Abstract
Autophagy is a cellular process to degrade long-lived or malfunctioning proteins and obsolete or damaged organelles. It maintains cellular homeostasis and helps cells survive stressful conditions. Tumor suppressors mostly positively regulate autophagy, whereas oncogene products usually inhibit autophagy. Alterations in key autophagy genes have also been shown to affect cancer development. However, the role of autophagy in cancer depends on the status of the cells and can either suppress or promote tumor growth. In the present review, we report on the current state of knowledge about the reciprocal regulation of autophagy and the potential role of autophagy played in cancer development and therapy.
Keywords: Autophagy, tumor suppressor, oncogene, cancer, cancer therapy, PTEN, p53, RB, E2F1, Bcl-2
Introduction
Macroautophagy (hereafter referred as “autophagy”) is an eukaryotic subcellular process in which membrane structures engulf cytoplasmic components, including long-lived or malfunctioning proteins and obsolete or damaged organelles, forming double-membraned vesicles called autophagosomes [1, 2]. Auto-phagosomes fuse with lysosomes to form single-membraned autolysosomes in which the contents are digested by hydrolases and recycled for biosynthesis in the cell [1, 2]. Autophagy is a critical means for cells to maintain homeostasis and execute differentiation and development functions [3]. The process also renders cells able to survive stressful conditions such as starvation or pathogen infection [3, 4]. Thus, in contrast to apoptosis, autophagy is mainly a cell survival process. Abnormalities in autophagy function are related to diverse diseases, including neurodegeneration, heart disease, and cancer [3-5].
Regarding to the role played by autophagy in cancer, mounting evidence suggests that autophagy is a tumor suppressor pathway. Tumor suppressor proteins, including phosphatase and tensin homolog (PTEN), p53, and retinoblastoma protein (RB) [3, 6], have been shown to positively regulate autophagy. By contrast, oncogene products, including Bcl-2 and the AKT/TOR pathway, have been found to inhibit autophagy [3]. Further supporting the concept, the inactivation of key autophagy genes, such as Beclin 1 and ATG4C, has been shown to result in the formation of spontaneous tumors [7] or to increase the formation of tumors by carcinogens in mice respectively [8]. However, the relationship between autophagy regulation and tumor is not clear cut; for example, cytoplasmic p53 has been found to inhibit autophagy [9]. In addition, although deficient autophagy has been linked to tumorigenesis [3], autophagy also enables cancer cells to survive stressful conditions such as hypoxia or cytotoxic therapies [3, 10], resulting in resistance to therapies and cancer recurrence.
In this review, we summarize the literature on the role played by tumor suppressors and oncogene products in the regulation of autophagy and the involvement of autophagic genes in cancer. We discuss the relevance of autophagy in cancer development and progression, and the potential role of autophagy in cancer therapy.
Oncogene products and tumor suppresors in autophagy regulation
PI3K-Akt-mTOR pathway
The phosphatidylinositol 3-kinase (PI3K)–Akt-mTOR pathway is perhaps the most commonly activated signaling pathway in human cancers [11]. The two most widely observed mechanisms of PI3K-Akt-mTOR alteration in human cancers are activation by receptor tyrosine kinases and somatic mutations in specific components of the signaling pathway [12]. This pathway is a key signal transduction system that integrates signals from growth factors, nutrients, and energy to regulate cell growth and proliferation via different cellular processes [11]. mTOR complex 1 (mTORC1) is a master regulator of autophagy that acts in the core autophagy molecular machinery [13]. It links upstream signals that inhibit autophagy through AKT. Under nutrient-rich conditions, mTORC1 phosphorylates ULK1 and ULK2 (two of the five human homologs of Atg1), and mAtg13, inhibiting the initiation of autophagy; however, upon inactivation of mTORC1 by nutrient starvation, mTORC1 dissociates, resulting in activation of ULK1 and ULK2, which initiates the autophagy cascade.
