1, DTX3L; KARS, Tubulin a-3C/D chain and β-actin) were also identified (Figure 2A, 2B).
Figure 1.
Purification of RECQL5-containing complexes. A. The strategy used for purifying RECQL5-containing complexes. B. Western blot detection of RECQL5 (top panel) and RPB1 (bottom panel) in nuclear extract (N) from HeLa S3 cells and in the P.5 (0.5 mM KCl) from P11 ion-exchange column chromatography. Lane 1, input nuclear extract; lanes 2-6: P.l, P.3, P.5, P.85 and P1.2 fractions eluted with 0.1, 0.3, 0.5, 0.85, and 1.2 M KCI, respectively.
Figure 2.
Results from immunoaffinity purification and mass spectrometry analysis. A. A photograph of a silver-stained SDS-PAGE gel showing the protein profile of products from antibody affinity chromatography experiments using either a pre-immued serum (lane 1) or anti-RECQL5 antibodies (lane 2). The positions of individual markers are indicated on the left (in kDa). The top and the bottom edges of the gel being processed for mass spectrometry analysis were indicated with a pair of arrowheads. The presumptive bands corresponding to individual components of RNAPII and SWI/SNF complex are also shown on the right of the gel image. B. A list of all polypeptides identified by mass spectrometry analysis.
Confirmation of RECQL5-RNAPII and RECQL5-SWI/SNF interactions
The result from the mass spectrometry analysis suggested that RECQL5 interacts with both the core complex of RNAPII and SWI/SNF chromatin -remodeling complex. Thus, additional IP experiments were carried out to verify these interactions. We found that several components of the SWI/SNF complex could be effectively pulled down by our anti-RECQL5 antibodies (Figure 2A). Similarly, the anti-RECQL5 antibodies also effectively pulled down RPB1 (Figure 2B), the largest subunit of RNAPII core complex. These data confirm that indeed RECQL5 physically interact with both SWI/SNF chromatin-remodelling complex and RNAPII core complex. In human cells, the RNAPII can exist as either a hypophosphorylated or a hyperphosphorylated, depending on the phosphorylation status of the C-terminal domain of RPB1, the largest subunit of RNAPII [35, 36]. The hyperphosphorylated RNAPII (RNAPIIO) migrates slower than its hypophosphorylated counterpart (RNAPIIA) on a SDS-PAGE gel and therefore could be distinguished from the hypophosphorylated form. The data presented in Figure 3 showed that RECQL5 could interact with RNAPIIA (Figure 3B). However, as the level of RNAPIIO in the test sample was very low, the data were not sufficient to indicate whether RECQL5 could also interact with RNAPIIO. In order to address this question, a series of pull-down experiments were carried out using three different anti-RPB1 antibodies (N20, 8WG16 and H14, respectively). N20 is a pan-RPB1 antibody and hence is used to detect both RNAPIIA and RNAPIIO, whereas 8WG16 and H14 can be used to specifically detect hypophosphorylated (RNAPIIA) and hyperphosphorylated (RNAPIIO) forms of RPB1, respectively. The results from these experiments show that RECQL5 interacts with both the hypophosphorylated and the hyperphosphorylated forms of RPB1, and therefore, both RNAPIIA and RNAPIIO.
Figure 3.
Results of immunoprecipitation experiments. In each case, the identities of individual samples are shown on top (i.e. IN, 5% input of total cell lysáte; or the name of the antigen for which specific antibody was used for the IP experiment), whereas the targets of the Western blots were shown on the right. A. IP of various components of SWI/SNF complex (BRG1, BAF170, SNF5) with anti-RECQL5 antibodies (middle), but not with pre-immune IgG (right). B. IP pull-down of RPB1 by anti-RECQL5 antibodies as detected by Western blotting using N20, a pan-antibody that can be used to detect both the hypo-and hyperphosphorylated form of the protein, but not by pre-immune IgG. Note that the RPB1 from this cell lysáte is predominantly hypophosphorylated. C. Reciprocal IP of RECQL5 using three different anti-RPB1 antibodies: N20, a pan-RPB1 antibody; 8WG16, an antibody specific for hypophosphorylated RPB1; H14, an antibody specific for hyperphosphorylated RPB1. Note that slower-migrating hyperphosphorylated RPB1 that was pulled down by N20 and H14, but not by 8WG16. Again, the pre-immune IgG was included as a negative control.
