Abstract
This study shows that state-of-the-art liquid chromatography (LC) and mass spectrometry (MS) can be used for rapid verification of identity and characterization of sequence variants and posttranslational modifications (PTMs) for antibody products. A candidate biosimilar IgG1 monoclonal antibody (mAb) was compared in detail to a commercially available innovator product. Intact protein mass, primary sequence, PTMs and the micro-differences between the two mAbs were identified and quantified simultaneously. Although very similar in terms of sequences and modifications, a mass difference observed by LC-MS intact mass measurements indicated that they were not identical. Peptide mapping, performed with data independent acquisition LC-MS using an alternating low and elevated collision energy scan mode (LC-MSE), located the mass difference between the biosimilar and the innovator to a two amino acid residue variance in the heavy chain sequences. The peptide mapping technique was also used to comprehensively catalogue and compare the differences in PTMs of the biosimilar and innovator mAbs. Comprehensive glycosylation profiling confirmed that the proportion of individual glycans was different between the biosimilar and the innovator, although the number and identity of glycans were the same. These results demonstrate that the combination of accurate intact mass measurement, released glycan profiling and LC-MSE peptide mapping provides a set of routine tools that can be used to comprehensively compare a candidate biosimilar and an innovator mAb.
Key words: biosimilar mAb, innovator mAb, molecular similarity, sequence variants, posttranslational modifications, N-linked glycosylation, chemical degradations, micro-heterogeneities, characterization, intact protein mass measurement, peptide mapping, glycan profiling, LC-MS, LC-fluorescence, MALDI MS
Introduction
Recombinant monoclonal antibodies (mAbs) are large, heterogeneous proteins that have emerged as therapeutics due to their predictable properties, controlled functions and long circulating life. MAbs represent a class of advanced, but expensive, medicines. With healthcare costs increasing to more than 16% of total gross domestic product in the US, lowering the cost of medicine is an economic and public health priority.1 There has recently been increasing interest in developing less costly biosimilar mAbs by both innovator and generic drug companies.2 Another driving force for the interest in biosimilars is the upcoming patent expiration for marketed protein products. In a recent workshop, the feasibility of the development and authorization of mAbs using European Medicines Agency's (EMA) biosimilar regulatory pathways was discussed.3
In Europe, EMA has established guidelines and defines a biosimilar as “a medicine which is similar to a biological medicine that has already been authorized.”4 A number of biosimilar products are already marketed in Europe, although none are mAbs5 and developing biosimilar products is challenging.6 Few biosimilar products have been approved in the US, where they are referred to as “follow-on protein products” or “follow-on biologics” (this article uses the term biosimilars throughout), due to restrictions in the legislative pathways used by the Food and Drug Administration to approve therapeutic agents. This situation may change as a consequence of healthcare legislation passed in March 2010 that contains specific provisions for biosimilars.7 The stated intent was to create an approval pathway similar to that used for small molecule generic drugs to potentially reduce healthcare costs and expand competition. Nonetheless, the bill was severely criticized by proponents of biosimilars because of the 12-year “data exclusivity” protection afforded biotherapeutics.8
Under these circumstances, innovators have an interest in ensuring that their products are well-characterized so that biosimilars undergo similar rigorous characterization. Conversely, biosimilar manufacturers must ensure that their product conforms as closely as possible to the existing product, reducing the need for expensive clinical trials and speed time to market. Therefore, all parties have an interest in performing comprehensive analysis of their products.
For innovative products as well as biosimilars, criteria for approval include quality, efficacy and safety. The objective of the biosimilar industry is to develop a product that is, as much as possible, similar to a marketed innovator product. Therefore, the quality, non-clinical and clinical development programs are designed to demonstrate similarity of a biosimilar to its innovator product in every aspect. A number of physicochemical and biological methods are required by regulatory authorities for characterization of mAbs.3,9 Participants in EMA's 2009 workshop questioned just how similar a biosimilar must be and commented on the difficulty of obtaining information on every atom in a product and the need for a biosimilar mAb to have the same distribution of antibody variants as the innovator product.3 It was emphasized that biosimilars must have the same amino acid sequence as the reference product, even though “both reference and biosimilar mAb products will be micro-heterogeneous mixtures of a large number of post-translationally modified molecular species.” Comments reported included the assertion that the strategy of comparability testing of reference products could be re-applied to the physicochemical testing of biosimilars and indeed that idea is applied in this paper. In the work presented here, primary sequence in a biosimilar candidate is shown to be different from that of the innovator product, showing that sequence variants can be discovered as part of routine analytical workflow.
Assessing the molecular similarity of a candidate biosimilar to the innovator product is a critical task during development of a biosimilar mAb. The biosimilar industry thus needs rapid and reliable analytical methods to establish molecular similarity required by regulators. Defining the molecular similarity of two IgG1 mAbs, each with an average molecular mass of 150 kDa, can be challenging. Unlike small molecule generic products, protein products generally exhibit micro-heterogeneities. Apart from primary sequence, it has been established that glycosylation can be critical for the biological function of mAbs.10–14 Product-related substances or impurities such as deamidated, isomerized or oxidized forms, or protein aggregates15–21 that may be introduced during cloning and production processes can affect both tertiary structure and antigen binding properties of mAbs.
In this study, molecular similarity between a candidate biosimilar mAb and its innovator product (trastuzumab) was assessed in detail by using a panel of advanced liquid chromatography (LC) and mass spectrometry (MS) technologies. Protein identity, sequence variants, glycosylation profile and other posttranslational modifications (PTMs) and impurities were rapidly characterized with the aid of software programs for automated LC-MS data processing and evaluation. Extensive comparison of N-linked glycosylation in the heavy chains was performed at the protein, peptide and free glycan levels using intact protein mass measurement, by peptide mapping with data independent acquisition LC-MS using an alternating low and elevated collision energy scan mode (LC-MSE), and by free glycan profiling.
Results
Revealing a mass difference in the biosimilar at the intact protein level.
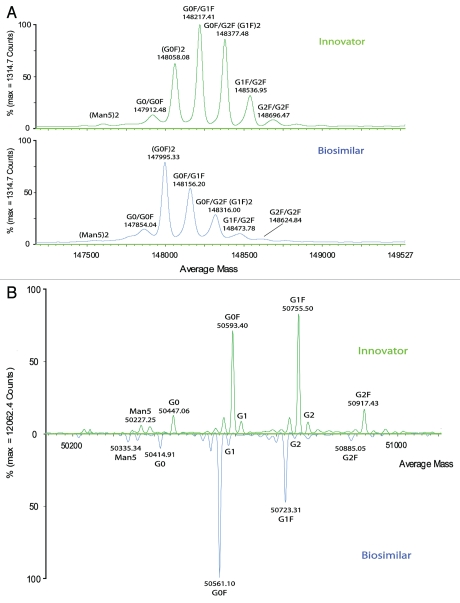
Like all IgG1 antibodies, trastuzumab has two light and two heavy chains connected by disulfide bonds. A consensus N-site in the Fc region of heavy chains is occupied with complex N-linked glycans.20 A combination of high resolution reverse-phase LC and accurate mass MS was used to measure the molecular masses of the intact antibody and single chains after reduction with dithiothreitol (DDT). A data processing application manager (BiopharmaLynx 1.2) automatically deconvoluted the collected MS spectra to average mass for each LC peak. The deconvoluted MS spectra of both intact and reduced forms of both innovator and biosimilar mAbs are shown in Figure 1. Both mAbs showed multiple peaks at the intact protein level (Fig. 1A), which are attributable to the glycoforms typically found in all IgG1 antibodies due to the N-linked glycosylation forms present in the conserved region of heavy chains.22 The glycosylation profiles were similar to those observed in a study that determined the lot-to-lot heterogeneity of trastuzumab.23 Details of the glycoforms of the two mAbs investigated here are discussed in the “Comprehensive Glycosylation Study” section.
