Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) and PARP-2 belong to a family of enzymes that, using NAD+ as a substrate, catalyze poly(ADP-ribosyl)ation of proteins. PARP-1 and PARP-2 catalytic activity is stimulated by DNA-strand breaks targeting mainly proteins involved in chromatin structure and DNA metabolism, providing strong support for a dual role of both PARP-1 and PARP-2 in the DNA damage response as DNA damage sensors and signal transducers to downstream effectors. The DNA damage response has important consequences for genomic stability and tumour development. In order to manipulate DNA damage responses to selectively induce tumour cell death, a considerable effort is centred on defining the molecular mechanisms that allow cells to detect, respond to, and repair DNA damage. PARP inhibitors that compete with NAD+ at the highly conserved enzyme active site are arisen as new potential therapeutic strategies as chemo- and radiopotentiation and for the treatment of cancers with specific DNA repair defects as single-agent therapies. In the present review, we highlight emerging information about the redundant and specific functions of PARP-1 and PARP-2 in genome surveillance and DNA repair pathways. Understanding these roles might provide invaluable clues to design new cancer therapeutic approaches. In addition, we provide an overview of ongoing clinical trials with PARP inhibitors and the value of PARP-1 and PARP-2 expression as prognostic biomarkers in cancer.
Keywords: Poly(ADP-ribose) polymerases, poly(ADP-ribosyl)ation, DNA repair, genomic instability, therapeutic approaches, prognostic markers, cancer
Introduction
Poly(ADP-ribose) polymerase-1 (PARP-1) and PARP-2 belong to a family of enzymes (PARP) that, using β-NAD+ as a substrate, synthesize and transfer ADP-ribose polymers onto glutamate, aspartate or lysine residues of acceptor proteins, modifying their functional properties. Poly(ADP-ribose) (PAR) molecules covalently attached to acceptor proteins vary greatly in size, up to several hundred ADP-ribose residues with branching and large negative charges [1,2]. This protein modification by poly(ADP-ribosyl) ation, first detected over 40 years ago in nuclear extracts [3], is a dynamic process as indicated by the short half-life of the ADP-ribose polymer, which is rapidly subjected to degradation by the poly(ADP-ribose) glycohydrolase (PARG) [4] and the poly(ADP-ribose) hydrolase 3 (ARH3) [5] enzymes (Figure 1). PARP family members share a conserved catalytic domain that contains the PARP signature motif, a highly conserved sequence that forms the active site [6,7]. Recently, a unified nomenclature referring to this family of proteins as ADP-ribosyl transferases (ARTs) has been proposed to recognize that not all family members have PARP activity and some are likely to function as mono(ADP-ribosyl) transferases (mARTs). PARPs 1-5 are bone fide PARPs containing a conserved gluta-mate (Glu-988 in PARP-1) that defines the PARP catalytic activity; PARPs 6-8, 10-12, and 14-16, which are confirmed or putative mARTs; and PARPs 9 and 13, which lack key NAD+-binding residues and the catalytic glutamate, and are likely inactive [8].
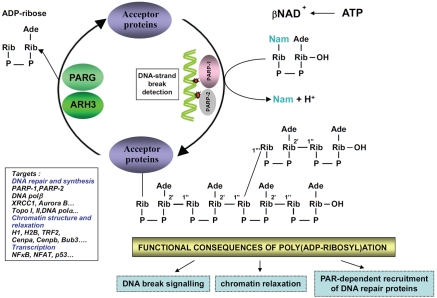
Figure 1.
Poly(ADP-ribosyl)ation reaction activated by DNA strand breaks. PARP-1 and PARP-2, rapidly recognizes DNA-strand breaks generated by genotoxic agents leading to their activation. Activated PARPs hydroiyse βNAD+, releasing nicotinamide (Nam) and one proton (H+) and catalyse the transfer of ADP-ribose moiety onto aminoacid residues of acceptor proteins. The proteins targeted are involved in numerous biological processes, including DNA repair, chromatin structure and transcription. Poly(ADP-ribosyl)ation of aceptor proteins has functional consequences such as DNA-break signalling, chromatin relaxation and recruitment of DNA repair proteins. The reaction is reversed by the activities of poly(ADP-ribose) giycohydroiase (PARG) and poly(ADP-ribose) hydrolase-3 (ARH3) that hydroiyse poly(ADP-ribose) into ADP-ribose units.
Among the members of the PARP family, PARP-1 and PARP-2 are, so far, the only known members whose activity is stimulated by DNA strand interruptions targeting mainly proteins involved in chromatin structure and DNA metabolism as well as PARP-1 and PARP-2 themselves [1,2]. Poly(ADP-ribosyl)ation mediated by PARP-1 and PARP-2 causes chromatin decondensation around damage sites, recruitment of repair machineries, and accelerates DNA damage repair, indicating a dual role of PARP-1 and PARP-2 in the DNA damage response as DNA damage sensors and signal transducers to down-stream effectors (Figure 1) [1,2]. DNA repair pathways and cell cycle control processes have important consequences for genomic stability and tumour development. Indeed a considerable effort is centred to manipulate DNA damage responses to selectively induce tumour cell death [9]. Radiotherapy and chemotherapeutic agents are the most prevalent cancer treatment by which DNA damage induces tumour cell death and there are efforts to understand and to improve the response to current cytotoxic chemotherapeutic agents. Accordingly, PARP inhibitors that compete with β-NAD+ at the highly conserved enzyme's active site are arisen as new potential therapeutic strategies as chemo- and radio-potentiation and for the treatment of cancers with specific DNA repair defects as single-agent therapies acting through the principle of synthetic lethality [10]. However, PARP-1 and PARP-2 have different targets both in DNA and in proteins, suggesting that they might play specific biological functions [2]. Indeed, it has previously shown that the genetic disruption of PARP-2, but not of PARP-1, in mice affects various differentiation processes, including spermatogenesis [11], adipogenesis [12], and the survival of thymocytes [13].
The aim of this review is to update the redundant and specific functions of PARP-1 and PARP-2 in genome surveillance and DNA repair pathways. A comprehensive understanding of the mechanistic involvement of PARP-1 and PARP-2 proteins in DNA repair and genomic instability is expected to provide invaluable clues to the rational development and exploitation of specific inhibitor drugs in a clinical setting and the design of new therapeutic approach in cancer. Ongoing clinical trials with PARP inhibitors and the value of PARP-1 and PARP-2 expression as prognostic biomarkers in cancer are also discussed.
PARP-1 and PARP-2: The two DNA-damage dependent PARP enzymes
The dramatic PAR formation stimulated by DNA-damage has been associated with PARP-1 and PARP-2 enzymatic activity, with PARP-1 being the most active protein, responsible for about 90% of cellular PAR formation observed under these conditions [14]. In fact, PARP-2 was discovered as a result of the presence of residual DNA-dependent PARP activity in PARP-1-deficient (Parp-1-/-) mouse embryonicfibroblasts (MEFs) [15].
The human PARP-1 (hPARP-1) protein (113 kDa) is a highly conserved nuclear protein organized into six domains, encoded by a gene located at position 1q41-42, which consists of 23 exons spanning ∼43 kb (Figure 2A) [1]. The amino-terminal DNA binding domain (DBD) contains two zinc fingers that define a DNA-break-sensing motif [16]. A third zinc finger motif has been identified in the PARP-1 C-domain, dispensable for DNA binding, but important for coupling damage-induced changes in the DBD to alterations in PARP-1 catalytic activity [17,18]. The B domain contains a nuclear localization signal and a caspase-3 cleavage site. The central automodification domain comprises a BRCA1 carboxy-terminal (BRCT) motif via which PARP-1 participates in protein-protein interactions. The C-terminal catalytic domain contains the PARP signature motif, a highly conserved sequence in PARP family proteins that forms the active site [1].