Bcl-2 family proteins
Another molecule at the core of autophagy regulation is Beclin 1 [13]. Beclin 1 is the mammalian ortholog of Atg6/Vps30 in yeast [14]. It interacts with class III PI3K (PI3KC3)/Vps34 to form the preliminary autophagic double-membrane, or autophagophore [15]. Beclin 1 is also involved in the recruitment of autophagy proteins–such as UVRAG (UV irradiation resistance-associated gene), Ambra1 (the activating molecule in Beclin 1-regulated autophagy), Bif-1 (Endophillin B1), and Barkor (Beclin 1-associated autophagy-related key regulator)–to the autophagophore for the initiation and maintenance of autophagy [16-19]. Beclin 1 was originally isolated and identified as a B cell lymphoma/leukemia 2 (Bcl-2) interacting protein [20]. It contains a Bcl-2 homology 3 (BH3) domain, conserved in many Bcl-2 family proteins, that is necessary for interaction with Bcl-2 and Bcl-XL [21, 22].
The Bcl-2 protein family plays a dual role in autophagy regulation. Antiapoptotic oncoproteins, such as Bcl-2, Bcl-XL, Bcl-w, and Mcl-1, can inhibit autophagy, whereas proapoptotic BH3-only proteins, such as BNIP3, Bad, Bik, Noxa, Puma, and BimEL, can induce autophagy [23]. In mice and humans, Beclin 1 constitutively interacts with the anti-apoptotic proteins. The interaction of Bcl-2/Bcl-XL with Beclin 1/Vps34 complex decreases PI3KC3/Vps34 activity, whereas the BH3 domain of BH3-only proteins such as Bad may competitively disrupt the inhibitory interaction of Beclin 1 and Bcl-2/Bcl-XL, leading to autophagy.
Recently, c-Jun N-terminal kinase 1 (JNK1), a mitogen–activated protein kinase, has been shown to mediate multisite phosphorylation of Bcl-2, thereby inhibiting its ability to bind Beclin 1 and facilitating autophagy during nutrient deprivation [24]. JNK has also been implicated as a tumor suppressor through its action of limiting Ras-induced transformation in vivo [25]. Furthermore, mutations in the gene for mitogen-activated kinase kinase 4, an upstream activator of JNK, are common in pancreatic, breast, colon, lung, and prostate cancers [26]. Even though JNK involvement in tumor progression remains controversial, the JNK activation of Beclin 1–mediated autophagy may indicate its role in tumor suppression. Another tumor suppressor that positively regulates Beclin 1 activity, DAPK (death-associated protein kinase), physically interacts with Beclin 1 and phosphorylates Beclin 1 on Thr119, a crucial position within the BH3 domain of Beclin 1, and thus promotes the dissociation of Beclin 1 from its inhibitor, BCL-XL, resulting in autophagy induction [27].
PTEN
The tumor suppressor PTEN is the second most frequently mutated tumor suppressor gene in human cancers [28, 29]. PTEN's primary function is to antagonize PI3K through its intrinsic lipid phosphatase activity by dephosphorylating PtdIns-3,4-P2 and PtdIns-3,4,5-P3 at the D3 position [30, 31]. Ectopic expression of wild-type PTEN has been shown to induce G1 growth arrest, anoikis, apoptosis, or autophagy, depending on the cell type [30, 32-35]. The molecular mechanisms involved in PTEN-induced autophagy are mainly mediated through its inhibition of the PI3K-Akt-mTOR signaling cascade [35]. FOXO3a, a downstream target of Akt inhibition [36], up-regulates the transcription of a panel of autophagic genes [37, 38], as well as PINK1/PARK6 (PTEN-induced putative kinase 1) [39]. PINK1 is recruited to mitochondria with Parkin and associates with LC3 to induce p62- and VDAC1-dependent mitophagy [40, 41].