Figure 1.
A comparison of deconvoluted masses (processed by BiopharmaLynx 1.2 using MaxEent1) between innovator and biosimilar mAbs. (A) intact mAbs; (B) mirror plot of heavy chain. (C) mirror plot of light chain.
When compared to a sample of the innovator product, the biosimilar mAb had glycoforms that were similar (attributable at the intact protein level), but each of the glycosylated peaks was offset by a consistent difference. Each peak had a mass difference ∼64 Da lower for the biosimilar candidate than the innovator product for each of the corresponding glycoforms.
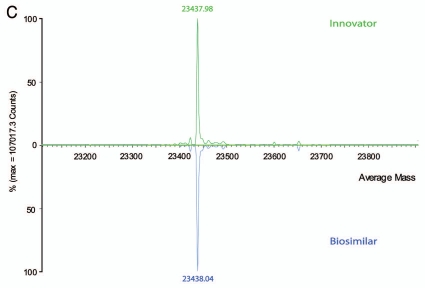
To make further analysis more straightforward, the samples were then reduced with DTT to remove complexity caused by the pairing of oligosaccharides at the intact protein level. Inspection of the individual heavy and light chains of reduced mAbs confirmed that N-linked glycoforms were located in the heavy chains as expected. The same deconvoluted mass (MH+, 23438.0 Da) was measured for the light chains of both mAbs (Fig. 1C); however, in the heavy chains, a consistent ∼32 Da lower mass per heavy chain (Fig. 1B) was observed for each major glycosylated peak of the biosimilar candidate compared with the innovator product. It is logical to hypothesize that the origin of the overall mass difference lay within the heavy chain of the candidate biosimilar mAb.
Peptide mapping to locate sequence variants and PTMs.
Peptide mapping is used to investigate differences in protein sequences and PTMs between the biosimilar candidate and the innovator product. Employing a data independent acquisition reverse-phase LC-MS with alternate low and elevated collision energy scanning (LC-MSE) also brings additional benefits for peptide maps.24 Unlike data dependent acquisition (DDA) LC-MS/MS, MSE generates fragments without precursor selection.25 Therefore all charge states of peptide precursors are fragmented in an approach unbiased by the user or by the precursor signal. The data provided by MSE acquisition allows for sequencing peptides above the limit of detection and for accurate quantification by MS signals, including low-abundance components. An LC-UV map was also collected in parallel for comparison purposes.
In a previous study,24 we established that peptide mapping with LC-MSE using tryptic digests of yeast enolase and alcohol dehydrogenase (ADH) was able to achieve high sequence coverage (97%) of targeted proteins, identify ADH sequence variants26–28 and characterize low-abundance protein impurities and site- specific modifications in a single LC-MS analysis. We applied the same peptide mapping method here, collecting two sets of data in parallel using LC-MSE acquisition. One data set contains low collision energy data (MS, effectively the accurate mass of peptide precursors) and the second data set the elevated collision energy data (MSE, all of the peptide fragments). The unbiased and reproducible nature of the data independent acquisition of LC-MSE permits accurate quantification over a wide dynamic range.29 Additionally, the same set of MS and MSE data is available for peptide sequence confirmation.24,25,29–31 In this study, the methodology was applied to tryptic digests of the biosimilar and innovator mAbs in order to compare their sequences and distinguish differences between them. An automated data processing and annotation procedure was applied with the same application manager (BiopharmaLynx 1.2) to the entire data collected. For peptide maps, the application manager automatically assigns peptide identity based on the accurate masses of charge-reduced and isotope-deconvoluted singly charged precursors and provides their MS signal intensities for accurate quantification. Meanwhile, singly charged fragment ion spectra of peptides are simultaneously provided for sequence confirmation.
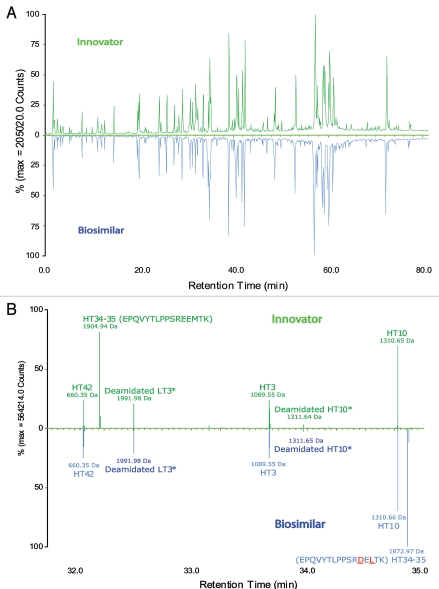
Figure 2 is a mirror plot view of the LC-MS peptide maps of the biosimilar and innovator mAbs presented either as total ion chromatograms (TICs, Fig. 2A) or charge-reduced and isotope-deconvoluted singly charged ions (MH+), processed by the application manager. The MS intensity is shown along with retention time (RT; subsets of the converted chromatogram as shown in Fig. 2B and C). Identified peptides were automatically annotated by the application manager with annotation scheme depicting whether they were tryptic peptides from the heavy chain (HTxx) or light chain (LTxx). Additionally, the converted singly charged MSE fragment spectra were available to confirm the amino acid sequences of assigned peptides and locate their modifications, as demonstrated in Figures 3 and 6.
Figure 2.
Mirror plots of LC-MS peptide maps from 4-hours tryptic digests of the innovator and biosimilar mAbs. (A) LC-MS (TIC) chromatograms; (B) a zoom view of charge-reduced, isotope-deconvoluted LC-MS chromatograms from 32.0–35.0 min. (C) a zoom view of charge-reduced, isotope-deconvoluted LC-MS chromatograms from 3.5–7.0 min. The zoom chromatograms were processed by BiopharmaLynx 1.2 and the peaks were annotated based on accurate mass MH+ of peptides in an automated mode.
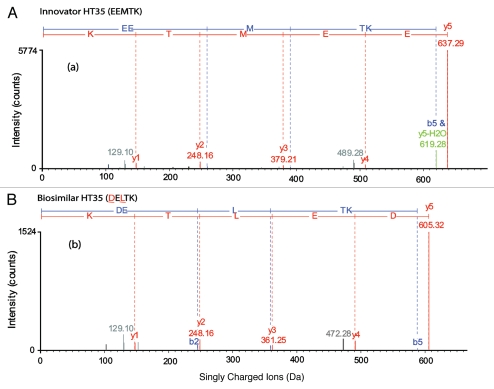
Figure 3.
Charge-reduced, isotope-deconvoluted MSE spectra (processed by BiopharmaLynx 1.2) of tryptic peptide HT35. (A) HT35 (EEMTK) of the innovator; (B) HT35 (DELTK) of the biosimilar mAb. The colors in the spectra represent: y ions (red); b ions (blue); y or b ions with a water or ammonia lost (green); unassigned ions (grey).
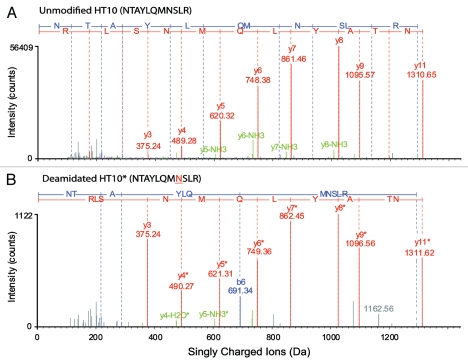
Figure 6.
Charge-reduced, isotope-deconvoluted MSE spectra (processed by BiopharmaLynx 1.2) of (A) unmodified and (B) N-deamidated tryptic peptide HT10 (NTAYLQMNSLR). The consistent 1-Da mass increase in y-serious ions from y4 in deamidated HT10 compared to unmodified HT10 clearly indicates the deamidated N site. The meaning of colors in the spectra is the same as described in Figure 3.