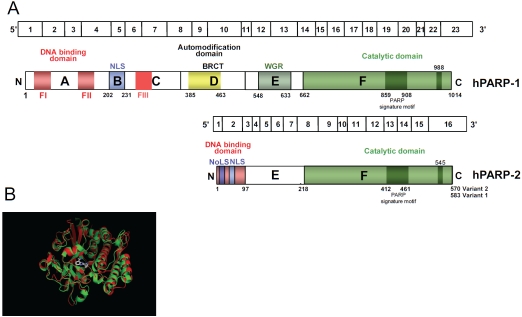
Figure 2.
Structural characteristics of human PARP-1 and PARP-2. (A) Schematic representation of human PARP-1 and PARP-2 gene organisation and protein domains. The region that is homologous to the PARP signature (residues 859-908 of PARP-1 and 412-461 of PARP-2 in variant 2) as well as the crucial residue for polymerase activity ( glu-tamic acid 988 of PARP-1 and glutamid acid 545 of PARP-2 in variant 2) are indicated as darkened green box within the catalytic domain. Fl, Fll: zinc fingers motifs; FIN: zinc ribbon domain; BRCT: BRCA1 C-terminus motif; WGR: domain with unknown function; NLS: nuclear localization signal; NoLS: nucleolar localization signal. (B) Superposition of the catalytic domain structures of human PARP-1 (red) and human PARP-2 (green) in complex with PARP inhibitor ABT-888 [159] (http://www.pdb.org).
Human PARP-2 (hPARP-2) is a nuclear protein of 62 kDa encoded by a gene located at position 14q11.2, that consists of 16 exons spanning about 13 kb (Figure 2A) [1]. Interestingly, two isoforms of the hPARP-2 protein generated by alternative splicing have been described, although its functional significance is unknown. The variant 2 lacks an internal segment of 13 amino-acid in the 5′ coding region, as compared to variant 1. The N-terminal domain of PARP-2 does not contain zinc-finger motifs but a highly basic DBD, and nuclear and nucleolar localization signals [1]. The PARP-2 DBD is structurally different from that of PARP-1 likely reflecting differences in the DNA structures recognized by each enzyme [15,19]. Accordingly, in contrast to PARP-1, PARP-2 binds less efficiently to DNA single-strand breaks (SSB) but instead recognizes gaps and flap structures [20]. A caspase-3 cleavage site defines the border between the DBD and domain E, homologous to the E domain of PARP-1. PARP-2 domain E acts both as the interacting interface with various partners and as an automodification domain [14]. A caspase-8 cleavage site marks the border between PARP-2 domains E and the C-terminal catalytic domain which display approximately 69% of similarity with the PARP-1 catalytic domain [15].
Analysis of the crystal structures of the catalytic domains of hPARP-1 and hPARP-2 (http://www.pdb.org) revealed a conserve structure and the mode of NAD+ cofactor binding is rather similar. Although the overall fold of PARP-2 catalytic domain is very similar to that of PARP-1, small structural feature differences between PARP-1 and PARP-2 catalytic domains could reflect specificities in the substrate proteins ADP-ribosylated by these enzymes (Figure 2B).
PARP-1 and PARP-2 as components of the DNAr damage response
The cellular genome is continuously exposed to different genotoxic agents, both exogenous (irradiation, genotoxic drugs, etc) and endogenous (reactive oxygen species, eroded te-lomeres, intermediates of immune and meiotic recombination, etc) that introduce damage to DNA. To combat this continuous threat to genomic integrity, cells have evolved mechanisms to detect DNA lesions, signal their presence and promote their repair. Concomitant to the repair of the DNA breaks, a rapid signalling cascade must be also coordinated at the lesion site that leads to the activation of cell cycle checkpoints and/or apoptosis. Cells defective in these mechanisms display accumulation of DNA damage that could lead to oncogenic chromosomal translocations and, eventually, to cancer [21]. PARP-1 and PARP-2, through their physical association with, or by the poly(ADP-ribosyl)ation of their partner proteins, are playing a dual role in the DNA damage response as DNA damage sensors and signal transducers to down-stream effectors (Figure 1).
Although Parp-1-/- cells and Parp-2-/- cells shown increased spontaneous genomic instability [22-24], Parp-2-/- mice do not exhibit a propensity for the development of spontaneous tumours [24,25] while Parp-1-/- mice develop spontaneous mammary and liver tumours only with long latency and at a low incidence [26,27]. However, both PARP-1 and PARP-2 deficiency accelerated spontaneous tumour development in p53 null mice, suggesting a synergistic functional interaction between PARPs proteins and p53 in tumour suppression through the role of PARP-1 and PARP-2 in the DNA damage response and genome integrity surveillance [23,24]. In addition, Parp-1-/- mice and Parp-2-/- mice are very sensitive to ionizing radiation and alkylating agents, although to different extents. Altogether, these data support a role for these proteins in the cellular response to DNA damage [25]. Indeed, different studies provided strong support for key shared functions of PARP-1 and PARP-2 in the cellular response to DNA damage: both proteins heterodimerize [25], share several common nuclear binding partners [28] and mice double deficient for PARP-1 and PARP-2 are not viable and die at the onset of gastrulation demonstrating the crucial role of poly(ADP-ribosyl)ation during embryonic development [25]. However, PARP-1 and PARP-2 have different targets both in DNA and in proteins, suggesting that they might also play specific functions in the response to DNA damage which are starting to be clarified [2].
PARP-1, PARP-2 and base-excision repair
In base-excision repair (BER), a damaged base is often recognized by a DNA glycosylase enzyme that mediates base removal, creating apurinic/apyrimidinic (AP) site. The repair of AP sites is initiated through strand incision by the AP endonuclease 1 (APE1) and polymerase and ligase proteins complete the repair [29]. The involvement of PARP-1 and PARP-2 in BER has long been recognized [1]. PARP-1 and PARP-2 were shown to accumulate with different kinetics at laser induced DNA damaged sites: while PARP-1 accumulated fast and transiently, PARP-2 showed a delayed and persistent accumulation at repair sites [30]. PARP-2 accumulation relies on the activity of PARP-1. Likewise, PARP-1 and PARP-2 interact with X-ray repair cross-complementing I (XRCC1), a crucial scaffold protein that interacts with and stimulates most of the SSBR/BER factors. Interestingly, the recruitment at damaged sites of XRCC1 was shown to be dependent on PARP-1 activity [31-33], but not on PARP-2 [30]. Taken together, these observations are in favour for an implication of PARP-2 at later steps of the repair process. This is strengthened by the fact that, as mentioned above, unlike PARP-1 which binds to SSB, PARP-2 has higher affinity for gaps or flaps, structures that correspond to more advanced repair intermediates. Thus, PARP-1 and PARP-2 have key but distinct roles in the spatial and temporal organization of SSBR/BER processes. In addition, both PARPs interact also with the other SSBR/BER factors DNA polymerase β and DNA ligase III [14]. Recently, Khodyreva et al. have demonstrated a new role for PARP-1 in the regulation of the BER process through its interaction at the AP site. PARP-1 interaction at the AP site could protect the site until APE1 becomes available to initiate strands incision and BER [34].
PARP-1, PARP-2, nucleotide excision repair and mismatch repair
Others DNA strand breaks repair pathways include the nucleotide excision repair (NER) pathway and the mismatch repair (MMR) pathway [29]. The NER pathway, which recognizes helix-distorting base lesions, is a multistep process that serves to repair a variety of DNA damage, including DNA lesions caused by ultraviolet (UV) radiation, mutagenic chemicals, or chemotherapeutic drugs [29]. UV-induced activation of PARP-1 has been reported and some evidence indicated a role of PARP-1 in the lesion recognition steps of the NER pathway, although the mechanistic details of this role remain elusive [35-37]. However, it is interesting to point out that while Parp-1-/- mice show increased susceptibility to carcinogenesis induced by alkylating agents [38], there is no such susceptibility regarding carcinogenesis induced by a hetero-cyclicamine, IQ (2-amino-3-methylimidazo[4,5-f] quinoline) and 4-nitroquinoline 1-oxide (4NQ0), both of which give rise to bulky DNA adducts [39,40]. Alkylation damage to DNA bases may be repaired mainly by BER, while bulky DNA adducts may be targeted by NER, suggesting in those experimental models a minor role of PARP-1 in NER.