However, several mTOR-independent mechanisms have been implicated in autophagy induced by PTEN. For instance, the nuclear accumulation of FOXO3a is also dependent on AMP-activated protein kinase (AMPK) [37], whose activation has also been observed in nuclear-PTEN-expressing cells [42]. Activated AMPK has been shown to induce autophagy through multiple mechanisms, including inhibition of mTOR via TSC2 (tuberous sclerosis 2) phosphorylation/activation [43], activation of p53 [44], and stabilization of p27/kip1 [45]. Whether nuclear PTEN-mediated AMPK activation depends on the Peutz-Jeghers syndrome protein (LKB1) remains to be elucidated, although LKB1 has been found to interact and phosphorylate PTEN [46]. Moreover, PTEN has been shown to mediate the activation of the RNA-dependent protein kinase–eukaryotic translation initiation factor 2α (eIF2α) pathway independently of its phosphatase activity [47]. Phosphorylated eIF2α is known to induce the expression of Atg12 and LC3 conversion from LC3-I to -II, both of which are essential factors for autophagy induction [48]. Alternatively, PTEN can potentially trigger autophagy through cross-talk with other tumor suppressors, such as stabilizing p53 [49] and restoration of RB activity [50].
p53
The TP53 gene is mutated in more than 50% of cases of human cancer, which makes it the most commonly mutated gene in cancers [51]. TP53 encodes for a well-known tumor suppressor protein, p53, which plays a critical role in the cellular responses to several stresses [52]. Since its discovery, p53 has been found to have an ever-increasing number of molecular functions. For that reason, it has been progressively viewed as not simply the “guardian of the genome,” but also as a pleiotropic regulator of multiple cellular responses [53]. Recent studies have revealed a regulation of autophagy by the p53 pathway [54, 55]; however, the role of p53 in autophagy seems to depend on the subcellular localization of the protein. Thus, nuclear p53 seems to facilitate autophagy, whereas cytoplasmic p53 seems to inhibit autophagy [56].
As a transcription factor, p53 can transactivate several targets that stimulate autophagy, mainly by inhibiting the negative regulator of autophagy mTOR [57]. These targets include PTEN, AMPK, TSC2, sestrin1, and sestrin2. AMPK phosphorylates TSC1 and TSC2 complex proteins, which in turn down-regulate mTOR activity [58]. Moreover, sestrin1 and sestrin2 can bind AMPK directly, inducing TSC2 phosphorylation and activation and subsequently inhibiting mTOR activity [59], leading to autophagy [60]. p53 has also been described as inducing autophagy directly through its transcriptional regulation of DRAM (damage-regulated autophagy regulator), a ly-sosomal protein that induces macroautophagy [61]. Finally, BH3-only proteins Bad, Bax, Bnip3, and Puma, all of which can be transactivated by p53, have been shown to exert proautophagic roles by destabilizing inhibitory interactions between Bcl-2/Bcl-XL and Beclin 1 [57, 62].
As mentioned above, in contrast to the nuclear actions of p53, cytoplasmic p53 seems to inhibit basal autophagy in unstressed cells. Knockout, knockdown, or pharmacological inhibition of p53 has been shown to induce autophagy in human, mouse, and nematode cells, indicating that p53 plays a role in controlling basal autophagy levels [9]. Moreover, the reintroduction of wild-type p53 into p53-null cells suppresses autophagy. This effect persists in enucleated cells, which indicates independence from nuclear translocation. Interestingly, some p53 mutants retain the ability to inhibit autophagy: p53 mutants that have lost their transactivation activity or that cannot bind DNA or Bcl-2 family members are still able to suppress basal autophagy [63]. In the face of chronic starvation, p53 enables reduced, yet sustainable autophagic flux to support cancer cell survival through posttranscriptionally downregulation of LC3 [64].
In summary, p53 exerts a bidirectional control of autophagy depending on its subcellular location. In response to several stresses, nuclear p53 induces autophagy mainly by regulating direct targets that block mTOR signaling or Bcl-2/Bcl-xL, whereas cytoplasmic p53 inhibits autophagy levels independently of its transcriptional activity.