Sequence variants.
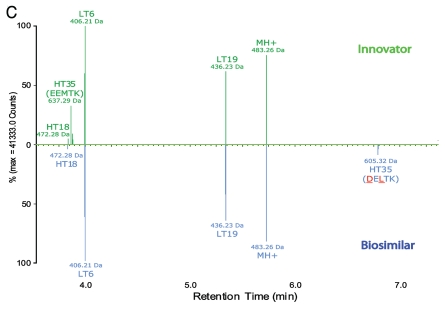
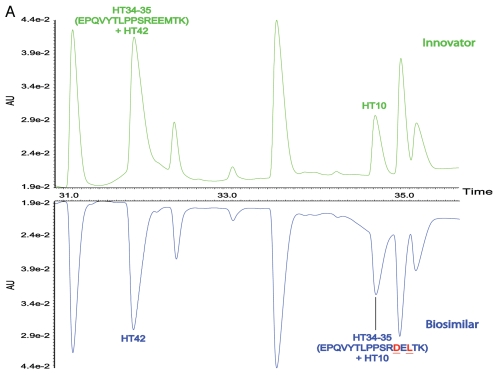
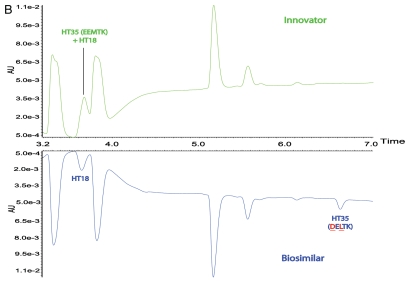
Although the differences between the LC-UV traces of the tryptic digests of the biosimilar and the innovator product were largely identical, varying only in peak shapes, the processed LC-MS maps showed two key differences that were not detectable in the UV traces. Peptides HT34–35 and HT35 of the innovator product were not identified in the map of the biosimilar (Fig. 2B and C). Conversely, two new masses at RT 34.87 and 6.79 min (also shown in Fig. 2B and C), were observed in the biosimilar map; these did not have corresponding matches in the innovator map.
To identify the two new masses in the biosimilar map and uncover their potential relationship with the unmatched HT34–35 and HT35 peptides of the innovator, we first examined the MSE spectra of HT34–35 and HT35 peptides in the innovator map and the fragments of the two unknown masses in the biosimilar. The automated assignment of the HT34–35 peptide (EPQVYTLPPSREEMTK with one miscleavage, RT = 32.21 min, MH+ = 1904.93 Da) and HT35 (EEMTK, RT = 3.86 min, MH+ = 637.28 Da) were both correct according to the fragment ion data (see Fig. 3A for the processed MSE spectrum of HT35); however, there was no matching fragment ion data for these two sequences in the biosimilar data. The difference was confirmed in the raw (unprocessed) data by the extracted ion chromatograms (XIC) of peptides HT34–35 and HT35. XIC peaks of precursors with peptide sequences EPQVYTLPPSREEMTK and EEMTK only appear in LC-MS data of innovator tryptic digests, but not in the biosimilar (data not shown).
The two new singly charged masses in the biosimilar were not matched to corresponding tryptic peptides in the innovator map. One had MH+ 1872.96 Da (at RT 34.87 min) and the other had MH+ 605.31 Da (at RT 6.79 min), showing a 31.97 Da mass difference from HT34–35 and HT35 of innovator mAb, respectively (Fig. 2B and C). The mass difference was consistent with the observation of intact mass measurements, i.e., the heavy chain of the biosimilar had ∼32 Da mass lower than the innovator mAb. Therefore, a molecular difference, potentially due to PTM or different composition of amino acid residues, must be present in HT34–35 and HT35 of the biosimilar compared with the innovator product. Taking into account that HT34–35 is a miscleavage of peptide HT35, the molecular and mass difference must logically be located in tryptic peptide HT35.
An examination of the processed MSE data of the biosimilar indicated that there were indeed clear fragments for the two new masses, but no confirmatory sequence assignment. Only partial sequence information of HT34–35 and HT35 could be derived. The sequence of the innovator mAb at peptide HT35 was found to be consistent with the published version,20 but that sequence was not consistent with the one available from DrugBank.32 In the heavy chain region of the latter sequence, a two amino acid difference was apparent from the sequence used by the application manager to match the innovator. Therefore, using the existing data already acquired, the fragmentation pattern could be re-matched with the alternative sequence obtained from the DrugBank. Figure 3B shows that a perfect match resulted when this alternative sequence was used, confirming that the sequence of HT35 in the biosimilar was DELTK.
That the alternative sequence variant was not present in the innovator product was easily checked. This was done by manually extracting precursor masses in the raw data with MH+ of 605.31 and 1872.96 Da to confirm the presence in the biosimilar, and absence in the innovator. Indeed, XIC peaks of precursors with peptide sequences EPQVYTLPPSRDELTK and DELTK only exist in LC-MS data of biosimilar tryptic digests (data not shown).
The identified sequence variants demonstrate that the biosimilar candidate is a different allotype compared with the innovator (trastuzumab) mAb.33,34 In Europe and the US, biosimilars must have the identical amino acid sequence as innovator protein therapeutics.3 Biological, non-clinical and clinical studies equivalent to those required for a novel product would likely be required for this candidate “biosimilar” mAb to be approved in these regions.35
It should be stressed that the above sequence difference can barely be observed in LC-UV tryptic maps of the two mAbs due to co-elution with other peptides. Except for peak shape, little difference of the LC peaks corresponding to HT34–35 was observed in the UV maps (Fig. 4). The innovator HT34–35 co-eluted with HT42 in the innovator map and the biosimilar HT34–35 co-eluted with HT10 in the biosimilar map. Similarly, HT35 also co-eluted with HT18 in the innovator map. Chromatographic co-elution of these peptides did not have an impact on either the MS information obtained, nor on the ability to quantify low-abundance impurities, such as modified peptides at substoichiometric concentrations, by MS signal because of the unbiased nature of the methodology.
Figure 4.
Mirror plots of two zoomed LC-UV chromatograms from 4-hour tryptic digests of the innovator and biosimilar mAbs, (A) from 30.5–36.0 min, (B) from 3.2–7.0 min.
Except for two small peptides HT28 (CK) and HT32 (AK), all peptide sequences were at least verified in 1-miscleavage tryptic peptide. All the verifications were confirmed by MSE spectra. Except for the difference on HT35, no other additional sequence difference was detected between the two mAbs.
Comprehensive glycosylation study.
The remaining differences in the glycosylation heterogeneities between the biosimilar candidate and the innovator product could also be related to the sequence variants. In a comprehensive study of glycosylation, three methods were applied: (1) Intact protein mass measurement and deconvolution, (2) Glycopeptide and glycoforms assignment within peptide maps and (3) MALDI-QTof MS characterization of released free glycans and LC-fluorescence quantification of 2-aminobenzamide (AB) labeled free glycans.
Intact protein mass analysis of glycosylation heterogeneities.
As shown in Figure 1A, at the intact protein level, the peaks of (G0F)2, G0F/G1F, (G1F)2, G1F/G2F and (G2F)2 could all be readily assigned based on mass differences of the sugars (see Fig. 5 for the structures of corresponding glycans). For each mAb, the five main peaks demonstrated a characteristic sequential mass difference of ∼162 Da, consistent with varying numbers of terminal galactose residues from the two core glycan structures. A peak corresponding to G0/G0F, which had a 146 Da mass difference from (G0F)2 due to incomplete occupancy of a fucosylation site on one of the core glycan structures, was observed in each mAb. In addition, a low-level Man5/Man5 form was detected in both mAbs.