Co-fractionation of RECQL5 with components of both SWI/SNF and RNAPII core complex
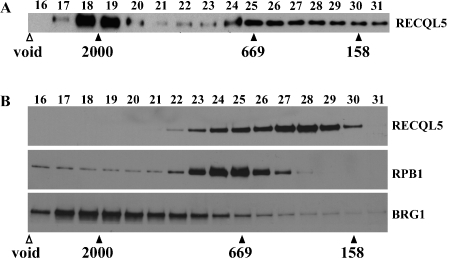
Several groups have recently reported the physical interaction of RECQL5 and RNAPII core complex [24–26]. Thus, our finding that RECQL5 interacts with both SWI/SNF complex and RNAPII core complex prompted us to ask whether RECQL5-SWI/SNF interaction reflects the presence of a previously unknown RECQL5-SWI/SNF complex in addition to the RECQL5-RNAPII complex or a RECQL5-SWI/SNF-RNAPII super complex, or both. To address this question, we examined the mass of the RECQL5-containing complex or complexes within the affinity-purified product by size exclusion column chromatography. We found that the bulk of RECQL5 in the purified product was eluted in fractions with a mass more than 2000 kDa (Figure 4A). These data indicate that our immunoaffinity purification has yielded primarily a RECQL5-containing complex or complexes of more than 2000 kDa.
Figure 4.
Analysis of RECQL5-containing complexes by size exclusion column chromatography. For each fractionation experiment, 200 μl of sample was applied to a Superose 6 (10/300) column and serial elutants (0.5 ml each) were collected. The first 15 fractions were considered void volume and discarded, whereas individual subsequent fractions with an estimated mass larger than 130 kDa (the expected size of monomeric RECQL5) were saved and analyzed by Western blotting. A. Western blot results with fractions derived from the product of the affinity purification using antibodies specific for RECQLS. Note the detection of a prominent RECQL5 specific signal in fractions 18 and 19. B. Same as in A except that the input was the P.5 fraction, i.e., the input for the immunoaffinity purification experiment and the membrane was incubated sequentially with antibodies against RECQLS (top panel), RPB1 (middle panel), and BRG1 (bottom panel), respectively. Positions of three protein size markers (158, 669, and 2000 kDa, respectively) are shown.
Intriguingly, however, when the P.5 fraction was subjected to the same size exclusion column, we found that the vast majority of the RECQL5-containing fractions are of molecular weights less than 800 kDa (Figure 4B, top panel), although RECQL5 could be detected in fractions of more than 2000 kDa when the Western blot was exposed much longer (data not shown). The level of RECQL5 in fractions higher than 30, in which monomeric RECQL5 with an estimated mass of 130 kDa is expected to be, was very low, indicating that the vast majority of RECQL5 in HeLa S3 cells does not exist as a monomeric form. Importantly, these data in conjunction with those shown in Figure 4A suggested that under the condition of our immunoaffinity purification procedure, the RECQL5-containing complex or complexes with high molecular weights were specifically pulled down. Moreover, RPB1 and BRG1, the largest subunit for RNAPII core complex and SWI/SNF complex, respectively, were detected in fractions with molecular weights over 2000 kDa (Figure 4B, middle and bottom panel, respectively). Together with the data from the mass spectrometry analysis, our data indicate that RECQL5 exists in either one or two super complexes that contain components of either SWI/SNF complex, RNAPII core complex, or both.