The MMR pathway plays an important role in repairing base-base mismatches and insertion/deletion loops that are formed during DNA replication [29]. MMR has important roles in both the predisposition to cancer and also the response to therapy. However, the role of PARP-1 and PARP-2, if any, in this pathway remain largely unknown.
PARP-1, PARP-2 and DNA double-strand breaks repair
Ataxia telangiectasia mutated (ATM) is an early signaling protein kinase that initiates the transduction cascade at DNA double-strand breaks (DSBs) sites. The early embryonic lethality of Parp-1-/-Atm-/- (Table 1) [41] and Parp-2-/-Atm-/-[42] mice (Table 2) is likely the consequence of the inefficient SSBR/BER of spontaneous lesions arising in highly proliferative embryonic cells due to the absence of PARP-1 or PARP-2, leading to the conversion of unrepaired SSB to DSB during replication. The absence of ATM then compromises the efficient processing of these DSB by repair processes. However, evidence is accumulating that PARP-1 and PARP-2 are playing a direct and important role in the DSB repair pathways.
Table 1.
Parp-1-/- mouse models
| Genotype | Development defects | Fertility defects | Spontaneous tumorigenesis | Refs. |
|---|---|---|---|---|
| Parp-1-/- | None | None | Mammary and livers tumours with long latency and at low incidence | [26,27] |
| Parp-1-/-Parp-2-/- | Early embryonic lethality | NA | NA | [25] |
| Parp-1-/-DNA-PK-/- | None | None | T-cell lymphoma | [143] |
| Parp-1-/-Ku80-/- | Early embryonic lethality | NA | NA | [27] |
| Parp-1-/-Ku80+/- | None | None | Hepatocellular carcinoma | [27] |
| Parp-1-/-Atm-/- | Early embryonic lethality | NA | NA | [41] |
| Parp-1-/-WRNDhel/Dhel | None | None | Early onset of different tumours | [144] |
| Parp-1-/-p53-/- | None | None | Early onset of lymphoma | [23, 145, 146] |
| Other carcinomas (breast, lung, prostate, skin, brain) | ||||
| Medulloblastomas | ||||
| Suppresion thymic lymphoma |
NA, not applicable
Table 2.
Parp-2-/- mouse models
| Genotype | Development defects | Fertility defects | Spontaneous tumorigenesis | Refs. |
|---|---|---|---|---|
| Parp-2-/- | Impaired thymopoiesis, adipogenesis and spermatogenesis | None | None | [11, 12, 13] |
| Parp-2-/-Parp-1-/- | Early embryonic lethality | NA | NA | [25] |
| Parp-2-/-Atm-/- | Early embryonic lethality | NA | NA | [42] |
| Parp-2-/-p53-/- | Partial embryonic lethality | None | Early onset of T cell lymphoma | [24] |
NA, not applicable
DSB repair can be mediated by two major repair pathways depending on the context of the DNA damage, non-homologous end-joining (NHEJ) or homologous recombination (HR) [29]. In NHEJ, the major repair pathway for DSBs in mammalian cells, DSBs are recognized by Ku proteins (Ku70 and Ku80) that then binds and activates the protein kinase DNA-PKcs, leading to recruitment and activation of end-processing enzymes, polymerases and DNA ligase IV. Functional interaction of PARP-1 with different NHEJ proteins has been described (Table 1), suggesting a role of PARP-1 in NHEJ. For instance, recent studies that investigated the interaction between PARP-1 and DNA-PK in the cellular response to ionizing radiation suggest that PARP-1 and DNA-PK cooperate within the same pathway to promote DSB repair [43]. In the mean time, the role of PARP-2 in NHEJ, remains elusive. A less-well-characterized Ku-independent NHEJ pathway called microhomology-mediated end-joining, which is biased toward microhomology usage, also exits [44]. This alternative NHEJ pathway has a significant contribution in the resolution of AID-induced DNA breaks during class switching recombination (CSR) [45]. Recently, it has been shown that PARP-1 is required for the alternative Ku-independent end-joining [46-49] and PARP-1, but not PARP-2, favours repair of switch regions through this microhomology-mediated pathway [50].
HR is a multistep process that requires several proteins and is generally restricted to S and G2 because it uses sister-chromatid sequences as the template to mediate faithful repair [29]. HR is initiated by SSB generation, which is promoted by various proteins including the Mre11-Rad50-NBS1 (MRN) complex. SSBs persisting into S-phase produce replication fork collapse, requiring BRCA1 and BRCA2-mediated HR repair for resolution [21]. PARP-1 and PARP-2 detect disrupted replication forks and attract Mrell for end processing that is required for subsequent recombination repair and restart of replication forks [51]. Recently, has also been reported that disruption of PARP-1 can inhibit HR by suppressing expression of BRCA1 and RAD51 [52].
PARP-1, PARP-2 and chromatin structure
It is becoming increasingly clear that chromatin structure is modulated in response to DNA damage and has an impact in the recognition of DNA strand breaks and accessibility to damage sites of the DNA-repair machinery [53]. Dynamic chromatin structures are governed in part by posttranslational modifications of histones and non-histone DNA-binding proteins [54]. Indeed, the earliest characterized effects of PARP-1 on the genome were the modulation of chromatin structure by poly(ADP-ribosyl)ation of histones providing the first clue to the function of poly (ADP-ribosyl)ation as an epigenetic modification [55-57]. Several laboratories identified glutamic acid residues in histone H1 and histone H2B to be modified by poly(ADP-ribosyl)ation [58-59]. Recently, it has also been shown that PARP-1, but not PARP-2, covalently modifies the tails of all four core histone on specific lysine residues [60]. In addition to histone modifications by poly (ADP-ribosyl)ation, non-histone chromosomal proteins, including HMGP and the heterochromatin proteins HP1a and HP1b have also been demonstrated to be poly(ADPribosyl)ated [61,62]. In addition to covalent modifications, a number of chromatin-modifying enzymes have been identified that are recruited to PARP-1-associated PAR in a non-covalent way, representing a new mechanism by which poly(ADP-ribosyl)ation orchestrates chromatin-related functions [63].
One of the best characterized examples of chromatin modulation in response to DNA damage is ATM/ATR/DNA-PK mediated phosphorylation of the histone variant H2AX on chromatin flanking DSB sites. This serves as a signal for the recruitment of DNA damage response factors plus other chromatin-modifying components which, together, are though to promote DSB repair and amplify DSB signalling [64]. The H2AX-associated factors promote both integration and dissociation of H2AX and exchange with conventional H2A histone. These factors include FACT (Spt16/SSRP1), DNA-PK and PARP-1. It has been shown that FACT, involved in the H2AX exchange process, is stimulated by phosphorylation and inhibited by ADP-ribosylation [65]. More recently, it has been shown that the chromatin-remodeling enzyme ALC1 (Amplified in Liver Cancer 1) is rapidly recruited to DNA damage sites via an interaction with poly(ADP-ribosyl)ated PARP-1, activating its ATPase and chromatin remodelling activities and catalyzing PARP-1-stimulated nucleosome sliding [66,67].
Likewise, through its role in chromatin remodelling PARP-1 also play a role in transcription regulation [68]. The deregulated expression of genes, which occur through both genetic and epigenetic mechanisms are known to promote tumorigenesis and tumour progression. Biochemical and in vivo studies showed that PARP-1 contributes to either the compaction or decondensation of the chromatin depending on the physiological conditions. For instances, it has been suggested that PARP-1 sets up a transient repressive chromatin structure at sites of DNA damage to block transcription and facilitate DNA repair [69]. On the other hand, PARP-1 localizes to the promoters of almost all actively transcribed genes [70], which suggests that it plays a role in promoting the formation of chromatin structures that are permissive to transcription. However, PARP-1 only regulates a subset of the genes to which it binds, and it has both positive and negative effects of transcription [70-72]. Thus, gene regulation by PARP-1 is a complex process that is likely to involve multiple mechanisms and be modulated by additional inputs. Meanwhile, the role of PARP-2 in transcription regulation remains largely elusive.