Figure 5.
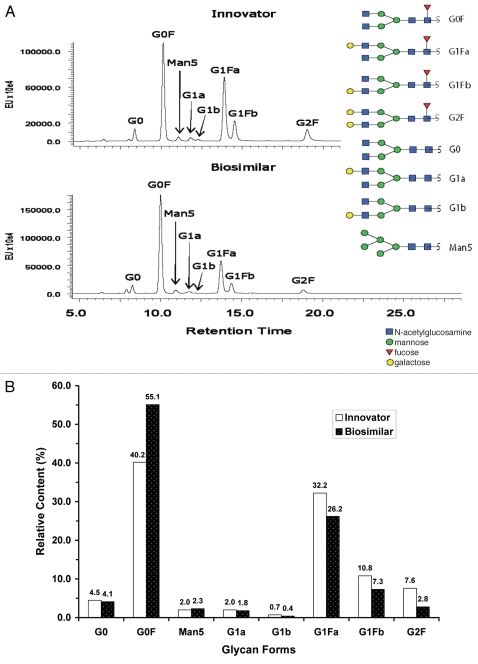
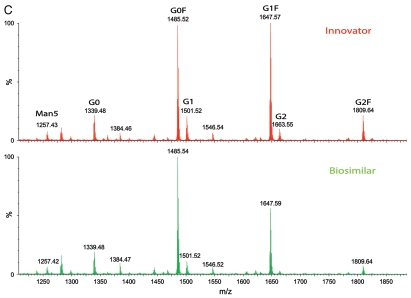
A comparison of LC-fluorescence separation and quantification of 2-AB labeled free glycans and MALDI MS profiling of unlabeled free glycans released from the innovator and biosimilar mAbs. (A) LC-fluorescence chromatograms of 2-AB labeled glycans. (B) Relative contents of 2-AB labeled glycans quantified by integrated peak area of LC-fluorescence chromatograms (C) MALDI MS profiling of unlabeled free glycans.
Similarly, the heavy chains show multiple masses (Fig. 1B) due to the attached biantennary oligosaccharides G0F, G1F and G2F, as well as low-level G0, G1, G2 and high mannose forms such as Man5. Additional details on glycosylation were investigated by peptide mapping and free glycan profiling. The intact/reduced heavy chain MS analyses of the two antibodies (Fig. 1A and B) revealed what appeared to be different glycosylation patterns. While G1F is the most abundant glycosylation form in the innovator product, G0F was the most abundant in the biosimilar (Fig. 1B). The ratios of G0F/G1F and G0F/G2F were significantly increased from the innovator to the biosimilar mAb. As expected, no glycosylation was observed on the light chains.
Peptide mapping assignment of N-linked glycopeptide and glycoforms.
Consistent with the previous study,20 N-glycosylation was only observed in an asparagine (N) residue located in the heavy chain tryptic peptide HT25 (EEQYNSTYR) for both mAbs. Nearly all (99%) of the peptide was glycosylated and a total of seven N-linked glycoforms were identified (see Fig. 5 for the corresponding glycan structures of the glycoforms). G0F, G1F and G2F were major glycoforms of glycosylated HT25, assigned automatically by the application manager by their accurate mass and confirmed by examining the MSE spectra, while G0, G1, G2 and Man5 were minor glycoforms determined by accurate masses. Although the number and identity of glycoforms identified from both biosimilar and innovator mAbs were the same, the pattern (or relative content) of the glycoforms was different. For example, the relative content of G0F was increased in the biosimilar compared with the innovator product, but decreased for G1F and G2F. This result was consistent with the intact protein mass measurements and free glycan profiling analyses.
Profiles of released glycans.
The 2-AB labeled glycans, released from mAbs by PNGase F, were separated using hydrophilic interaction chromatography (HILIC) LC with fluorescence detection for quantification. The glycan mass profiling and structure elucidation were performed by MALDI QTof MS and MS/MS in a separate experiment without 2-AB labeling. Seven major glycans (G0F, G1F, G2F, G0, G1, G2 and Man5; Fig. 5) were identified and quantified for both antibodies, and the result was consistent with peptide mapping data showing seven glycoforms in the N-linked glycopeptide HT25. Further, two isomeric forms of G1F and G1 were separated and quantified by HILIC chromatography. LC-fluorescence quantification data (Fig. 5B, based on integrated peak areas in Fig. 5A) again show that the glycan proportions in the antibodies were different. The relative ratios of G0F/G1F and G0F/G2F were significantly increased in the innovator compared with the biosimilar mAb. While G1F was the most abundant glycan structure in the innovator product as demonstrated by both MALDI-MS measurements of unlabeled free glycans (Fig. 5C) and LC-fluorescence quantification (the sum of G1Fa and G1Fb in Fig. 5B), G0F became the most abundant in the biosimilar. These glycan pattern changes were confirmed by all three orthogonal methods applied in this study.
Differences in other PTMs.
Besides N-linked glycosylation, other PTMs were also characterized as part of the routine peptide mapping by LC-MSE. The methodology provides three pieces of additional information for all modifications:24 (1) MSE is able to locate modification sites and determine modification type; (2) the intensities of precursors can be used to estimate the concentration of PTMs and (3) differences in modification levels can be used to compare samples.
An example of the identification of a deamidated and an unmodified HT10 (NTAYLQMNSLR) is shown in Figure 2B. Sequences were verified by MSE spectra and the modification site of deamidated HT10 was located (Fig. 6). The relative concentration of deamidated HT10 could be directly related to unmodified HT10 in each mAb and between the two mAbs by MS signal intensities.
Thirteen N-deamidation sites, three aspartic acid (D)-isomeri-zation sites and two methionine (M)-oxidation sites were located in 15 tryptic peptides (11 on heavy chain and 4 on light chain). All the modifications were confirmed by MSE spectra, and the modification sites were located by y- or b-series ions (as illustrated in Fig. 6). The modification sites and level (%) in both the biosimilar and innovator mAbs are listed in Table 1.
Table 1.