Direct interaction between RECQL5 and RNAPII core complex
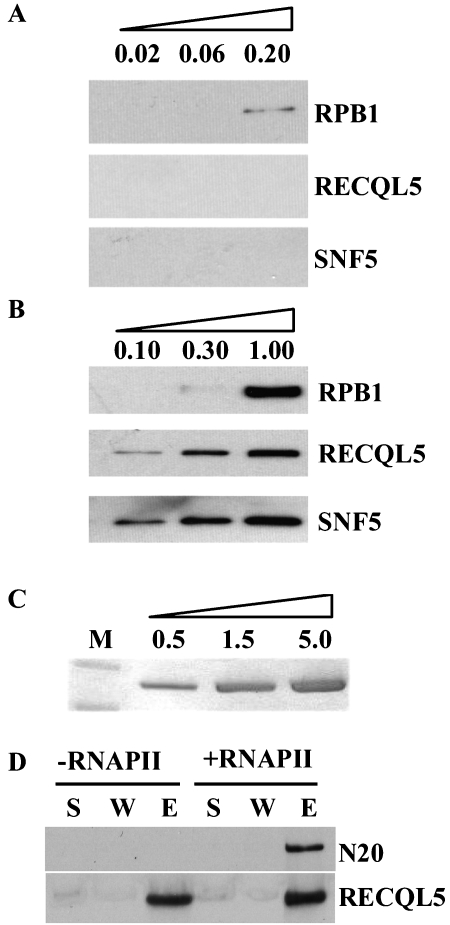
RNAPII holo-enzyme contains SWI/SNF [29, 37]. As both RNAPII core complex and RNAPII holoenzyme complex are routinely purified and used for various types of in vitro reconstituted transcription assay, we examined whether RECQL5 was co-purified as an integral component in either of these two RNAPII preparations. Analysis of previously purified RNAPII core complex and RNAPII holo-enzyme preparations [30] by Western blot revealed that RECQL5 could not be detected in RNAPII core complex preparation by standard Western blotting (at 0.2 microgram core RNAPII, Figure 5A). In contrast, RECQL5 was readily detectable in the RNAPII holoenzyme preparation even when the amount of RPB1 was undetectable under our western blot condition (at 0.10 microgram RNAPII holoenzyme, Figure 5B). Similarly, SNF5, a component of SWI/SNF complex, was detected in the RNAPII holoenzyme preparation but not in RNAPII core complex (Figure 5A, 5B). These data therefore show that at least a fraction of RNAPII holo-enzyme contains RECQL5 and/or SWI/SNF complex.
Figure 5.
Direct physical interaction between RECQL5 and RNAPII. A. Western blot analysis of purified RNAPII core complex with antibodies specific for RPB1 (top panel), RECQL5 (middle panel), or SNF (bottom panel). Note the absence of RECQL5 and SNF5 signals. The amount of RNAPII core complex used in each experiment is indicated on the top of the lane (in microgram). B. Same as A, except that purified RNAPII holoenzyme was analyzed. Note the detection of both RECQL5 and SNF even with the lowest amount of RNAPII holoenzyme. The amount of RNAPII holoenzyme used in each experiment is indicated on the top of the lane (in microgram). C. A photograph of a Coomassie-stained PAGE gel showing the single band of recombinant RECQL5 protein purified from insect cells. D. Results of Western blot experiments showing the affinity capturing of RPB1 from purified RNAPII core complex with beads coated with anti-RECQL5 antibodies when recombinant RECQL5 protein and RNAPII core complex were added (+RNAPII); but not when only recombinant RECQL5 protein was added (-RNAPII). S, supernatant; W, wash; E, elutant. N20 was used to detect RPB1 (top panel), while anti-RECQL5 antibodies were also used to detect RECQL5 as a control (bottom panel) to assess the relative amount of RECQL5 in individual samples.
We have shown earlier that RECQL5 could interact with the hyperphosphorylated form of RPB1 (Figure 3C), suggesting that RECQL5 can interact directly with RNAPII core complex even in the absence of SWI/SNF complex. To test this hypothesis, we examined whether purified recombinant RECQL5 protein could interact with purified RNAPII core complex. Recombinant RECQL5 protein was produced using the Bacu-lovirus system in insect cells (Figure 5C). When purified RECQL5 was incubated with purified RNAPII core complex, it enabled the capturing of RPB1 onto the agarose beads that are precoated with anti-RECQL5 antibodies (Figure 5D), providing direct proof that RECQL5 interact directly with RNAPII core complex.