Recent studies have begun to link PARP-1-dependent poly(ADP-ribosyl)ation with DNA methylation, a stable epigenetic mark that can be passed to daughter cells upon cell division and is associated with the repression of gene expression [73,74]. The chromatin insulator CTCF plays an essential role in the effects of PARP-1 on DNA methylation. CTCF is an activator of PARP-1 automodification that in turn inhibits DNA methyltransferase Dnmt1 activity with consequences on the methylation state of both genomic DNA and in CpG island regions [75]. Recently, Krishnakumar and Kraus have also shown that PARP-1 regulates chromatin structure and transcription through the histone de-methylase KDM5B-dependent pathway [76].
Other mechanisms link PARP-1 and PARP-2 with genome surveillance and cancer
Defects in other biological processes such as chromosome segregation and loss of telomeres could lead to genomic instability, a hallmark of most cancer [77].
PARP-1, PARP-2 and chromosome segregation
Segregation of sister chromosomes during the metaphase to anaphase transition is a dramatic event that results in the inheritance of a complete set of chromosomes by each daughter cell undergoing cell division. In essence, duplicated chromosomes are condensed and then lined up at the metaphase plate, where the sister chromatids are subsequently pulled apart by microtubules attached to the kinetochores [78]. This process requires the temporal and spatial coordination of a myriad of proteins in order that genomic stability is maintained over successive rounds of cell division. Indeed, chromosomal missegregation and centrosome amplification frequently occur in cancer cells [79].
PARP-1 and PARP-2 associate with functional mammalian centromeres in a cell-cycle dependent manner and interacts with the kinetochore proteins centromere protein A (CENPA), centromere protein B (CENPB) and mitotic spindle checkpoint protein BUB3 [80,81]. Interestingly, BUB3 is suggested to act as a regulator of the Anaphase-Promoting Complex or Cyclosome (APC/C) complex which is largely associated with cell cycle progression and sister chromatid separation [82]. Recently, it has been shown that PARP-1 interacts with eight of the twelve proteins belonging to the APC/C complex, suggesting a role of PARP-1 in mitotic progression [28]. Unlike PARP-1, which binds to a broad centromeric-pericentromeric heterochromatic region [83], PARP-2 appears to transiently associate with the outer kinetochore at centromeres in prometaphase and metaphase cells [81]. Interestingly, this centromeric accumulation of PARP-2 is increased when microtubule dynamics are disrupted, behaviour reminiscent of that observed with spindle checkpoint proteins. In line with this observation, Parp-2-/- cells exhibit DNA-damage-induced kinetochore defects resulting in chromosome mis-segregation in mitotic cells [25]. In addition, Parp-2-/- male mice display meiotic chromosome mis-segregation, which is related to defective centromeric hetero-chromatin and/or abnormal spindle configurations [11]. All together, these observations argue for essential roles of PARP-1 and/or PARP-2 in accurate chromosome segregation through the maintenance of centromeric heterochromatin structure and/or mitotic spindle integrity.
PARP-1, PARP-2 and telomeres
Telomeres are specialised DNA–protein complexes that cap the end of chromosomes to protect them from being recognised as DSBs needing repair [84]. Human telomeres consist of double stranded tandem repeats of the hexanucleotide sequence TTAGGG and a protective, specific protein complex (shelterin/telosome) with associated nontelomere-specific proteins [85,86]. Telomeres can fold into t-loops that may result from the invasion of the 30 overhang into duplex DNA [87] or into G-quadruplex (G4) DNA, an unusual DNA conformation based on guanine quartets [88].
The existing evidence of the involvement of PARP-2 in telomere integrity comes from the identification of a physical and functional interaction of PARP-2 with telomeric repeat binding factor 2 (TRF2), a key player in telomere protection through its ability to interact with DNA-damage signalling and repair factors [89-91]. PARP-2 regulates the DNA-binding activity of TRF2 via both a covalent heteromodification of the dimerisation domain of TRF2 and a non-covalent binding of poly(ADP-ribose) to the TRF2 DNA-binding domain. Both possible ways of TRF2 regulation act to open the t-loop structure in response to DNA damage to facilitate access of the repair machinery. Accordingly, primary Parp-2-/- MEFs show normal telomere length and telomere capping but display a spontaneously increased frequency of chromosome ends lacking detectable T2AG3 repeats [89]. Together, these observations describe PARP-2, through its regulation of TRF2, as an additional central component of telomere integrity. PARP-1 also interacts with TRF2 and controls TRF2 DNA-binding activity in response to DNA damage [92].
Recently, it has been reported that upon telomere damage induced by the G-quadruplex ligand RHPS4 [93], PARP1, but not PARP2, is recruited at the telomeres and forming several ADP-ribose polymers that co-localize with the telomeric repeat binding factor 1 protein. This process is inhibited by PARP inhibitors, suggesting the beneficial effect of PARP inhibitors in telomere-based therapy [94].
Targeting PARP-1 and PARP-2 in cancer
PARP inhibitors first emerged 30 years ago as potential anticancer drugs, showing an exquisite cytotoxicity in proliferating cells, but only after treatment with genotoxic agents [95]. Three generations of inhibitors later, increased potency and suitable pharmacokinetic properties have allowed preclinical studies to evaluate the benefit of these inhibitors in cancer [96]. This academic and industrial effort has made PARP inhibitors headway in clinical trials (Table 3). However, current PARP inhibitors target the catalytic site of PARP enzymes which is highly similar amongst PARPs family members and no isoform-specific PARP inhibitors are available [10, 96].
Table 3.
Clinical trials with PARP inhibitors in cancer
| Inhibitor | Company | Indications | In combination with | Clinical phase | Route | Refs |
|---|---|---|---|---|---|---|
| AZD2281 (olaparib) | AstraZeneca | Breast neoplasms in BRCA1/2 mutation carriers | - | II | Oral | [102, 147-149] |
| Ovarian neoplasms in BRCA1/2 mutation carriers | - | II | ||||
| Avanced or metastatic solid tumours | - | I | ||||
| Avanced serous ovarian cancer | Paclitaxel and Carboplatin | II | ||||
| Solid tumours | Cisplatin and Gemcitabine | I | ||||
| Ovarian cancer, triple-negative breast cancer | - | I | ||||
| Malignant solid tumours | Topotecan | I | ||||
| Melanoma neoplasm | Dacarbazine | I | ||||
| Avanced solid tumours | Bevacizumab | I | ||||
| ABT-888 (Veliparib) | Abbott | Metastatic breast cancer in BRCA1/2 mutation carriers | Temozolomide | II | Oral | [150-152] |
| Non-hematologic malignancies and metastatic melanoma | Temozolomide | I | ||||
| Adult refractory solid tumours and lymphomas | - | I | ||||
| Adult refractory solid tumours and lymphomas | Metronomic Cyclophosphamide | I | ||||
| BSI-201 (Iniparib) | Sanofi-Aventis | Non-small cell lung cancer stage IV | Gemcitabine and Cisplatin | II | Intravenous | [153, 154] |
| BRCA1/2 associated advanced epithelial ovarian, Fallopian tube or primary peritoneal cancer | - | II | ||||
| Triple-negative metastatic breast cancer | Gemcitabine and Cisplatin | III | ||||
| Platinum-sensitive recurrent ovarian cancer | Gemcitabine and Carboplatin | II | ||||
| AG014699 | Pfizer | Advanced solid tumours | Temozolomide | I | Intravenous | [97, 155] |
| Ovarian cancer, breast cancer in BRCA1/2 mutation carriers | - | II | ||||
| CEP-8983/9722 | Cephalon | Advanced solid tumours | Temozolomide | I | Subcutaneous | [156] |
| MK-4827 | Merck | Advanced solid tumours, BRCA1/2 mutant tumours | - | I | Oral | [157] |
| INO-1001 | Inotek | Stage III or IV melanoma | Temozolomide | Ib | Intravenous | [158] |
So far, PARP inhibitors have two therapeutic applications in cancer: (i) as chemo/radio-potentiator and (ii) as a stand-alone therapy for tumour types that are already deficient in certain types of DNA repair mechanisms (Table 3). In the first application, the combination of PARP inhibitors with DNA damaging chemotherapeutics or radiation could compromise the cancer cell DNA repair mechanisms, resulting in genomic dysfunction and cell death [96]. Indeed, the first phase I clinical trial of a PARP inhibitor was carried out between 2003 and 2005 with AGO14699 in combination with the methylating agent temozolomide in patients with advanced solid tumours [97]. Phase I, Phase II and phase III clinical trials with other PARP inhibitors in combination with chemotherapeutic agents are ongoing (Table 3).