Posttranslational modifications (PTMs) other than glycosylation characterized from the candidate biosimilar and innovator mAbs
| Relative content in mAb** | |||||||
| Protein | Peptide | Modifications | Sequence and modification site | RT (min) | Mass (Da) | Innovator (%) | Biosimilar (%) |
| Heavy Chain | HT6 | No | IYPTNGYTR | 19.77 | 1083.53 | 55.77 | 52.02 |
| Heavy Chain | HT6* | N-deamidation | IYPTNGYTR | 20.07/21.37 | 1084.51 | 42.62 | 46.32 |
| Heavy Chain | HT6* | N-Succinimide | IYPTNGYTR | 22.65 | 1067.51 | 1.61 | 1.66 |
| Heavy Chain | HT10 | No | NTAYLQMNSLR | 34.78 | 1039.64 | 92.96 | 92.53 |
| Heavy Chain | HT10* | N-deamidation | NTAYLQMNSLR | 33.96/36.23 | 1040.62 | 5.83 | 6.13 |
| Heavy Chain | HT10* | N-deamidation | NTAYLQMNSLR | 35.76/37.11 | 1040.62 | 1.21 | 1.34 |
| Heavy Chain | HT12 | No | WGGDGFYAMDYMGQGTLVTVSSASTK | 60.13 | 2873.25 | 83.54 | 84.8 |
| Heavy Chain | HT12* | D-ismerization | WGGDGFYAMDYMGQGTLVTVSSASTK | 60.57 | 2873.25 | 16.46 | 15.2 |
| Heavy Chain | HT15 | No | DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL YSLSSVVTVPSSSLGTQTYIC#NVNHKPSNTK |
72.46 | 6172.31 | 93.43 | 93.61 |
| Heavy Chain | HT15* | N-deamidation | DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL YSLSSVVTVPSSSLGTQTYIC#NVNHKPSNTK |
72.92 | 6173.59 | 6.57 | 6.39 |
| Heavy Chain | HT21 | No | DTLMISR | 25.97 | 834.43 | 95.61 | 89.67 |
| Heavy Chain | HT21* | M-oxidation | DTLMISR | 21.58 | 850.42 | 4.39 | 10.33 |
| Heavy Chain | H23 | No | FNWYVDGVEVHNAK | 40.65 | 1676.79 | 91.66 | 91.16 |
| Heavy Chain | H23 | D-isomerization | FNWYVDGVEVHNAK | 39.77 | 1676.79 | 2.92 | 2.98 |
| Heavy Chain | HT23* | N-deamidation | FNWYVDGVEVHNAK | 41.6/42.35 | 1677.77 | 5.42 | 5.86 |
| Heavy Chain | HT26 | No | VVSVLTVLHQDWLNGK | 59.46 | 1807 | 62.66 | 61.69 |
| Heavy Chain | HT26* | N-deamidation | VVSVLTVLHQDWLNGK | 60.15/60.53 | 1807.98 | 28.14 | 29.48 |
| Heavy Chain | HT26* | N-Succinimide | VVSVLTVLHQDWLNGK | 61.7 | 1790.98 | 9.2 | 8.83 |
| Heavy Chain | HT36 | No | NQVSLTC#LVK | 39.12 | 1160.62 | 97.92 | 97.9 |
| Heavy Chain | HT36* | N-deamidation | NQVSLTC#LVK | 40.97/42.14 | 1161.6 | 2.08 | 2.1 |
| Heavy Chain | HT37 | No | GFYPSDIAVEWESNGQPENNYK | 48.88 | 2543.12 | 60.55 | 61.07 |
| Heavy Chain | HT37* | N-deamidation | GFYPSDIAVEWESNGQPENNYK | 48.67/49.52 | 2544.1 | 21.29 | 20.93 |
| Heavy Chain | HT37* | N-deamidation | GFYPSDIAVEWESNGQPENNYK | 49.34 | 2544.1 | 8.7 | 8.5 |
| Heavy Chain | HT37* | N-Succinimide | GFYPSDIAVEWESNGQPENNYK | 50.21 | 2527.1 | 9.55 | 9.5 |
| Heavy Chain | HT38 | No | TTPPVLDSDGSFFLYSK | 53.23 | 1872.91 | 98.08 | 98.32 |
| Heavy Chain | HT38* | D-ismerization | TTPPVLDSDGSFFLYSK | 52.56 | 1872.91 | 1.92 | 1.68 |
| Heavy Chain | HT41 | No | WQQGNVFSC#SVMHEALHNHYTQK | 39.1 | 2800.26 | 94.69 | 95.39 |
| Heavy Chain | HT41* | N-deamidation | WQQGNVFSC#SVMHEALHNHYTQK | 39.82 | 2801.24 | 5.31 | 4.61 |
| Light Chain | LT1 | No | DIQMTQSPSSLSASVGDR | 35.06 | 1877.88 | 99.65 | 99.41 |
| Light Chain | LT1* | M-oxidation | DIQMTQSPSSLSASVGDR | 28.95 | 1893.87 | 0.35 | 0.59 |
| Light Chain | LT3 | No | ASQDVNTAVAWYQQKPGK | 31.38 | 1989.99 | 72.82 | 71.77 |
| Light Chain | LT3* | N-deamidation | ASQDVNTAVAWYQQKPGK | 30.02/32.51 | 1990.97 | 27.18 | 28.23 |
| Light Chain | LT11 | No | SGTASVVC#LLNNFYPR | 60.39 | 1796.89 | 97.06 | 96.87 |
| Light Chain | LT11* | N-deamidation | SGTASVVC#LLNNFYPR | 52.38/62.44 | 1797.87 | 2.94 | 3.13 |
| Light Chain | LT14 | No | VDNALQSGNSQESVTEQDSK | 20.19 | 2134.96 | 99.02 | 98.99 |
| Light Chain | LT14* | N-deamidation | VDNALQSGNSQESVTEQDSK | 20.93 | 2135.94 | 0.98 | 1.1 |
modified peptides;
Calculated from precursor intensity according to reference 24; C#, Carbamidomethyl C.
Rates of N-deamidation and D-isomerization are dependent on a number of factors, including the nature of sample solution, pH, temperature, primary sequence and 3-dimentional structure,36–39 and generally increase with the time of sample storage. It is also well-known that artificial deamidation can be introduced during sample preparation,40 particularly during tryptic digestion in basic pH solutions such as ammonium bicarbonate or tris buffer. Methods to differentiate N-deamidation that occurred prior to and during sample preparation,41 and to minimize artificial modifications of mAb digests42 have been reported. In this study, a RapiGest-assisted 4-hour tryptic digestion protocol was applied for preparation of tryptic digests of the two mAbs. This method achieves a full digestion and has a relatively short digestion time compared with standard overnight tryptic digestion protocols.40
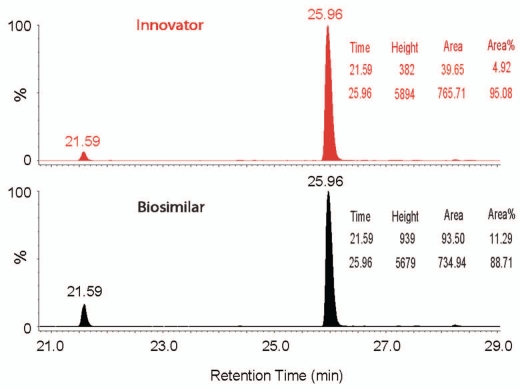
The N-deamidation and D-isomerization were comparable for each corresponding site in the two mAbs; however, the amount of oxidation on the 2 M-sites was doubled in the biosimilar compared to the innovator product. For example, only about 4% M-oxidized HT21* (DTLMISR) was detected in the innovator, but more than 10% M-oxidation was detected in HT21* of the biosimilar mAb. Quantification of oxidized and unmodified HT21 using XIC peak area (Fig. 7), demonstrated the same difference. M-oxidized HT21* co-eluted with other peptides, again indicating the practical utility of LC-MSE methodology.
Figure 7.
A comparison of XIC chromatograms of oxidized (21.6 min) and unmodified (26.0 min) tryptic peptide HT21 (DTLMISR) in the innovator and biosimilar mAbs. The relative quantification of oxidized and unmodified HT21 (based on peak area) is inserted.
Discussion
Over 20 mAbs have been approved and marketed19,43 and data on potency assays and safety for these products are available. A wealth of experience in the development of mAbs is also available. This makes it possible to develop less expensive biosimilar mAbs based on the philosophy of similarity. However, it is important to note that a biosimilar product may potentially exhibit a range of differences compared to the innovator product. The molecular equivalence needs to be assessed using a panel of analytical methods, and this task is not trivial. Although a number of physicochemical and biological methods are available for characterization of mAbs,3,9,44 the ability to routinely compare a biosimilar mAb to the corresponding innovator product on a molecular level has been challenging. Here, three LC and MS based approaches on a common platform were successfully applied to define the molecular similarity between a biosimilar and innovator mAb.
Intact protein mass measurement was chosen for a fast evaluation of obvious differences between the biosimilar and the innovator protein therapeutics. Acquired mass spectra (Fig. 1A–C) indicated that the innovator and biosimilar mAbs have different glycosylation patterns. In addition, analysis of the reduced protein indicated that there was a mass difference between the heavy chains of the two mAbs (Fig. 1B). While MS analysis of intact protein is fast and useful, it cannot specify the exact nature of the differences. In addition, intact mass measurement has limitations for detection and characterization of chemical degradations with small or no mass difference such as N-deamidation, D-isomerization and M-oxidation that were investigated in this study. The detailed study of modifications and variants presented here was performed with LC-MSE peptide mapping.