RB-E2F1 pathway and CDKIs (cyclin-dependent kinase inhibitors)
RB was the first identified tumor suppressor. Originally, the tumor suppressor function of RB was mainly attributed to the central role it plays in cell cycle regulation, in which RB arrests cells in the G1 phase through binding and inhibiting E2F transcription factors [65]. However, it is now believed that RB has many cellular functions in addition to serving as a cell cycle brake, including controlling cellular differentiation during embryogenesis and, in adult tissues, regulating apoptotic cell death, maintaining permanent cell cycle arrest, and preserving chromosomal stability [66]. Inactivation of RB is found in many types of cancers [67]. In tumors with wild-type RB gene, RB is frequently inactivated by the lack of expression of activators, such as the p16INK4a (p16) protein, and overexpression of repressors, including cyclin D1 and Cdk4 [67]. RB is also the target of DNA tumor viruses that cause cancer. For instance, in cervical carcinoma and squamous cell carcinoma of the head and neck, the tumors are initiated in part by inactivating RB through expression of the E7 oncoprotein by human papillomavirus [68, 69].
The first clue that RB is linked to autophagy appeared in a report showing that p27 (Kip 1), an RB activator [70], induces autophagy when it is overexpressed in glioma cells [71]. In addition, when cells undergo glucose starvation, p27 is phosphorylated at Thr 198 downstream of the LKB1-AMPK energy-sensing pathway [45]. The phosphorylation of p27 stabilizes p27 and permits cells to survive growth factor withdrawal and metabolic stress through autophagy. p27 and p16, which are both RB activators, have been shown to induce autophagy in an RB-dependent manner [6]. Moreover, restoration of RB expression in RB-null cells causes autophagy. However, when E2F1, the main repression target of RB function, coexpresses with RB in the cells, it antagonizes RB-mediated autophagy, inducing apoptosis; and RB mutants unable to bind E2F cannot induce autophagy. Thus, RB induces autophagy through repressing E2F1 activity. As supporting evidence of this process, Bcl-2, which is transactivated by E2F1 [72] and inhibits autophagy through binding Beclin 1 [73], is down-regulated by RB activity. Therefore, just as these two proteins play opposite roles in the regulation of the cell cycle and apoptosis [74, 75], they act against each other in the regulation of autophagy as well [6, 76].
However, E2F1 transactivates more than 2000 genes, and the role of E2F1 in regulating transcription is highly dependent on context [77]. In addition, RB interacts with more than 200 proteins [67]. Thus, the function of RB-E2F1 in autophagy regulation could be contextual. For example, when cells undergo hypoxia, RB seems to negatively modulate hypoxia-inducible factor-mediated autophagy by attenuating Bnip3 induction by E2F1 [78]. A weak E2F1 transactivation of autophagy-related genes has also been observed in a drug-induced E2F1-expressing system [79]. It has recently been reported that autophagy is activated during senescence and that it may mediate the acquisition of the senescence phenotype [80]. Thus, given the critical role played by CDKIs and RB during cellular senescence [81], there might be a link between the autophagy mediated by these molecules and senescence.
Autophagy genes as tumor suppressors
Additional evidence of the connection between autophagy and cancer is that alterations in autophagy genes result in susceptibility to tumorigenesis. A seminal study to identify the role of autophagy genes in cancer development discovered that the beclin 1 locus is located at 17p21 and that this region is deleted in up to 75% of ovarian cancers and 50%–70% of breast cancers [82]. Furthermore, beclin 1 null mice die as early embryos, and although heterozygous mice survive, they have a 4-fold increase in spontaneous tumor formation over wild-type mice [83]. Further studies have indicated that positive regulators of autophagy are located at or near sites common to genetic instability or loss of heterozygosity [84]. For instance, colon and gastric cancers with microsatellite instability frequently have frame-shift mutations in the autophagy genes UVRAG, ATG2B, ATG5, ATG9B, and ATG12 [85, 86]. UVRAG and Bif-1, which enhance the binding of Beclin 1 with PI3KC3 to form the complex required for nucleation of autophagic vesicles, have been shown to have tumor suppression activity [16, 18]. Loss of UVRAG correlates with increased tumorigenesis, and the UVRAG-/- tumorigenic phenotype can be rescued by the administration of exogenous UVRAG [16]. Bif-1 null mice develop normally, with the exception of an enlarged spleen, and have an increased incidence of spontaneous tumor formation. 82.8% of Bif-1 null mice developed lymphoma, compared with 14.3% of their wild-type counterparts [18]. Moreover, overexpression of ULK3 (Unc 51–like kinase 3), one of the five human homologs of Atg1 (a serine/threonine protein kinase that is an upstream regulator of autophagy under mTORC1 regulation), leads to premature senescence and is up-regulated in response to DNA damage [80]. In Crohn disease, which predisposes patients to cancer [87, 88], genome-wide association scanning has revealed variants of ATG16L1 [89] and IRGM [90]. ATG16L1 encodes Atg16L1 to complex with Atg5-Atg12 for autophagosome elongation [91]. IRGM, which belongs to the p47 immunity-related GTPase family, induces autophagy and thereby control of intracellular Mycobacterium tuberculosis in human macrophages [92]. In addition, Atg4C, a cysteine proteinase involved in cleavage of the C-terminal glycine of LC3B [93], has been shown to increase fibrosarcoma tumorigenesis after chemical carcinogenesis treatment, although ATG4C is not required for normal mouse development [8].