A major breakthrough in the field of PARP inhibitors coming out in 2005 when two independent groups demonstrated the sensitivity of BRCA1 and BRCA2-deficient cell lines toward PARP inhibitors, supporting for the first time the potential use of PARP inhibitors as single therapeutic agents in cancer cell types with deficiency in certain types of DNA repair mechanisms [98,99]. This approach is based on the concept that PARP inhibition will lead to an increase in SSB will eventually lead to DSB via replication fork collapse [100], and the repair of these DSB will be compromised in tumour cells that have lost BRCA1 and BRCA2, critical components of the HR pathway, leading to chromosomal aberrations and instability of the genome resulting in cell death (Figure 3). This synthetic lethal approach, defined as the situation when mutation in one gene will result in cell susceptibility (i.e. loss of a PARP enzyme or a BRCA protein) but the loss of both is lethal [101], seems to be a promising approach in the development of cancer treatment. Different clinical trials have been initiated to test the efficacy of this approach. Indeed, a trial with the orally active PARP inhibitor olaparib showed clinical benefit in BRCA1 or BRCA2-mutant tumours [102,103]. In addition, any tumour with deficiency in other homologous recombination pathway proteins will be sensitive to PARP inhibitors. For instance, recent results have shown that cells harbouring PTEN (phosphatase and tensin homologue) mutations are sensitive to PARP inhibitors [104-106]. Similarly, PALB2-deficient cells are also sensitive to PARP inhibitors [107]. In addition, it had been shown that ATM deficiency sensitizes mantle cell lymphoma cells to PARP inhibitors [108].
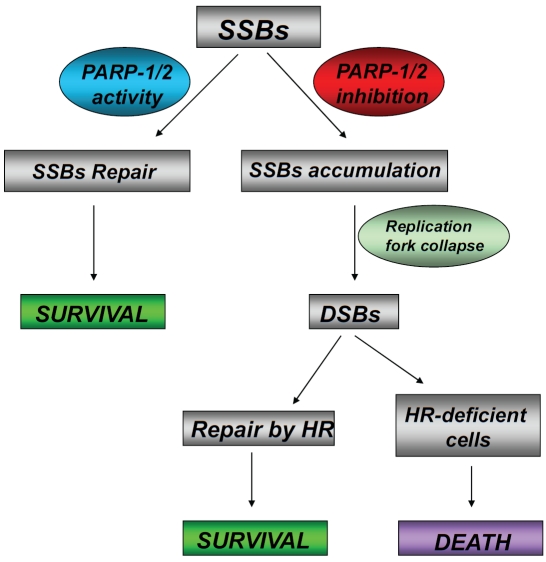
Figure 3.
A model for the selective effects of PARP inhibitors on HR-deficient cells. PARP-1 and PARP-2 promote the repair of DNA single-strand breaks (SSBs) that result from various genotoxic insults such as oxidative damage by base excision repair (BER). When PARP-1 and PARP-2 are inhibited, these lesions are unresolved and large numbers of DNA SSBs persist and are encountered by DNA replication forks. These lead to replication fork arrest associated with DNA double-strand breaks (DSBs). Such DSBs are effectively repaired by homologous recombination (HR) in normal cells but not in HR-deficient cells (i.e. BRCA1- or BRCA2-deficient cells). Accordingly, PARP inhibition in these HR-deficient cells causes a high degree of genomic instability and cell death.
As PARP inhibitors move as therapeutic drugs in cancer, several major challenges should be addressed: (i) To develop isoform-specific PARP inhibitors; (ii) To understand the specific involvement of the PARP-1 and the PARP-2 proteins in the DNA damage response and genome surveillance that will provide a basis for the rational exploitation of isoform-specific PARP inhibitors; (iii) To examine the potential long-term effects of PARP inhibitors as PARP-1 and PARP-2 have been implicated in tumour suppression [23,24]; (iv) To elucidate the details of the DNA damage response pathways to overcame PARP inhibitor resistance [109] due to reactivation of BRCA1 or BRCA2 by secondary mutations [110,111]. High-resolution crystal structures of inhibitors bound to PARP catalytic sites (Figure 2A) are essential for an in-depth understanding of the binding mode of these compounds, evaluation of the risks and mechanisms of their potential side effects, and optimization of compound selectivity and specificity.
PARP-1 and PARP-2 as prognostic biomarkers in cancer
PARP-1 over-expression both at mRNA and protein levels has been observed in various human tumour types and frequently correlated with a poor outcome, while the expression of PARP-2 in cancer samples and its linkage with evolution of the disease is largely unknown. For instance, increased expression of PARP-1 has been reported in Ewing's sarcomas [112], malignant lymphomas [113], the early stage of colorectal carcinogenesis [114], intestinal adenomas of patients with familial adenomatous polyposis [115], hepatocellular carcinoma [116], nonatypical and atypical endometrial hyperplasia [117], breast, uterine, lung, and ovarian cancers [118]. Interestingly, no significant differences in PARP-2 expression were observed between normal tissues and breast, uterine, lung, and ovarian cancers [118].
In a recent meta-analysis performed in a large public retrospective gene expression data set from breast cancers, PARP-1 mRNA expression correlated with high grade, medullary histological type, tumour size, worse metastasis-free survival and overall survival [119]. In cutaneous malignant melanomas over-expression of PARP-1 correlated with recurrence and/or progression of the disease [120]. Similarly, PARP-1 overexpression in ovarian serous carcinomas was correlated with poor outcome [121]. Furthermore, it has also been reported a positive correlation between PARP-1 protein expression and response to neoadjuvant chemotherapy [122, 123]. Altogether, these data indicated that PARP-1 expression level may serve as a promising new biological marker of aggressive tumour behaviour with prognostic value.
Polymorphisms in the promoter region of PARP-1 gene may influence PARP-1 protein expression. A microsatellite polymorphism consisting of a variable number of CA nucleotide repeat has been identified in the PARP-1 promoter [124]. Additionally, 4 sequence variations have been identified in the 5′ flanking sequence of the PARP-1 gene: C410T, poly(A)n, C1362T, and G1672A [125]. However, Zaremba et al. did not find any correlation between the level of PARP-1 expression and length of the CA-repeats in several tumor cell lines [126]. In addition, the T2444C single-nucleotide polymorphism (SNP) that results in an amino-acid substitution V762A in the PARP-1 activity domain [127] reduces PARP-1 catalytic activity by 30-40% [128,129]. This variant form has been found to be associated with prostate cancer, oesophageal, lung and thyroid cancer [128, 130, 131]. Two additional SNP that results in M129T and E251K substitutions have been described in human germ cell tumor cell lines although its relevance remains unknown [132]. Overexpression of PARP-1 in tumours could be also associated with a genomic gain/amplification of PARP-1 gene. For instance, it has been reported an association between mRNA overexpression and gain/amplification at the PARP-1 locus in breast cancer [119].
Interestingly, in human tumour cell lines there was no significant correlation between PARP activity, PARP-1 protein expression and/or a polymorphism in the DNA sequence encoding the enzyme active site, suggesting the complexity of PARP-1 regulation [126]. However, it has been observed that PARP-1 is hyperactivated in replicating BRCA2-defective cells, suggesting that the presence of PAR polymers could be used to identify HR-defective cells that are sensitive to PARP inhibitors [133].