Peptide mapping of enzymatic (often tryptic) digests of large proteins such as mAbs is challenging in general because of the large number of peptides present in the digest that must be separated by LC. LC-MS was critical to identify the differences between the two mAb samples. The UV chromatogram did not reveal distinct differences between the two peptide maps due to the limitation of UV detection for differentiating co-eluting peptides (Fig. 4A and B). In contrast, the LC-MS chromatograms (Fig. 2B and C) show unambiguously the presence of unique peptides in both peptide maps, even though they co-eluted with other components.
We previously developed an LC-MSE peptide mapping method that can simultaneously identify and quantify low-abundance impurities, sequence variants and site-specific modifications.24 The MSE acquisition provided raw data that was rapidly processed to confirm the sequences of detected peptides, including sequence variants (Fig. 3) and PTMs (Fig. 6). In this peptide mapping methodology, data processing and interpretation were facilitated with an application manager having built-in algorithms for comparison, which automated and accelerated data analysis by removing tedious data interpretation work.
While the reverse-phase LC-MS based accurate mass intact protein measurements detected that the biosimilar mAb had a mass ∼64 Da less compared with the innovator product (Fig. 1A), and defined that the mass difference was located on the heavy chains (∼32 Da each; Fig. 1B), peptide mapping with reverse-phase LC-MSE identified the difference as a sequence variant of peptide HT35 (Figs. 2 and 3). Heavy chain sequences with two different amino acids on HT35 (D359 and L361 instead of E359 and M361) produced a 31.97 Da lower mass for the biosimilar compared with the innovator product. No other sequence differences were detected between the two mAbs.
The same glycoforms, but different patterns, were observed by both intact mass measurements (Fig. 1A and B) and peptide mapping experiments. While intact mass measurement can locate glycoforms in the heavy chain, peptide mapping determined only N-linked glycoforms on N300 of peptide HT25 (EEQYNSTYR) as expected. Again, as expected, G0F, G1F and G2F were the major glycoforms, but their relative amounts were different between the two mAbs. For example, G1F was the most abundant glycoform in the innovator product, but G0F was the most abundant glycoform in the biosimilar mAb.
While both intact MS analysis and peptide maps clearly reveal differences in the relative glycosylation patterns for the investigated antibodies, a HILIC LC-fluorescence method for extensive profiling of 2-AB labeled glycans (released by PNGase F) was performed for accurate quantification of the released glycans (Fig. 5B), further confirming the differences. The main difference between the innovator and the biosimilar measured by this methodology was in the relative proportions of G0F and G1F. In addition to accurate and sensitive quantification, another advantage of the method is the ability to resolve isomers of glycans such as G1Fa and G1Fb, and G1a and G1b (Fig. 5A). MALDI QTof MS measurement (Fig. 5C) of unlabeled free glycans was helpful for elucidation of the free glycan structures.
Peptide mapping with LC-MSE also identified and quantified common PTMs in the two mAbs (Table 1), such as deamidation, isomerization and oxidation. Although comparable N-deamidation and D-isomerization were characterized between the two mAbs, increased M-oxidation was identified in the biosimilar compared with the innovator product. For example, only ∼4% oxidation of M255 on HT21 (DTLMISR) of the innovator mAb was measured, but the M-oxidation was more than doubled (∼10%) for the biosimilar mAb (Fig. 7).
In conclusion, peptide mapping with LC-MSE, combined with intact mass measurements and free glycan profiling, can provide a comprehensive comparison of a candidate biosimilar mAb to an innovator product. Intact protein mass measurements can compare molecular mass and heterogeneities such as glycoforms and non-product impurities. Peptide mapping with LC-MSE is able to locate and verify the heterogeneities and impurities. In addition, LC-MSE can also systematically compare other differences due to PTMs with small, e.g., +0.98 Da of N-deamidation, or no mass differences, e.g., 0.0 Da of D-isomerization because the isomers have different retention times. Even small differences in glycosylation have been detected as part of the routine peptide mapping characterization. The identity and relative content of individual glycan structures were compared with free glycan profiling. The observation of increased M-oxidation also indicates that the operating conditions of the production process for the biosimilar may lead to greater oxidation.
Finally, the confirmation of a sequence variant (two amino acid residue difference on HT35) of the biosimilar mAb in this study can immediately be used as a guide to correct clone selection, saving significant time and money in biosimilar development. Knowledge of structural differences, which determine if biological, non-clinical and clinical studies should be initiated, is critical for development of biosimilar mAbs. Therefore the characterization of molecular similarity between a biosimilar and an innovator product should be done as early as possible. Routine bioinformatics and LC-MS tools described here can characterize the differences between two mAbs in days. BiopharmaLynx automates and removes the tedious LC-MS data processing in LC-MS peptide mapping and intact protein mass measurements. This has traditionally been done manually or using proteomics software that is not optimized for biopharmaceutical workflows.
Materials and Methods
Samples and materials.
The candidate biosimilar and innovator (trastuzumab) IgG1 antibodies were donated by a generous collaborator from an emerging Asian market. Iodoacetamide (IAM), dithiothereitol (DTT), ammonium formate, ammonium bicarbonate (NH4HCO3), trifluroacetic acid (TFA), peptide N-glycosidase (PNGase F) and GlycoProfile 2-ab-labeling Kit were purchased from Sigma Chemical Co., (St. Louis, MO, USA), Sequence-grade trypsin from Promega Corp., (Madison, WI, USA), Formic acid (FA) from EM sciences (Gibbstown, NJ, USA), Optima-grade acetonitrile (ACN) from Fisher Scientific (Pittsburg, PA, USA), and RapiGest SF, MassPREP Glycoanalysis Kit containing RapiGest SF, 96-well microElution HILIC solid phase extraction (SPE) plate and MassPREP MALDI matrix 2,5-dihydroxybenzoic acid (DHB) were obtained from Waters Corp., (Milford, MA, USA). A Millipore Mili-Q purification system (Bedford, MA, USA) was used to prepare deionized water (18 ΩM cm) for HPLC mobile phases and in all sample processing procedures.
Instrumentation.
Separations were performed on Waters ACQUITY UPLC™ system equipped with a tunable ultraviolet (TUV; for peptide mapping) or fluorescence (for released glycan profiling) detector. All MS measurements were implemented on a Waters SYNAPT HDMS system equipped with both an ESI and MALDI sources. The systems were controlled by MassLynx™ 4.1 software and all LC-MS experiments were implemented in online configurations.
Intact protein mass measurements.
The antibody samples were denatured with RapiGest SF and reduced with DTT. The intact and reduced antibody samples were analyzed by reverse-phase LC-MS. The reverse-phase desalting separations of intact and reduced mAbs were performed on a Waters MassPREP™ Micro Desalting Column (2.1 × 5 mm) using a 1.5 min gradient (5–90% B) for intact and a 10 min gradient (5–50% B) for reduced antibody. The mobile phase A was 0.1% formic acid in water, while mobile phase B contained 0.1% formic acid in acetonitrile. The flow rate and column temperature were maintained at 200 µL/min and 80°C, respectively, throughout the run. Mass spectrometric analysis was carried out in positive ion V mode. The desolvation gas and source temperatures were set to 350°C and 120°C, respectively. The capillary and cone voltages were set at 3,000 and 40 V, respectively. All the other voltages were optimized to provide optimal signal intensity. Trap and transfer collision energies were set at 6 and 4 V, respectively. The instrument was calibrated in the m/z range of 600–5,000 using CsI. The deconvolution of ESI mass spectra of intact and reduced mAbs was performed by Biopharmalynx 1.2 using MaxEnt 1 algorithm. For deconvolution, the m/z range from 1,500–3,500 (intact mAb) and 1,100–2,400 (reduced mAb) were used with the following MaxEnt 1 parameters: mass range from 130,000–160,000 Da; minimum intensity ratio left and right, 50%; width at half height for uniform Gaussian model, 1.4 (low m/z) and 1.5 (high m/z); number of iterations, 10 (intact mAb) and mass range from 20,000 to 60,000 Da; minimum intensity ratio left and right, 30%; width at half height for uniform Gaussian model, 0.4 (low m/z) and 0.8 (high m/z); number of iteration, 10 (reduced mAb).