Discussion
We reported here biochemical data for both RECQL5-RNAPII and RECQL5-SWI/SNF interactions. The finding that RECQL5 interacts with RNAPII core complex is consistent with recent reports that show a direct physical interaction between RECQL5 and RNAPII [24–26]. This unique interaction between RECQL5 and RNAPII suggests a potential role of RECQL5 in RNAPII transcription or an RNAPII-related process(es). A recent study showed that RECQL5 had an inhibitory effect on both initiation and elongation of RNAPII-mediated transcription from naked DNA templates in an in vitro reconstituted transcription assay [27]. Interestingly, our immunoaffinity purification experiment using a newly raised anti-RECQL5 antibody had led to the specific purification of a RECQL5-containing complex or complexes of more than 2000 kDa from the P.5 fraction of a Pll ion exchange chromatography-fractionated nuclear extract from HeLa S3 cells (Figure 4A). Importantly, mass spectrometry analysis of this affinity purification product revealed the presence of components of both SWI/SNF chromatin-remodeling complex and RNAPII core complex. The existence of a high molecular weight complex within which RECQL5 and RPB1 could physically interact has been reported previously. But the nature of this complex has not been determined [24]. Thus, this is the first report on the existence of a super complex that contains both RECQL5 and components of SWI/SNF chromatin-remodeling complex.
The co-purification of components from both SWI/SNF complex and RNAPII core complex by immunoaffinity purification using anti-RECQL5 antibodies suggests the existence of a RECQL5-containing super complex that also contains components of both SWI/SNF complex and RNAPII core complex, although it remains possible that RECQL5 interacts with SWI/SNF complex and RNAPII core complex in separate entities within this immunoaffinity purification product. However, we also found that both RECQL5 and SNF5 (a component of SWI/SNF complex) were present in RNAPII holoenzyme that was previously purified based on a widely used Flag-tagged RPB3 strategy (Figure 5, [29]). In addition, human SWI/SNF complex has been shown to co-purify and co-immunoprecipitate with RNAPII holoenzyme [37–39]. More recently, it has also been shown that RECQL5 could be pulled down in antibody-affinity purification experiments using Flag-tagged components of the mediator complex as baits [40], providing additional support that RECQL5 is present at the pre -initiation complex on at least some RNAPII-transcribed genes. Together, these data strongly suggest that we have in fact purified a previously unknown RECQL5-containing super complex that includes most, perhaps all components of both SWI/SNF chromatin-remodelling complex and RNAPII core complex. In addition, these data provide a clue to the temporal arrangement of this RECQL5-SWI/SNF-RNAPII interaction, i.e. during the assembly of the PIC complex. It should be noted that purification of a presumptive promoter-bound PIC complex from a nuclear extract preparation is not unexpected since PIC complex is present in nuclear extract.
In yeast, chromatin modulators, including SWI/SNF complex, play an important role in the initiation of transcription by facilitating the assembly of transcription complex at individual nucleosome-bound promoters [41]. The recruitment of SWI/SNF by gene-specific regulators to human genes has also been reported [42–44]. Our data showed that within the range of molecular weight analyzed, all but one component (i.e. BAF53) of an SWI/SNF BAF complex were effectively pulled down by our anti-RECQL5 antibodies. The inability to detect BAF53 by our analysis is expected since BAF53 was excluded from our mass spectrometry analysis after the gel slice containing the 50 kDa IgG heavy chain was discarded (see Materials and Methods). Thus, our data indicate that RECQL5 interacts specifically with the BAF subtype among the BRGl-containing SWI/SNF complexes. In this regard, it can be envisaged that RECQL5 can either affect the recruitment of SWI/SNF chromatin-remodeling complex to specific promoters to aid in the assembly of a bulky RNAPII transcription machinery on individual nucleosome-bound promoters or the activity of these chromatin modulators once they are recruited to individual promoters. In addition, it remains to be determined whether such a RECQL5-SWI/SNF-RNAPII interaction is restricted to a specific subset of genes or represents a general phenomenon during the assembly of PIC in human cells. In any case, it is likely that RECQL5 has a role in transcriptional regulation of at least a subset of genes. Interestingly, recent studies have shown that BRGF1-dependent chromatin-remodeling complexes play key roles in proliferation and differentiation in many types of cells and tissues [45]; and BRG1 is essential for embryonic development in mice [46]. In addition, BGR1 containing SWI/SNF complex occupies the promoters of a specific subset of genes in mouse embryonic stem (ES) cells and play an important role in maintaining the homeostasis of these ES cells [47, 48]. In addition, it should also be noted that RECQL5 could interact with RNAPIIo as well. Thus, it remains possible that RECQL5 may also have a role in RNAPII transcription elongation. Future experiments are required to address these important issues and to formally assess the in vivo role of RECQL5 in RNAPII transcription and/or related process(es).
