Abstract
Dynamics of nucleosomes and spontaneous unwrapping of DNA are fundamental property of the chromatin enabling access to nucleosomal DNA for regulatory proteins. Probing of such dynamics of nucleosomes performed by single molecule techniques revealed a large scale dynamics of nucleosomes including their spontaneous unwrapping. Dissociation of nucleosomes at low concentrations is a complicating issue for studies with single molecule techniques. In this paper, we tested the ability of 3-[(3-Cholamidopropyl)dimethylammonio]-l-propanesulfonate (CHAPS) to prevent dissociation of nucleosomes. The study was performed with mononucleosome system assembled with human histones H2A, H2B, H3 and H4 on the DNA substrate containing sequence 601 that provides the sequencespecific assembly of nucleosomes. We used Atomic Force Microscopy (AFM) to directly identify nucleosomes and analyze their structure at the nanometer level. These studies showed that in the presence of CHAPS at millimolar concentrations, nucleosomes, even at sub-nanomolar concentrations, remain intact over days compared to a complete dissociation of the same nucleosome sample over 10 min in the absence of CHAPS. Importantly, CHAPS does not change the conformation of nucleosomes as confirmed by the AFM analysis. Moreover, 16 µM CHAPS stabilizes nucleosomes in over one hour incubation in the solution containing as low as 0.4 nM in nucleosomes. The stability of nucleosomes is slightly reduced at physiological conditions (150 mM NaCl), although the nucleosomes dissociate rapidly at 300 mM NaCl. The sequence specificity of the nucleosome in the presence of CHAPS decreased suggesting that the histone core translocates along the DNA substrate utilizing sliding mechanism.
Keywords: Chromatin, nucleosome structure, AFM, single molecule analysis, DNA, CHAPS
Introduction
Dynamics of nucleosomes is one of fundamental problems underlining molecular mechanism of regulation of chromatin activity. DNA is tightly packed within chromosomes but free DNA is required for accomplishing all genetic processes [1]. Accessibility of DNA within nucleosomes, fundamental units of chromatin, is provided by remodeling systems [2], but this process can be facilitated by inherent dynamics of nucleosomes [3]. Single molecule approaches of various types were instrumental in characterization of the nucleosomes dynamics and stability using single molecule florescence [4–10], single molecule probing [11, 12] and Atomic Force Microscopy (AFM) imaging [13–16]. Note in this regard recent studies with the use of time-lapse observations enabling direct observation of the dynamics and unwrapping of nucleosomes [17–19].
The stability of nucleosomes is an important issue for numerous biophysical studies and single molecule studies in particular. Reconstituted chromatin remains stable at low ionic strength, but dissociates if the concentration of nucleosomes is in the nanomolar range [8, 9, 20–24]. At the same time, single molecule approaches are typically performed at very low concentration of nucleosomes, and spontaneous dissociation of nucleosomes impedes these studies. It was recently shown that the stability of mononucleosomes was increased in the presence of non-ionic detergent Nonidet P-40 (NP-40) [8]. However, the dissociation occurs at elevated ionic strengths, so the nucleosomes are unstable at conditions close to physiological ones [8, 21, 25]. In a number of works, a zwitterionic detergent, 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesultonate (CHAPS), was successfully used for stabilization of various protein-DNA complexes (e.g., [26]). In the early work [26], 0.5 % CHAPS was able to stabilize complexes between DNA and DNA-binding factors such as AP-1, SPI, GATA-1 and α-regulated factor ISGF3 involved in the regulation of constitutive, tissue-specific and inducible genes. Importantly, proteins in complexes stabilized by CHAPS retain their biochemical activity [27–30]. Given such attractive properties of CHAPS, we have tested whether it is capable to stabilize nucleosomes in diluted solutions including the sub-nanomolar range.
Here, we report that CHAPS indeed prevents dissociation of mononucleosomes diluted to sub-nanomolar concentrations. The particles remain intact over days compared to a complete dissociation over a 10 min period in the absence of CHAPS. Importantly, the conformation of nucleosomes analyzed by high-resolution AFM remains unchanged. In addition, nucleosomes in 0.4 nM concentration remain stable over one hour incubation in the solution containing as low as 16 µM CHAPS. The stability of nucleosomes is slightly reduced at physiological conditions, although the nucleosomes dissociate rapidly at 300 mM NaCl. The sequence specificity of the nucleosome in the presence of CHAPS decreased enabling the translocation of the histone core along the DNA substrate. This and other effects of CHAPS are discussed.