PARP-1 overexpression may promote tumour progression by different mechanisms that still need to be fully elucidated. For instance, PARP-1 has been linked to inflammation and cancer through its role in the regulation of NFkB transcriptional activation [134,135] which is elevated in a wide spectrum of cancers and is correlated with malignancy and progression [136,137]. Indeed, it has been shown that PARP-1 play an important role in the link of DNA damage-induce nuclear events to cytoplasmic IKK activation which in turn permits NFkB activation to avert programmed cell death [138,139]. It has also been reported a direct implication of PARP-1 function in angiogenesis [140] and stable depletion of PARP-1 reduces in vivo melanoma growth and increases chemosensitivity, associated to a diminished neovasculature formation within the tumour [141]. On the other hand, as indicated above, cells with defects in DSB repair such as BRCA-deficient cells are more dependent on PARP-1 and BER to maintain genomic integrity [100]. Additionally, PARP-1 overexpression might promote tumour cell survival by coactivating hypoxia-inducible factor-1 (HIF-1)-dependent gene expression [142].
Conclusion
Although PARP-1 and PARP-2 both participate in similar biological processes controlling genome integrity, biochemical and structural studies increasingly predict that both proteins might act at different steps and/or interact with distinct protein partners. Therefore, one major challenge for the future will be to identify the specific functions of PARP-1 and PARP-2 in the genome surveillance and DNA damage response. Understanding these specific functions is expected to provide a basis for the rational development and exploitation of isoform-specific PARP inhibitors, the design of new therapeutic approaches and the identification of new target molecules.
Acknowledgments
The J.Y. laboratory is supported by grants from Spanish Ministerio de Ciencia e Innovacion (grants SAF2008-01572), Generalitat de Catalunya (SGR/524), and Fundacion Mutua Madrilena.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 5.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose)glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 6.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 7.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP ribosyl) transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 10.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Ménissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Ménissier-de Murcia J. Poly(AaDP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J Biol Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 13.Yélamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, Almarza E, Aparicio P, Ménissier-de Murcia J. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 15.Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Ménissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 16.Gradwohl G, Ménissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. The second zinc-finger domain of poly (ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci U S A. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly (ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 18.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics. 1999;57:442–445. doi: 10.1006/geno.1999.5799. [DOI] [PubMed] [Google Scholar]
- 20.Berghammer H, Ebner M, Marksteiner R, Auer B. pADPRT-2: a novel mammalian polymerizing (ADP-ribosyl)transferase gene related to truncated pADPRT homologues in plants and Caenorhabditis elegans. FEBS Lett. 1999;449:259–263. doi: 10.1016/s0014-5793(99)00448-2. [DOI] [PubMed] [Google Scholar]
- 21.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trucco C, Oliver FJ, de Murcia G, Ménissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong WM, Hande MP, Lansdorp PM, Wang ZQ. DNA strand break-sensing molecule poly(ADP-Ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol Cell Biol. 2001;21:4046–4054. doi: 10.1128/MCB.21.12.4046-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolás L, Martínez C, Baró C, Rodríguez M, Baroja-Mazo A, Sole F, Flores JM, Ampurdanés C, Dantzer F, Martin-Caballero J, Aparicio P, Yelamos J. Loss of poly(ADP-ribose) polymerase-2 leads to rapid development of spontaneous T-cell lymphomas in p53-deficient mice. Oncogene. 2010;29:2877–2883. doi: 10.1038/onc.2010.11. [DOI] [PubMed] [Google Scholar]
- 25.Ménissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Amé JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong WM, Yang YG, Cao WH, Galendo D, Frappart L, Shen Y, Wang ZQ. Poly(ADP-ribose) polymerase-1 plays a role in suppressing mammary tumourigenesis in mice. Oncogene. 2007;26:3857–3867. doi: 10.1038/sj.onc.1210156. [DOI] [PubMed] [Google Scholar]
- 27.Tong WM, Cortes U, Hande MP, Ohgaki H, Cavalli LR, Lansdorp PM, Haddad BR, Wang ZQ. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 2002;62:6990–6996. [PubMed] [Google Scholar]
- 28.Isabelle M, Moreel X, Gagné JP, Rouleau M, Ethier C, Gagné P, Hendzel MJ, Poirier GG. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol.Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khodyreva SN, Prasad R, Nina ES, Sukhanova MV, Kutuzov MM, Liu Y, Hou EW, Wilson SH, Lavrik OI. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc Natl Acad Sci U S A. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vodenicharov MD, Ghodgaonkar MM, Halappanavar SS, Shah RG, Shah GM. Mechanism of early biphasic activation of poly(ADP-ribose) polymerase-1 in response to ultraviolet B radiation. J Cell Sci. 2005;118:589–599. doi: 10.1242/jcs.01636. [DOI] [PubMed] [Google Scholar]
- 36.Ghodgaonkar MM, Zacal N, Kassam S, Rainbow AJ, Shah GM. Depletion of poly(ADP-ribose) polymerase-1 reduces host cell reactivation of a UV-damaged adenovirus-encoded reporter gene in human dermal fibroblasts. DNA Repair (Amst) 2008;7:617–632. doi: 10.1016/j.dnarep.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Tao GH, Yang LQ, Gong CM, Huang HY, Liu JD, Liu JJ, Yuan JH, Chen W, Zhuang ZX. Effect of PARP-1 deficiency on DNA damage and repair in human bronchial epithelial cells exposed to Benzo(a)pyrene. Mol Biol Rep. 2009;36:2413–2422. doi: 10.1007/s11033-009-9472-z. [DOI] [PubMed] [Google Scholar]
- 38.Tsutsumi M, Masutani M, Nozaki T, Kusuoka 0, Tsujiuchi T, Nakagama H, Suzuki H, Konishi Y, Sugimura T. Increased susceptibility of poly (ADP-ribose) polymerase-1 knockout mice to nitrosamine carcinogenicity. Carcinogenesis. 2001;22:1–3. doi: 10.1093/carcin/22.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Gunji A, Uemura A, Tsutsumi M, Nozaki T, Kusuoka O, Omura K, Suzuki H, Nakagama H, Sugimura T, Masutani M. Parp-1 deficiency does not increase the frequency of tumors in the oral cavity and esophagus of ICR/129Sv mice by 4-nitroquinoline 1-oxide, a carcinogen producing bulky adducts. Cancer Lett. 2006;241:87–92. doi: 10.1016/j.canlet.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Ogawa K, Masutani M, Kato K, Tang M, Kamada N, Suzuki H, Nakagama H, Sugimura T, Shirai T. Parp-1 deficiency does not enhance liver carcinogenesis induced by 2-amino-3-methylimidazo[4,5-f]quinoline in mice. Cancer Lett. 2006;236:32–38. doi: 10.1016/j.canlet.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 41.Ménisser-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A, de Murcia G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol. 2001;21:1828–1832. doi: 10.1128/MCB.21.5.1828-1832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell J, Smith GC, Curtin NJ. PolyfADP-Ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int J Radiat Oncol Biol Phys. 2009;75:1520–1527. doi: 10.1016/j.ijrobp.2009.07.1722. [DOI] [PubMed] [Google Scholar]
- 44.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 46.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 47.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly (ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol. 2006;356:257–265. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010;38:6065–6077. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert I, Dantzer F, Reina-San-Martin B. Parpl facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mrell-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc Natl Acad Sci U S A. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 55.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huletsky A, de Murcia G, Muller S, Hengartner M, Ménard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 57.Realini CA, Althaus FR. Histone shuttling by poly (ADP-ribosylation) J Biol Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- 58.Ogata N, Ueda K, Hayaishi O. ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J Biol Chem. 1980;255:7610–7615. [PubMed] [Google Scholar]