Autophagy, cancer development and cancer therapy
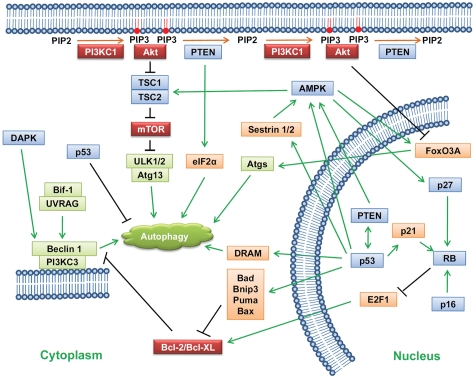
In the case of cancer development and cancer therapy, autophagy is a double-edged sword. Tumor suppressors usually positively regulate autophagy, whereas oncogene products inhibit autophagy. Alterations in key autophagy genes have been found to be relevant to cancer development (Figure 1). However, there is a lack of direct evidence showing that autophagy deficiency is required for cancer initiation. Moreover, the mechanisms underlying how autophagy suppresses cancer have not been well established. One hypothesis is that autophagy may function as a housekeeping pathway to exert quality control on organelles, proteins, and DNA. Mathews et al. observed that in autophagy-deficient and apoptosis-incompetent tumor cells, metabolic stress leads to the accumulation of p62, elevated expression of endoplasmic reticulum chaperones, damaged mitochondria, and reactive oxygen species [94], resulting in genome instability and leading to tumorigenesis [95]. Another possibility is that the combined impairment of apoptosis and autophagy promotes necrotic cell death under hypoxia and metabolic stress, both of which have been associated with inflammation and accelerated tumor growth [96]. Generally, chronic tumor inflammation favors protumor immunity, such as infiltration of protumor inflammatory mediators (e.g., macrophages) [97, 98]. Thus, autophagy may suppress cancer by buffering metabolic stress when apoptosis is inhibited, to prevent necrotic cell death [96]. Moreover, as mentioned above, autophagy has been recently identified as a new effector mechanism of senescence [80]. Autophagy is activated during oncogene- and DNA damage–induced senescence; inhibition of autophagy delays the senescence phenotype, including senescence-associated secretion. Thus, autophagy-mediated senescence may limit the proliferation of damaged cells and prevent them from developing into tumors.
Figure 1.
Schematic representation of the regulation of autophagy. Tumor suppressors (blue) except for cytoplasmic p53 are among the factors positively regulating autophagy, whereas oncogene products (red) inhibit autophagy. Some autophagy genes (green), such as Bif-1, UVRAG, and Beclin 1, are mutated in cancers.
However, autophagy can also contribute to tumor survival. The high proliferation rate of cancer cells within a tumor usually causes metabolic stress and hypoxia, such as in poorly vascularized solid tumors. In apoptosis-defective cells, inhibition of autophagy confers sensitivity to metabolic stress [96]. In fact, autophagy is observed in unvascularized, metabolically stressed regions of tumors [96]. In addition, autophagy may promote tumor cell survival during dissemination and metastasis during which the detachment of cells from the extracellular matrix may cause cell death, namely anoikis. In this regard, autophagy has been reported to increase epithelial cell survival during anoikis, including in detached cells harboring antiapoptotic lesions [99].