Materials and methods
Preparation of nucleosomal DNA
DNA for nucleosome assembly similar to the previous studies was generated by PCR using plasmid pGEM3Z-601 as a template, which codes for a high-affinity nucleosome positioning sequence [31].
Histone octamer assembly and purification
Histone octamers were assembled as described in [32]. Octamers were separated from tetramer and dimer fractions with size-exclusion chromatography (SEC) with Superdex 200 PC 3.2/30 column (GE Healthcare) at 4°C. SEC fractions were analyzed for purity and histone stoichiometry using SDS-PAGE. Fractions containing histones H2A, H2B, H3 and H4 in approximately equal ratios were pooled and concentrated by centrifugation at 10,000 g.
Nucleosome refolding
Nucleosomes were prepared as described [32, 33]. Briefly, histone octamers and DNA containing the nucleosome positioning sequence were mixed in equimolar concentrations in 2 M NaCl and kept for 30 min at room temperature. A series of dilutions was prepared by using 10 mM Tris HCI to produce final concentrations of 1 M, 0.67 M, and 0.5 M NaCl. Diluted samples were kept at 4°C for 1 h before dialysis against one change of volume of 0.2 M NaCl overnight. Nucleosomes were concentrated using Microcon centrifugal filter device, MWCO 10,000 at 7,000g for 10 min at 4°C and dialyzed against one change of 200 ml of buffer containing 10 mM HEPES-NaCl, pH 7.5, and 1 mM EDTA for 3h at 4°C.
Atomic force microscopy
Freshly cleaved mica was modified with 167 µM solution of 1-(3-aminopropyl)-silatrane (APS) for 30 min at room temperature to make APS-mica as described [32, 34, 35]. The nucleosome stock solution was diluted into 10 mM Tris-HCl, pH 7.5, 4 mM, MgCl2 buffer and 5 µl of the solution were deposited on APS-treated mica for 3 minutes, washed with deionized water and dried under argon flow. AFM images were collected on NanoScope IIId system (Veeco/Digital Instruments, Santa Barbara, CA) as described in [32, 35].
Measurement of nucleosome parameters
The samples deposited on APS mica were analyzed with Femtoscan Online software. The following five parameters were measured: length of each DNA arm, angle between arms (interarm angle), height of nucleosome core particle and diameter as width of nucleosome core particle at half height [32]. The length of DNA was measured with FemtoScan Online software. The length of wrapped DNA was measured by subtracting the sum of both DNA arms from the length of uncomplexed DNA. The contour length of the DNA substrate was obtained by measuring naked DNA molecules appearing on the images along with nucleosomes. The mean value for the contour length was assigned to the maximum of Gaussian fitting for the histogram obtained for the dataset not less than 100 molecules (Figure S1 supporting information). The measured value yields for the length conversion coefficient value of 0.33 nm/bp. The number of DNA turns was calculated from the length measurements on the crystallographic data according to which 147 bp DNA makes 1.65 turns around the histone core.
Results
Time-dependent dissociation of nucleosomes in diluted solutions
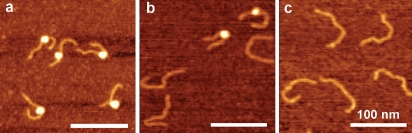
The nucleosome sample stock solution (240 nM) stored at 0°C was diluted to the concentration 0.4 nM with 10 mM HEPES (pH 7.5), 4 mM MgCl2 and the diluted sample was deposited onto APS-mica after the incubation at 0°C during specific times for AFM imaging. Figure 1 shows the typical AFM images of nucleosomes (0.4 nM) corresponding to the incubation for 0 min, 5 min and 30 min. The initial sample (Figure 1a) primarily consists of nucleosomes appearing as filaments with bright round features (blobs) and DNA arms at both sides of the particle. The incubation during 5 min (Figure 1b) leads to a substantial loss of nucleosome particles accompanied with accumulation of naked DNA. There are two nucleosomes in addition to three naked DNAs in this frame compared to the previous image that has only nucleosomal particles with no free DNA. Additional incubation for 30 min leads to almost complete dissociation of nucleosomes, so primarily naked DNA appears on the AFM images (Figure 1c). The AFM images were used to calculate the percentage of nucleosomes for each sample and the dependence of the percentage of the nucleosomes on the incubation time is shown in Figure 2a. This plot shows that the number of nucleosomes decays exponentially with a characteristic time of 9.7 min. Note that the initial sample (zero incubation time) contained ~ 80% nucleosomes (as shown in Figure 1a) contains ~20% of free DNA.