- 59.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 60.Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanuma S, Johnson GS. ADP-ribosylation of nonhistone high mobility group proteins in intact cells. J Biol Chem. 1983;258:4067–4070. [PubMed] [Google Scholar]
- 62.Quenet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F. The histone subcode: poly(ADPribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1alpha. FASEB J. 2008;22:3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- 63.Quénet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 64.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18:8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 65.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 66.Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly (ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP -ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, co-regulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 71.Carrillo A, Monreal Y, Ramírez P, Marin L, Parrilla P, Oliver FJ, Yélamos J. Transcription regulation of TNF-alpha-early response genes by poly (ADP-ribose) polymerase-1 in murine heart endothelial cells. Nucleic Acids Res. 2004;32:757–766. doi: 10.1093/nar/gkh239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caiafa P, Zampieri M. DNA methylation and chromatin structure: the puzzling CpG islands. J Cell Biochem. 2005;94:257–265. doi: 10.1002/jcb.20325. [DOI] [PubMed] [Google Scholar]
- 75.Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–1880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–774. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 78.Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- 79.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 80.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J Biol Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 81.Saxena A, Wong LH, Kalitsis P, Earle E, Shaffer LG, Choo KH. Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum Mol Genet. 2002;11:2319–2329. doi: 10.1093/hmg/11.19.2319. [DOI] [PubMed] [Google Scholar]
- 82.van Leuken R, Clijsters L, Wolthuis R. To cell cycle, swing the APC/C. Biochim Biophys Acta. 2008;1786:49–59. doi: 10.1016/j.bbcan.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 83.Earle E, Saxena A, MacDonald A, Hudson DF, Shaffer LG, Saffery R, Cancilla MR, Cutts SM, Howman E, Choo KH. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum Mol Genet. 2000;9:187–194. doi: 10.1093/hmg/9.2.187. [DOI] [PubMed] [Google Scholar]
- 84.Donate LE, Blasco MA. Telomeres in cancer and ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:76–84. doi: 10.1098/rstb.2010.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 86.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 87.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 88.Oganesian L, Graham ME, Robinson PJ, Bryan TM. Telomerase recognizes G quadruplex and linear DNA as distinct substrates. Biochemistry. 2007;46:11279–11290. doi: 10.1021/bi700993q. [DOI] [PubMed] [Google Scholar]
- 89.Dantzer F, Giraud-Panis MJ, Jaco I, Amé JC, Schultz I, Blasco M, Koering CE, Gilson E, Ménissier-de Murcia J, de Murcia G, Schreiber V. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright WE, Shay JW. Telomere-binding factors and general DNA repair. Nat Genet. 2005;37:116–118. doi: 10.1038/ng0205-116. [DOI] [PubMed] [Google Scholar]
- 91.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, 3rd, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 92.Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol Biol Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D'Incalci M, Blasco M, Chiorino G, Bauwens S, Horard B, Gilson E, Stoppacciaro A, Zupi G, Biroccio A. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salvati E, Scarsella M, Porru M, Rizzo A, Iachettini S, Tentori L, Graziani G, D'Incalci M, Stevens MF, Orlandi A, Passeri D, Gilson E, Zupi G, Leonetti C, Biroccio A. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29:6280–6293. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- 95.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 96.Ferraris DV. Evolution of poly(ADP-ribose) poly-merase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 97.Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A, Curtin N, Boddy A, McHugh P, Newell D, Harris A, Johnson P, Steinfeldt H, Dewji R, Wang D, Robson L, Calvert H. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 99.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 100.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 102.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly (ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 103.Hutchinson L. Targeted therapies: PARP inhibitor olaparib is safe and effective in patients with BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol. 2010;7:549. doi: 10.1038/nrclinonc.2010.143. [DOI] [PubMed] [Google Scholar]
- 104.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dedes KJ, Wetterskog D, Mendes-Pereira AM, Natrajan R, Lambros MB, Geyer FC, Vatcheva R, Savage K, Mackay A, Lord CJ, Ashworth A, Reis-Filho JS. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001538. 53ra75. [DOI] [PubMed] [Google Scholar]
- 106.McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70:5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buisson R, Dion-Côté AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williamson CT, Muzik H, Turhan AG, Zamò A, O'Connor MJ, Bebb DG, Lees-Miller SP. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Issaeva N, Thomas HD, Djureinovic T, Jaspers JE, Stoimenov I, Kyle S, Pedley N, Gottipati P, Zur R, Sleeth K, Chatzakos V, Mulligan EA, Lundin C, Gubanova E, Kersbergen A, Harris AL, Sharma RA, Rottenberg S, Curtin NJ, Helleday T. 6-thioguanine selectively kills BRCA2-defective tumors and overcomes PARP inhibitor resistance. Cancer Res. 2010;70:6268–6276. doi: 10.1158/0008-5472.CAN-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 111.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prasad SC, Thraves PJ, Bhatia KG, Smulson ME, Dritschilo A. Enhanced polyfadenosine diphosphate ribose) polymerase activity and gene expression in Ewing's sarcoma cells. Cancer Res. 1990;50:38–43. [PubMed] [Google Scholar]
- 113.Tomoda T, Kurashige T, Moriki T, Yamamoto H, Fujimoto S, Taniguchi T. Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma. Am J Hematol. 1991;37:223–227. doi: 10.1002/ajh.2830370402. [DOI] [PubMed] [Google Scholar]
- 114.Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, Imamura A, Imai K, Shi-nomura Y. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42:2374–2381. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 115.Idogawa M, Yamada T, Honda K, Sato S, Imai K, Hirohashi S. Poly(ADP-ribose) polymerase-1 is a component of the oncogenic T-cell factor-4/ beta-catenin complex. Gastroenterology. 2005;128:1919–1936. doi: 10.1053/j.gastro.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 116.Quiles-Perez R, Muñoz-Gámez JA, Ruiz-Extremera A, O'Valle F, Sanjuán-Nuñez L, Martín-Alvarez AB, Martín-Oliva D, Caballero T, Muñoz de Rueda P, León J, Gonzalez R, Muntané J, Oliver FJ, Salmerón J. Inhibition of poly adenosine diphosphate-ribose polymerase decreases hepatocellular carcinoma growth by modulation of tumor-related gene expression. Hepatology. 2010;51:255–266. doi: 10.1002/hep.23249. [DOI] [PubMed] [Google Scholar]
- 117.Ghabreau L, Roux JP, Frappart PO, Mathevet P, Patricot LM, Mokni M, Korbi S, Wang ZQ, Tong WM, Frappart L. Poly(ADP-ribose) polymerase-1, a novel partner of progesterone receptors in endometrial cancer and its precursors. Int J Cancer. 2004;109:317–321. doi: 10.1002/ijc.11731. [DOI] [PubMed] [Google Scholar]
- 118.Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherma BM. Upregulation of Poly(ADP-ribose) Polymerase-1 (PARP-1) in triple-Negative Breast Cancer and Other Primary human tumor types. Genes & Cancer. 2010;1:812–821. doi: 10.1177/1947601910383418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gonçalves A, Finetti P, Sabatier R, Gilabert M, Adelaide J, Borg JP, Chaffanet M, Viens P, Birnbaum D, Bertucci F. Poly(ADP-ribose) polymerase-1 mRNA expression in human breast cancer: a meta-analysis. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-1199-y. in press. [DOI] [PubMed] [Google Scholar]
- 120.Csete B, Lengyel Z, Kadar Z, Battyani Z. Polyfadenosine diphosphate-ribose) polymerase-1 expression in cutaneous malignant melanomas as a new molecular marker of aggressive tumor. Pathol Oncol Res. 2009;15:47–53. doi: 10.1007/s12253-008-9086-0. [DOI] [PubMed] [Google Scholar]
- 121.Brustmann H. Polyfadenosine diphosphate-ribose) polymerase expression in serous ovarian carcinoma: correlation with p53, MIB-1, and outcome. Int J Gynecol Pathol. 2007;26:147–153. doi: 10.1097/01.pgp.0000235064.93182.ec. [DOI] [PubMed] [Google Scholar]