Increased autophagy is often observed in cancers during chemotherapy or radiation. The majority of the literature has reported a protective role of autophagy, although it has also been shown to facilitate cell death. Specific inhibition of autophagy with RNAi has been shown to accelerate cell death during cancer treatment [100]. Although there may be an off-target effect, a battery of chemical inhibitors of autophagy, such as 3-methyladenine, bafilomycin A1, and chloriquine, enhance cytotoxic cancer therapy [101]. In addition, in cells that have escaped therapy or at metastatic sites, autophagy-mediated senescence may also contribute to tumor dormancy [102]. These cells may recover and reenter the cell cycle to cause a cancer recurrence [103, 104]. Therefore, prompt disruption of autophagy pathway could be an efficient approach to extend the therapeutic benefits and reduce the incidence of cancer recurrence.
Nonetheless, in certain cancer treatments, autophagy is required for inducing robust cell death in tumor cells [101]. Because autophagy is a process to destroy cellular structures in the cytoplasm, depending on the intensity and persistence of the stress, this destruction by autophagy may pass a threshold to cause a catastrophe in the cell, leading to cell death. Moreover, caspase activity has also been reported to contribute to this type of cell death [105-107]. Thus, dissection of the transition of autophagy to autophagic cell death and the cross-talking between autophagy and apoptosis components may shed fresh light on more efficacious interventions in cancer therapy.
Concluding remarks
Autophagy plays an essential role in maintaining cellular homeostasis and protecting cells from stressful conditions. It is not surprising to see the involvement of autophagy in cancer. However, the role of autophagy in tumorigenesis and cancer therapy appears to be contextual. Since autophagy is mainly a survival mechanism to maintain the well-being of the cell, depending on the state of the cell, autophagy could suppress or promote cancer development and growth. In normal cells, autophagy clean up damaged organelles and malfunctioned proteins to prevent the cells from becoming cancerous. In cancer cells, autophagy may help them survive stresses, resulting in enhanced malignancy or recurrence after therapy.
Tumor suppressors regulating autophagy complements their tumor suppression function through the control of cell cycle, cell growth and apoptosis. Their regulation of autophagy also explains some of the functions contrary to their tumor-suppressing role. For instance, tumor suppressors p53 and RB have been shown to help cancer cells survive stress [64, 75, 108]. Through maintaining better autophagic homeostasis and adjusting the rate of autophagy to changing circumstances, wildtype p53 benefit some cancer cells from the resultant increased fitness under limited nutrient supply [64]. Moreover, loss of RB in cancer cells leads to increased sensitivity to therapy [75, 109, 110] and activation of RB via transfer of p16 increases chemoresistance [111].
Therefore, research into autophagy regulation and its relevance in tumorigenesis and cancer therapy may provide new avenues for cancer prevention and therapeutic strategies. For instance, periodic stimulation of autophagy could be beneficial for cancer prevention. The status of the genes involved in autophagy regulation should be taken into consideration when developing personalized cancer therapy with autophagy-inducing medicines to optimize the therapeutic effect in patients.