Peptide mapping analysis.
Tryptic digests of antibodies were prepared using sequence-grade trypsin (1:50 w/w) by incubating at 37°C in 50 mM NH4HCO3 solution containing 0.1% RapiGest SF (pH ∼7.5) for 4 hours. Before digestion, proteins are denatured by heating at 80°C for 15 min, and reduced with DTT and alkylated with IAM. Formic acid (0.5% v/v) was used to degrade RapiGest SF and quench trypsin enzymatic reactions. ACN (to 30% v/v) was used to dissolve precipitate due to a RapiGest SF degradation product. The digests were diluted to 0.36 µg/µl solution containing 5% ACN and 0.1% FA prior to reverse-phase LC-MSE analysis. 3.6 µg (in 10 µl) protein tryptic digest was injected for each LC run.
Peptide mixture was separated on a 2.1 × 150 mm BEH300 1.7 µm column at 65°C using a 90-min gradient (1–40% B). The LC mobile phases and flow rate were the same as in above intact protein mass analyses. LC-eluted peptides were detected by MS with an alternating low collision energy (4 V) and elevated collision energy (ramping from 20 to 40 V) ESI+ acquisition mode to obtain the precursor ions (MS) and their fragmentation data (MSE), respectively. Scan time was 0.5 second (1 s total duty cycle). A capillary voltage of 3.0 kV, source temperature of 100°C, cone voltage of 35 V, cone gas flow of 10 L/h were maintained during the analyses. An auxiliary pump was used to spray a solution of 100 fmol/µl Glu1-fibrinopeptide B (GFP) in 50/50 ACN/water containing 0.1% FA for mass accuracy (lockmass channel), with a flow rate of 20 µl/min and sampling every 1 min. The system was tuned for a minimum resolution of 10,000 and calibrated using a 100 fmol/µl GFP infusion.
The collected LC-MS data were processed by BiopharmaLynx 1.2, using fully tryptic cleavage rules45 for both heavy chain and light chain sequences of the innovator mAb. Cysteine carbamidomethylation (+57.02 Da) was set as a fixed modification, while N-deamidation (+0.98 Da for aspartic and isoaspartic acid products, and −17.03 Da for succinimide intermediate) and M-oxidation (+15.99 Da) as variable modifications. Glycan structures of human mAbs46,47 were also set as variable modifications. The mass tolerance for both precursors and fragments was set to less than 30 ppm. The identified peptides were confirmed by MSE spectra with at least three fragment ions.
Free glycan profiling.
After denaturing with RapiGest SF and reduction with DDT, N-linked glycans of the antibodies were deglycosylated by adding PNGase F and incubating at 37°C overnight. Released glycans were extracted using HILIC microE-lution SPE in a 96-well format plate using a protocol as described before.48 The glycans were subsequently labeled with 2-aminonbensamide (2-AB) using a Sigma GlycoProfile 2-AB labeled Kit. Again, HILIC SPE plate was applied to remove excess 2-AB labeling agent.
The 2-AB labeled glycans were subsequently separated on a HILIC column (2.1 × 150 mm, BEH amide, 1.7 µm) at 40°C and detected by an ACQUITY UPLC fluorescence detector. Separations were performed at a 45-min gradient (72–62% B) and 0.2 ml/min flow rate using 100 mM ammonium formate, pH 4.5 as mobile phase A and 100% ACN as mobile phase B.
The released free glycans were also characterized without labeling using MALDI QTof MS in a positive ion mode to obtain mass profiling and structure elucidation. The operating conditions were described in a previous study.48 Simglycan (v. 2.75) from Premier Biosoft International was used for the identification.
Acknowledgements
The authors thank Drs. Scott Berger and John Gebler for helpful discussion and support.
Abbreviations
- mAb
monoclonal antibody
- HT
heavy chain tryptic peptide
- LT
light chain tryptic peptide
- PTM
posttranslational modification
- LC
liquid chromatography
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- LC-MSE
dada independent acquisition LC-MS with an alternating low and elevated collision energy scan mode
- ESI
electrospray ionization
- MALDI
matrix-assisted laser desorption/ionization
- UV
ultra violet
- RT
retention time
- XIC
extracted ion chromatography
- HILIC
hydrophilic interaction chromatography
- DTT
dithiothereitol
- FA
formic acidexpiration
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/11986
Note Added to Proof
It has been brought to our attention that the level of deamidation reported for many of the peptides in the innovator antibody are much higher than previously reported for this innovator antibody (Table 1, Heavy Chain peptide HT6 and Light Chain peptide LT3 examples and reference 20 for previously reported data). We acknowledge that the digestion protocol was not optimized to minimize sample preparation-induced deamidation and believe that the high levels reported for certain peptides may not be reflective of the true level of deamidation in the sample prior to digestion. The main conclusion of the paper, namely that there was a sequence difference between the two samples, is still valid.
References
- 1.The Henry J. Kaiser Family Foundation U.S. Healthcare Costs, author. www.kaiseredu.org/topics_im.asp?imID=1&parentID=61&id=358.
- 2.Hughes B. Gearing up for follow-on biologics. Nat Rev Drug Discov. 2009;8:181. doi: 10.1038/nrd2847. [DOI] [PubMed] [Google Scholar]
- 3.Reichert JM, Beck A, Iyer H. European Medicines Agency workshop on biosimilar monoclonal antibodies, July 2, 2009, London UK. mAbs. 2009;1:394–416. doi: 10.4161/mabs.1.5.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Questions and Answers on Biosimilar Medicines (Similar Biological Medicinal Products); London, 22 October 2008; Doc Ref EMEA/74562/2006 Rev 1. www.ema.europa.eu/pdfs/human/pcwp/7456206en.pdf.
- 5.Procedures for Marketing Authorization Volume 2A, Chapter 1, November 2005. http://ec.europa.eu/enterprise/pharmaceuticals/eudralex/vol-2/a/vol2a_chap1_2005-11.pdf.
- 6.Waltz E. Western biotechs ponder follow-on possibilities. Nat Biotechnol. 2008;26:962–963. doi: 10.1038/nbt0908-962. [DOI] [PubMed] [Google Scholar]
- 7.Healthcare Reform Draws Mixed Reviews. http://pharmtech.findpharma.com/pharmtech/articleDetail.jsp?id=662434; Patricia Van Arnum.
- 8.GPhA Statement on House Passage of Health Reform Bill. www.gpha.online.org/media/press-releases/2010/gpha-statement-house-passage-health-reform-bill.