Figure 1.
AFM-images of nucleosomes imaged (a) immediately after dilution of the stock solution, (b) 5 min incubation and (c) 30 min incubation at the temperature 0°C. Scale bars are 100 nm.
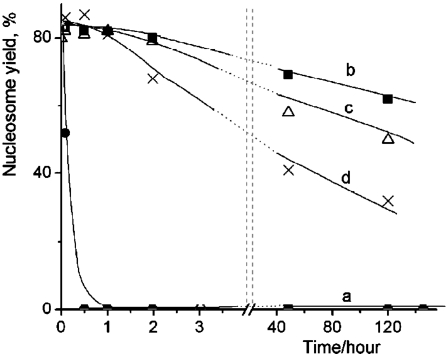
Figure 2.
Time dependent yields of nucleosomes in 0.4 nM nucleosome solutions at 0°C containing different CHAPS concentrations: a – 0 mM CHAPS (solid circles); b – 4.8 mM CHAPS (solid squares); c – 1.6 mM CHAPS (triangles); d – 16 µM CHAPS (crosses).
Dissociation of nucleosomes in the presence of CHAPS
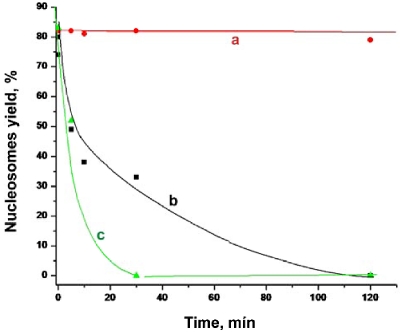
Similar experiments were performed for the nucleosome sample diluted in the same buffer solution, but containing CHAPS. In contrast with fast dissociation of nucleosomes in the previous experiments, the AFM images of nucleosomes incubated in the presence of 4.8 mM CHAPS for several hours are indistinguishable from the original sample and were similar to ones shown above in Figure 1a. AFM imaging was performed for the samples incubated for different times up to 120 hours; the yield of nucleosomes for each incubation time was calculated from these images and the data are plotted in Figure 2b. This graph shows that there is no noticeable dissociation of nucleosomes after 1 hour of incubation. Even the 2-hour incubation period leads to the drop of only few percent of initial nucleosomes. Importantly, more than 70% of initial numbers of nucleosomes remain intact after incubation for 120 hours.
We tested whether a lower concentration of CHAPS works. Graph in Figure 2c shows the results obtained at three times less concentration of CHAPS. The difference between this sample and the previous one is only noticeable after the long-term incubation times. There is a loss of the nucleosomes after the incubation for 120 hours, but more than 50% of initial nucleosomes remain intact by that time. Surprisingly, the stabilization effect of CHAPS is still high at concentration of CHAPS as low as 16 μM (Figure 2d) that is 100 times less than the one shown in the previous data set. According to this plot, there is no noticeable loss of nucleosomes after the 1 hour incubation period.
Effect of CHAPS on the structure of nucleosomes
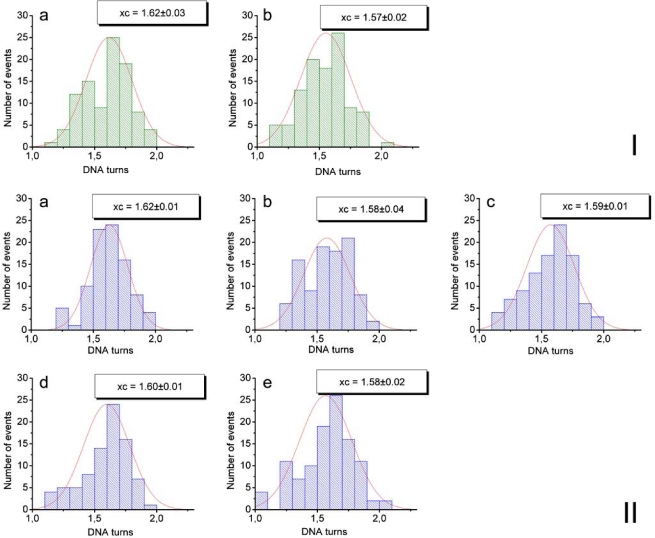
To answer the question of whether CHAPS changes the structure of nucleosomes, we analyzed the morphology of nucleosomes using approaches described earlier [32]. One of the parameters characterizing the structure of nucleosomes is the number of DNA turns around the histone octamer (nucleosomal turns). We have previously shown [17, 32] that AFM imaging is capable of measuring the number of nucleosomal turns using three different parameters of nucleosomes, the lengths of the DNA arms, the angle between the arms and the volume of the nucleosome blobs. Here, we primarily used the first parameter, the lengths of the DNA arms. Briefly, the lengths of the arms were measured directly from the AFM images such as ones shown in Figure 1, the values for each nucleosome were subtracted from the mean length of naked DNA measured on the same images to yield the length of nucleosomal DNA. The numbers of turns was calculated from this value taking into account that 147 bp of nucleosomal DNA make 1.65 nucleosomal turns [36]. The data assembled as histograms are shown in Figure S2. The results of such analysis for the samples obtained with and without CHAPS are summarized in Table 1. This table shows that the number of DNA turns in nucleosomes incubated in the presence of CHAPS, even for 48 hours, is the same as in the initial sample and is not distinguishable from the sample incubated in the absence of CHAPS. These data suggest that CHAPS stabilization effect does not increase the number of nucleosomal turns.