Acknowledgments
This work was supported by the National Cancer Institute (R01-CA-090879 and NIH/P50CA127001 to J. Fueyo), Brain Tumor Society (to J. Fueyo), and The University of Texas MD Anderson Cancer Center (Institutional Research Grant to J. Fueyo). We thank Virginia M. Mohlere (Department of Scientific Publications, M. D. Anderson) for editorial assistance.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo Y, Kondo S. Autophagy and cancer therapy. Autophagy. 2006;2:85–90. doi: 10.4161/auto.2.2.2463. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 9.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2009;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 15.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 17.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encepha litis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 22.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.0040025. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy NJ, Sluss HK, Jones SN, Bar-Sagi D, Flavell RA, Davis RJ. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes Dev. 2003;17:629–637. doi: 10.1101/gad.1062903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng DH, Perry WL, 3rd, Hogan JK, Baumgard M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T, Hu R, Jammulapati S, Janecki T, Leavitt A, Mitchell JT, Pero R, Sexton D, Schroeder M, Su PH, Swedlund B, Kyriakis JM, Avruch J, Bartel P, Wong AK, Tavtigian SV, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 27.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyro-sine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 30.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 32.Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/ Akt pathway. Proc Natl Acad Sci U S A. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies MA, Lu Y, Sano T, Fang X, Tang P, LaPushin R, Koul D, Bookstein R, Stokoe D, Yung WK, Mills GB, Steck PA. Adenoviral transgene expression of MMAC/PTEN in human glioma cells inhibits Akt activation and induces anoikis. Cancer Res. 1998;58:5285–5290. [PubMed] [Google Scholar]
- 34.Lu Y, Lin YZ, LaPushin R, Cuevas B, Fang X, Yu SX, Davies MA, Khan H, Furui T, Mao M, Zinner R, Hung MC, Steck P, Siminovitch K, Mills GB. The PTEN/MMAC1/TEP tumor suppressor gene decreases cell growth and induces apoptosis and anoikis in breast cancer cells. Oncogene. 1999;18:7034–7045. doi: 10.1038/sj.onc.1203183. [DOI] [PubMed] [Google Scholar]
- 35.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 36.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Chiacchiera F, Simone C. Inhibition of p38alpha unveils an AMPK-FoxO3A axis linking autophagy to cancer-specific metabolism. Autophagy. 2009;5:1030–1033. doi: 10.4161/auto.5.7.9252. [DOI] [PubMed] [Google Scholar]
- 38.Matrone A, Grossi V, Chiacchiera F, Fina E, Cappellari M, Caringella AM, Di Naro E, Loverro G, Simone C. p38alpha is required for ovarian cancer cell metabolism and survival. Int J Gynecol Cancer. 2010;20:203–211. doi: 10.1111/igc.0b013e3181c8ca12. [DOI] [PubMed] [Google Scholar]
- 39.Mei Y, Zhang Y, Yamamoto K, Xie W, Mak TW, You H. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc Natl Acad Sci U S A. 2009;106:5153–5158. doi: 10.1073/pnas.0901104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 41.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Liu JL, Sheng X, Hortobagyi ZK, Mao Z, Gallick GE, Yung WK. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol Cell Biol. 2005;25:6211–6224. doi: 10.1128/MCB.25.14.6211-6224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC path way by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5′-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- 45.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 46.Mehenni H, Lin-Marq N, Buchet-Poyau K, Reymond A, Collart MA, Picard D, Antonarakis SE. LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Hum Mol Genet. 2005;14:2209–2219. doi: 10.1093/hmg/ddi225. [DOI] [PubMed] [Google Scholar]
- 47.Mounir Z, Krishnamoorthy JL, Robertson GP, Scheuner D, Kaufman RJ, Georgescu MM, Koromilas AE. Tumor suppression by PTEN requires the activation of the PKR-eIF2alpha phosphorylation pathway. Sci Signal. 2009;2 doi: 10.1126/scisignal.2000389. ra85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 49.Li AG, Piluso LG, Cai X, Wei G, Sellers WR, Liu X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol Cell. 2006;23:575–587. doi: 10.1016/j.molcel.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 50.Paramio JM, Navarro M, Segrelles C, Gomez-Casero E, Jorcano JL. PTEN tumour suppressor is linked to the cell cycle control through the retinoblastoma protein. Oncogene. 1999;18:7462–7468. doi: 10.1038/sj.onc.1203151. [DOI] [PubMed] [Google Scholar]
- 51.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 53.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 54.Galluzzi L, Morselli E, Kepp O, Maiuri MC, Kroemer G. Defective autophagy control by the p53 rheostat in cancer. Cell Cycle. 2009;9:250–255. doi: 10.4161/cc.9.2.10493. [DOI] [PubMed] [Google Scholar]