- 9.Beck A, Bussat MC, Zorn N, Robillard V, Klinguer-Hamour C, Chenu S, et al. Characterization by liquid chromatography combined with mass spectrometry of monoclonal anti-IGF-1 receptor antibodies produced in CHO and NS0 cells. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;819:203–218. doi: 10.1016/j.jchromb.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 10.Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. 2008;358:1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jefferis R. Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends Pharmacol Sci. 2009;30:356–362. doi: 10.1016/j.tips.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 14.Sheridan C. Commercial interest grows in glycan analysis. Nat Biotechnol. 2007;25:145–146. doi: 10.1038/nbt0207-145. [DOI] [PubMed] [Google Scholar]
- 15.Kroon DJ, Baldwin-Ferro A, Lalan P. Identification of sites of degradation in a therapeutic monoclonal antibody by peptide mapping. Pharm Res. 1992;9:1386–1393. doi: 10.1023/a:1015894409623. [DOI] [PubMed] [Google Scholar]
- 16.Manning MC, Patel K, Borchardt RT. Stability of protein pharmaceuticals. Pharm Res. 1989;6:903–918. doi: 10.1023/a:1015929109894. [DOI] [PubMed] [Google Scholar]
- 17.Paborji M, Pochopin NL, Coppola WP, Bogardus JB. Chemical and physical stability of chimeric L6, a mouse-human monoclonal antibody. Pharm Res. 1994;11:764–771. doi: 10.1023/a:1018948901599. [DOI] [PubMed] [Google Scholar]
- 18.Beck A, Wagner-Rousset E, Bussat MC, Lokteff M, Klinguer-Hamour C, Haeuw JF, et al. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol. 2008;9:482–501. doi: 10.2174/138920108786786411. [DOI] [PubMed] [Google Scholar]
- 19.Beck A, Wurch T, Corvaia N. Therapeutic antibodies and derivatives: from the bench to the clinic. Curr Pharm Biotechnol. 2008;9:421–412. doi: 10.2174/138920108786786420. [DOI] [PubMed] [Google Scholar]
- 20.Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B Biomed Sci Appl. 2001;752:233–245. doi: 10.1016/s0378-4347(00)00548-x. [DOI] [PubMed] [Google Scholar]
- 21.Vlasak J, Bussat MC, Wang S, Wagner-Rousset E, Schaefer M, Klinguer-Hamour C, et al. Identification and characterization of asparagine deamidation in the light chain CDR1 of a humanized IgG1 antibody. Anal Biochem. 2009;392:145–154. doi: 10.1016/j.ab.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 22.Rothman RJ, Warren L, Vliegenthart JF, Hard KJ. Clonal analysis of the glycosylation of immunoglobulin G secreted by murine hybridomas. Biochemistry. 1989;28:1377–1384. doi: 10.1021/bi00429a065. [DOI] [PubMed] [Google Scholar]
- 23.Damen CW, Chen W, Chakraborty AB, van Oosterhout M, Mazzeo JR, Gebler JC, et al. Electrospray ionization quadrupole ion-mobility time-of-flight mass spectrometry as a tool to distinguish the lot-to-lot heterogeneity in N-glycosylation profile of the therapeutic monoclonal antibody trastuzumab. J Am Soc Mass Spectrom. 2009;20:2021–2033. doi: 10.1016/j.jasms.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Xie H, Gilar M, Gebler JC. Characterization of protein impurities and site-specific modifications using peptide mapping with liquid chromatography and data independent acquisition mass spectrometry. Anal Chem. 2009;81:5699–56708. doi: 10.1021/ac900468j. [DOI] [PubMed] [Google Scholar]
- 25.Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 26.Wills C, Jornvall H. The two major isozymes of yeast alcohol dehydrogenase. Eur J Biochem. 1979;99:323–331. doi: 10.1111/j.1432-1033.1979.tb13260.x. [DOI] [PubMed] [Google Scholar]
- 27.Jornvall H. Differences between alcohol dehydeogenases. European Journal of Biochemistry. 1977;72:443–452. doi: 10.1111/j.1432-1033.1977.tb11268.x. [DOI] [PubMed] [Google Scholar]
- 28.Bennetzen J, Hall B. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982;257:3018–3025. [PubMed] [Google Scholar]
- 29.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–1200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty AB, Berger SJ, Gebler JC. Use of an integrated MS—multiplexed MS/MS data acquisition strategy for high-coverage peptide mapping studies. Rapid Commun Mass Spectrom. 2007;21:730–7344. doi: 10.1002/rcm.2888. [DOI] [PubMed] [Google Scholar]
- 32. drugbank.ca. www.drugbank.ca/drugs/DB00072.
- 33.Jefferis R, Lefranc MP. Human immunoglobulin allotypes: possible implications for immunogenicity. MAbs. 2009;1:332–338. doi: 10.4161/mabs.1.4.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magdelaine-Beuzelin C, Vermeire S, Goodall M, Baert F, Noman M, Assche GV, et al. IgG1 heavy chain-coding gene polymorphism (G1m allotypes) and development of antibodies-to-infliximab. Pharmacogenet Genomics. 2009;19:383–387. doi: 10.1097/FPC.0b013e32832a06bf. [DOI] [PubMed] [Google Scholar]
- 35.Wang YM, Chow AT. Development of biosimilars—pharmacokinetic and pharmacodynamic considerations. J Biopharm Stat. 20:46–61. doi: 10.1080/10543400903280357. [DOI] [PubMed] [Google Scholar]
- 36.Brennan TV, Clarke S. Spontaneous degradation of polypeptides at aspartyl and asparaginyl residues: effects of the solvent dielectric. Protein Sci 1993. 2:331–338. doi: 10.1002/pro.5560020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelius D, Rehder DS, Bondarenko PV. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal Chem. 2005;77:6004–6011. doi: 10.1021/ac050672d. [DOI] [PubMed] [Google Scholar]
- 38.Kosky AA, Razzaq UO, Treuheit MJ, Brems DN. The effects of alpha-helix on the stability of Asn residues: deamidation rates in peptides of varying helicity. Protein Sci. 1999;8:2519–2523. doi: 10.1110/ps.8.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson NE, Robinson AB. Prediction of primary structure deamidation rates of asparaginyl and glutaminyl peptides through steric and catalytic effects. J Pept Res. 2004;63:437–448. doi: 10.1111/j.1399-3011.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 40.Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Deamidation of—Asn-Gly—sequences during sample preparation for proteomics: Consequences for MALDI and HPLC-MALDI analysis. Anal Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- 41.Gaza-Bulseco G, Li B, Bulseco A, Liu HC. Method to differentiate asn deamidation that occurred prior to and during sample preparation of a monoclonal antibody. Anal Chem. 2008;80:9491–9498. doi: 10.1021/ac801617u. [DOI] [PubMed] [Google Scholar]
- 42.Ren D, Pipes GD, Liu D, Shih LY, Nichols AC, Treuheit MJ, et al. An improved trypsin digestion method minimizes digestion-induced modifications on proteins. Anal Biochem. 2009;392:12–21. doi: 10.1016/j.ab.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 43.Reichert JM. Monoclonal antibodies as innovative therapeutics. Curr Pharm Biotechnol. 2008;9:423–430. doi: 10.2174/138920108786786358. [DOI] [PubMed] [Google Scholar]
- 44.Srebalus Barnes CA, Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom Rev. 2007;26:370–388. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 45.Xie H, Griffin TJ. Trade-off between high sensitivity and increased potential for false positive peptide sequence matches using a two-dimensional linear ion trap for tandem mass spectrometry-based proteomics. J Proteome Res. 2006;5:1003–1009. doi: 10.1021/pr050472i. [DOI] [PubMed] [Google Scholar]
- 46.Sinha S, Pipes G, Topp EM, Bondarenko PV, Treuheit MJ, Gadgil HS. Comparison of LC and LC/MS methods for quantifying N-glycosylation in recombinant IgGs. J Am Soc Mass Spectrom. 2008;19:1643–1654. doi: 10.1016/j.jasms.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M. Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 um sorbent. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:403–408. doi: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Yu YQ, Gilar M, Kaska J, Gebler JC. A rapid sample preparation method for mass spectrometric characterization of N-linked glycans. Rapid Commun Mass Spectrom. 2005;19:2331–2336. doi: 10.1002/rcm.2067. [DOI] [PubMed] [Google Scholar]