Acknowledgement
We are grateful to Drs. Peter Harte for discussion and his comments on the manuscript. This study was supported by the following grants: 01 -E-109 (G.L.) from the Kinship Foundation; and R01 CA112094 (G.L.), R01NS049103 (H.L.), R01CA103867, R01CA124760 (C.-M. C.) from the US National Institutes of Health.
References
- [1].Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- [2].Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- [3].Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kornberg RD, Lorch Y. Twenty five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- [5].Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007:446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- [6].Mavrich TN, Jiang C, loshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [9].Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- [10].Nakayama H NK, Nakayama R, Irino N, Naka-yama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195:7. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- [11].Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008 doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- [12].Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- [13].Oakley TJ, Hickson ID. Defending genome integrity during S-phase: putative roles for RecQ helicases and topoisomerase III. DNA Repair (Amst) 2002;1:175–207. doi: 10.1016/s1568-7864(02)00002-2. [DOI] [PubMed] [Google Scholar]
- [14].Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- [15].Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eu-karyotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- [16].Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–1655. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. Embo J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQöbeta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hu Y RS, Sehorn M G, Lu X, Bussen W, Zheng L, Stark J M, Barnes E L, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5 Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zheng L KR, Mihaljevic B, Schwendener S, Sartori A A, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hu Y LX, Barnes E, Yan M., Lou H., Luo G. Recql5 and Blm RecQ DNA helicases have non-redundant roles in suppressing crossover. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hu Y LX, Zhou G, Barnes E L, Luo G. Recql5 Plays an Important Role in DNA Replication and Cell Survival after Camptothecin Treatment. Mol. Biol. Cell. 2009;20 doi: 10.1091/mbc.E08-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Izumikawa KYM, Hayano T, Tachikawa H, Koma-tsu W, Shimamoto A, Futami K, Furuichi Y, Shinkawa T, Yamauchi Y, Isobe T, Takahashi N. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]
- [25].Aygün O SJ, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci USA. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Islam MN FDr, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms-Interacting with RNA polymerase II and Acting as a helicase. Mol Cell Biol. 2010 doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].AygünO XX, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009;284:23197–23203. doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chi T, Yan Z, Xue Y, Wang W. Purification and functional analysis of the mammalian SWI/SNF-family of chromatin-remodeling complexes. Methods Enzymol. 2004;377:299–316. doi: 10.1016/S0076-6879(03)77018-9. [DOI] [PubMed] [Google Scholar]
- [29].Wu SY, Thomas MC, Hou SY, Likhite V, Chiang CM. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J Biol Chem. 1999;274:23480–23490. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- [30].Kershnar E, Wu SY, Chiang CM. Immunoaf-finity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multipro-tein complexes. J Biol Chem. 1998;273:34444–34453. doi: 10.1074/jbc.273.51.34444. [DOI] [PubMed] [Google Scholar]
- [31].Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- [32].Wu SY, Chiang CM. Expression and purification of epitope-tagged multisubunit protein complexes from mammalian cells. Curr Protoč Mol Biol. 2002 doi: 10.1002/0471142727.mb1613s60. Chapter 16: Unit 16 13. [DOI] [PubMed] [Google Scholar]
- [33].Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nud Recept Signal. 2008;6 doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- [35].Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- [36].Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- [37].Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- [38].Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- [41].Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- [42].Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- [43].Kowenz-Leutz E, Leutz A. A C/EBP beta iso-form recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- [44].Lee CH, Murphy MR, Lee JS, Chung JH. Targeting a SWI/SNF-related chromatin remodeling complex to the beta-globin promoter in erythroid cells. Proc Natl Acad Sci U S A. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodel-lers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- [46].Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Cham-bon P, Crabtree G, Magnuson T. A Brgl null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- [47].Kidder BL, Palmer S, Knott JG. SWI/SNF-Brgl regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- [48].Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddi-son PJ. Smarccl/Bafl55 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]