- 55.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363–369. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 59.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 61.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 62.Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 64.Scherz-Shouval R, Weidberg H, Gonen C, Wilder S, Elazar Z, Oren M. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc Natl Acad Sci U S A. 2010;107:18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 66.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 68.Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- 69.Perez-Ordonez B, Beauchemin M, Jordan RC. Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol. 2006;59:445–453. doi: 10.1136/jcp.2003.007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 71.Komata T, Kanzawa T, Takeuchi H, Germano IM, Schreiber M, Kondo Y, Kondo S. Antitumour effect of cyclin-dependent kinase inhibitors (p16(INK4A), p18(INK4C), p19(INK4D), p21(WAF1/CIP1) and p27(KIP1)) on malignant glioma cells. Br J Cancer. 2003;88:1277–1280. doi: 10.1038/sj.bjc.6600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee HY, Hu M, Spurgers KB, Glass TL, Koul D, Liu TJ, McDonnell TJ, Yung WK. Transfer of E2F-1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61:6693–6697. [PubMed] [Google Scholar]
- 73.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8:532–535. doi: 10.4161/cc.8.4.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nat Rev Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 76.Jiang H, Martin V, Alonso M, Gomez-Manzano C, Fueyo J. RB-E2F1: molecular rheostat for autophagy and apoptosis. Autophagy. 2010;6:1216–1217. doi: 10.4161/auto.6.8.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson DG, Degregori J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Curr Mol Med. 2006;6:731–738. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 78.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxiainduced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 80.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas DM, Yang HS, Alexander K, Hinds PW. Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol Ther. 2003;2:124–130. [PubMed] [Google Scholar]
- 82.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 83.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang C, Jung JU. Autophagy genes as tumor suppressors. Curr Opin Cell Biol. 2009;22:226–233. doi: 10.1016/j.ceb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39:1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 86.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217:702–706. doi: 10.1002/path.2509. [DOI] [PubMed] [Google Scholar]
- 87.Papaconstantinou I, Theodoropoulos G, Gazouli M, Panoussopoulos D, Mantzaris GJ, Felekouras E, Bramis J. Association between mutations in the CARD15/NOD2 gene and colorectal cancer in a Greek population. Int J Cancer. 2005;114:433–435. doi: 10.1002/ijc.20747. [DOI] [PubMed] [Google Scholar]
- 88.Noffsinger A, Unger B, Fenoglio-Preiser CM. Increased cell proliferation characterizes Crohn's disease. Mod Pathol. 1998;11:1198–1203. [PubMed] [Google Scholar]
- 89.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 90.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12- Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 92.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 93.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeNardo DG, Johansson M, Coussens LM. Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev. 2008;27:11–18. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 98.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4 (+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 101.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2009;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gewirtz DA. Autophagy, senescence and tumor dormancy in cancer therapy. Autophagy. 2009;5:1232–1234. doi: 10.4161/auto.5.8.9896. [DOI] [PubMed] [Google Scholar]
- 103.Fehm T, Mueller V, Marches R, Klein G, Gueckel B, Neubauer H, Solomayer E, Becker S. Tumor cell dormancy: implications for the biology and treatment of breast cancer. APMIS. 2008;116:742–753. doi: 10.1111/j.1600-0463.2008.01047.x. [DOI] [PubMed] [Google Scholar]
- 104.Goss P, Allan AL, Rodenhiser DI, Foster PJ, Chambers AF. New clinical and experimental approaches for studying tumor dormancy: does tumor dormancy offer a therapeutic target? APMIS. 2008;116:552–568. doi: 10.1111/j.1600-0463.2008.001059.x. [DOI] [PubMed] [Google Scholar]
- 105.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 106.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, Piris MA, Dash R, Barral PM, Rodriguez-Peralto JL, Ortiz-Romero P, Tuting T, Fisher PB, Soengas MS. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–114. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 109.Stengel KR, Dean JL, Seeley SL, Mayhew CN, Knudsen ES. RB status governs differential sensitivity to cytotoxic and molecularlytargeted therapeutic agents. Cell Cycle. 2008;7:1095–1103. doi: 10.4161/cc.7.8.5737. [DOI] [PubMed] [Google Scholar]
- 110.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grim J, D'Amico A, Frizelle S, Zhou J, Kratzke RA, Curiel DT. Adenovirus-mediated delivery of p16 to p16-deficient human bladder cancer cells confers chemoresistance to cisplatin and paclitaxel. Clin Cancer Res. 1997;3:2415–2423. [PubMed] [Google Scholar]