- 122.von Minckwitz G, Muller B, Loibl S, Blohmer JU, duBois A, Huober J, Kandolf R, Budczies J, Denkert C. PARP is expressed in all subtypes of early breast cancer and is a predictive factor for response to neoadjuvant chemotherapy. Eur J Cancer. 2010;8(Suppl):188. [Google Scholar]
- 123.Loibl S, Mueller B, Von Minckwitz G, Blohmer JU, Bois Ad Huober JB, Fend F, Budczies J, Denkert C. PARP expression in early breast cancer and its predictive value for response to neoadyuvant chemotherapy. J Clin Oncol (Meeting Abstracts) 2010;28:10511. [Google Scholar]
- 124.Fougerousse F, Meloni R, Roudaut C, Beckmann JS. Dinucleotide repeat polymorphism at the human poly (ADP-ribose) polymerase gene (PPOL) Nucleic Acids Res. 1992;20:1166. doi: 10.1093/nar/20.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kato N, Morita H, Sugiyama T, Kurihara H, Tsubaki S, Nabika T, Kitamura K, Yamori Y, Yazaki Y. Evaluation of the poly(ADP-ribose) polymerase gene in human stroke. Atherosclerosis. 2000;148:345–352. doi: 10.1016/s0021-9150(99)00284-1. [DOI] [PubMed] [Google Scholar]
- 126.Zaremba T, Ketzer P, Cole M, Coulthard S, Plummer ER, Curtin NJ. Poly(ADP-ribose) polymerase-1 polymorphisms, expression and activity in selected human tumour cell lines. Br J Cancer. 2009;101:256–262. doi: 10.1038/sj.bjc.6605166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cottet F, Blanché H, Verasdonck P, Le Gall I, Schächter F, Bürkle A, Muiras ML. New polymorphisms in the human poly(ADP-ribose) polymerase-1 coding sequence: lack of association with longevity or with increased cellular poly (ADP-ribosyl)ation capacity. J Mol Med. 2000;78:431–440. doi: 10.1007/s001090000132. [DOI] [PubMed] [Google Scholar]
- 128.Lockett KL, Hall MC, Xu J, Zheng SL, Berwick M, Chuang SC, Clark PE, Cramer SD, Lohman K, Hu JJ. The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res. 2004;64:6344–6348. doi: 10.1158/0008-5472.CAN-04-0338. [DOI] [PubMed] [Google Scholar]
- 129.Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem Biophys Res Commun. 2007;354:122–126. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- 130.Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q, Lin D, He F. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378–4384. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- 131.Zhang X, Miao X, Liang G, Hao B, Wang Y, Tan W, Li Y, Guo Y, He F, Wei Q, Lin D. Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res. 2005;65:722–726. [PubMed] [Google Scholar]
- 132.Ogino H, Nakayama R, Sakamoto H, Yoshida T, Sugimura T, Masutani M. Analysis of poly(ADP-ribose) polymerase-1 (PARP1) gene alteration in human germ cell tumor cell lines. Cancer Genet Cytogenet. 2010;197:8–15. doi: 10.1016/j.cancergencyto.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 133.Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, Issaeva N, Sleeth K, Sharma RA, Helleday T. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 134.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 135.Oliver FJ, Ménissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rayet B, Gélinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 137.Annunziata CM, Stavnes HT, Kleinberg L, Berner A, Hernandez LF, Birrer MJ, Steinberg SM, Davidson B, Kohn EC. Nuclear factor kappaB transcription factors are coexpressed and convey a poor outcome in ovarian cancer. Cancer. 2010;116:3276–3284. doi: 10.1002/cncr.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 139.Hinz M, Stilmann M, Arslan SÇ, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-CIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-KB activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 140.Tentori L, Lacal PM, Muzi A, Dorio AS, Leonetti C, Scarsella M, Ruffini F, Xu W, Min W, Stoppacciaro A, Colarossi C, Wang ZQ, Zhang J, Graziani G. Poly(ADP-ribose) polymerase (PARP) inhibition or PARP-1 gene deletion reduces angiogenesis. Eur J Cancer. 2007;43:2124–2133. doi: 10.1016/j.ejca.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 141.Tentori L, Muzi A, Dorio AS, Bultrini S, Mazzon E, Lacal PM, Shah GM, Zhang J, Navarra P, Nocentini G, Cuzzocrea S, Graziani G. Stable depletion of poly (ADP-ribose) polymerase-1 reduces in vivo melanoma growth and increases chemosensitivity. Eur J Cancer. 2008;44:1302–1314. doi: 10.1016/j.ejca.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 142.Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, Keller S, Gassmann M, Hottiger MO. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res. 2008;6:282–290. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- 143.Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997;17:479–482. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- 144.Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am J Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tong WM, Ohgaki H, Huang H, Granier C, Kleihues P, Wang ZQ. Null mutation of DNA strand break-binding molecule poly(ADP-ribose) polymerase causes medulloblastomas in p53(-/-) mice. Am J Pathol. 2003;162:343–352. doi: 10.1016/S0002-9440(10)63825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Conde C, Mark M, Oliver FJ, Huber A, de Murcia G, Ménissier-de Murcia J. Loss of poly(ADP-ribose) polymerase-1 causes increased tumour latency in p53-deficient mice. EMBO J. 2001;20:3535–3543. doi: 10.1093/emboj/20.13.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M, Zevenhoven J, Lau A, Boulter R, Cranston A, O'Connor MJ, Martin NM, Borst P, Jonkers J. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M. Carmichael JOral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 149.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, Lu K, Schmutzler RK, Matulonis U, Wickens M, Tutt A. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 150.Penning TD, Zhu GD, Gandhi VB, Gong J, Liu X, Shi Y, Klinghofer V, Johnson EF, Donawho CK, Frost DJ, Bontcheva-Diaz V, Bouska JJ, Osterling DJ, Olson AM, Marsh KC, Luo Y, Giranda VL. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52:514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 151.Palma JP, Wang YC, Rodriguez LE, Montgomery D, Ellis PA, Bukofzer G, Niquette A, Liu X, Shi Y, Lasko L, Zhu GD, Penning TD, Giranda VL, Rosenberg SH, Frost DJ, Donawho CK. ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–7290. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- 152.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, Ji J, Monks A, Low JA, Chen A, Murgo AJ, Collins J, Steinberg SM, Eliopoulos H, Giranda VL, Gordon G, Helman L, Wiltrout R, Tomaszewski JE, Doroshow JH. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.O'Shaughnessy J, Osborne C, Pippen J, Yoffe M, Patt D, Monaghan G, Rocha C, Ossovskaya V, Sherman B, Bradley C. Efficay of BSI-201, a poly(ADP-ribose) polymerase-1 (PARP-1) inhibitor, in combination with gemcitabine/ carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial. J Clin Oncol. 2009;27:3. [Google Scholar]
- 154.Liang H, Tan AR. Iniparib, a PARP1 inhibitor for the potential treatment of cancer, including triple-negative breast cancer. IDrugs. 2010;13:646–656. [PubMed] [Google Scholar]
- 155.Drew Y, Mulligan EA, Vong WT, Thomas HD, Kahn S, Kyle S, Mukhopadhyay A, Los G, Hostomsky Z, Plummer ER, Edmondson RJ, Curtin NJ. Therapeutic Potential of Poly(ADP-ribose) Polymerase Inhibitor AG014699 in Human Cancers With Mutated or Methylated BRCA1 or BRCA2. J Natl Cancer Inst. 2010 doi: 10.1093/jnci/djq509. in press. [DOI] [PubMed] [Google Scholar]
- 156.Miknyoczki S, Chang H, Grobelny J, Pritchard S, Worrell C, McGann N, Ator M, Husten J, Deibold J, Hudkins R, Zulli A, Parchment R, Ruggeri B. The selective poly(ADP-ribose) polymerase-1(2) inhibitor, CEP-8983, increases the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan but does not potentiate myelotoxicity. Mol Cancer Ther. 2007;6:2290–2302. doi: 10.1158/1535-7163.MCT-07-0062. [DOI] [PubMed] [Google Scholar]
- 157.Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C, Lamartina S, Monteagudo E, Ontoria JM, Orsale MV, Palumbi MC, Pesci S, Roscilli G, Scarpelli R, Schultz-Fademrecht C, Toniatti C, Rowley M. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and -2 mutant tumors. J Med Chem. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- 158.Bedikian AY, Papadopoulos NE, Kim KB, Hwu WJ, Homsi J, Glass MR, Cain S, Rudewicz P, Vernillet L, Hwu P. A phase IB trial of intravenous INO-1001 plus oral temozolomide in subjects with unresectable stage-III or IV melanoma. Cancer Invest. 2009;27:756–763. doi: 10.1080/07357900802709159. [DOI] [PubMed] [Google Scholar]
- 159.Karlberg T, Hammarström M, Schütz P, Svensson L, Schüler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]