Figure S2.
Statistical histograms fitted by Gaussian distributions which were plotted based on the measurements of DNA arms lengths in nucleosome particles (NCP), deposited from 0.4 nM NOP solutions in the presence or absence of CHAPS on APS-mica. Number of DNA turns was estimated as the maximum of Gaussian distribution (the most probable value). Set I – NCP solution not containing CHAPS: a – just after its preparation (1.62 DNA turns), b – after 5 min of its storage at 0°C (1.57 DNA turns). Set II – NCP solution containing 1.6 mM CHAPS: a – just after the the preparation (1.62 DNA turns), b – after 5 min of its storage at 0°C (1.58 DNA turns), c – 1 hour (DNA turns 1.59), d – 2 hours (1.6 DNA turns), e – 48 hours (1.58 DNA turns).
Table 1.
Number of DNA turns wrapped around histone octamer in nucleosomes incubated with and without CHAPS at different times at 0°C
| 0 min | 5 min | 30 min | 1h | 2h | 48 h | |
|---|---|---|---|---|---|---|
| 0.4 nM NCP solution | 1.62±0.03 | 1.57±0.02 | - | - | - | - |
| 0.4 nM NCP solution with 1.6 mM CHAPS | 1.62±0.01 | 1.58±0.04 | 1.59±0.01 | 1.59±0.01 | 1.60±0.01 | 1.58±0.02 |
Effect of ionic strength on the nucleosome stability in the presence of the CHAPS
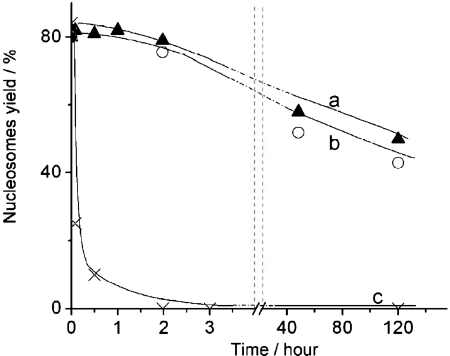
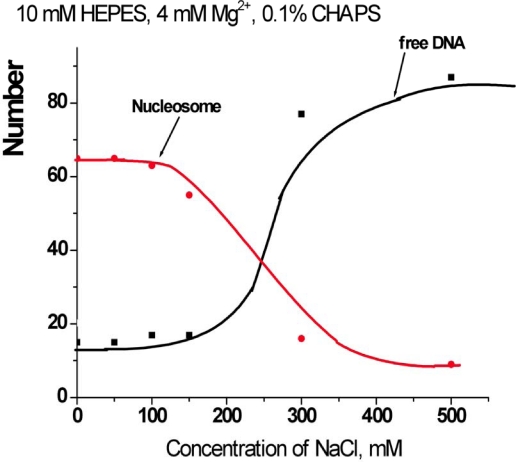
The experiments described above were performed at low ionic strength conditions in buffer containing 10 mM HEPES (pH 7.5), 4 mM MgCl2. To investigate the effect of ionic strength on the stabilization effect of CHAPS, we performed a similar AFM study in the presence of 150 and 300 mM NaCl, measuring the yield of nucleosome. The time-dependence data are summarized in Figure 3. The dissociation rate of nucleosomes remains low in the presence of 150 mM NaCl. Moreover, there is no substantial difference between the original sample and the one incubated at 150 mM NaCl. However, the dissociation is very fast if more salt (300 mM NaCl) is added. Such measurements were performed at other concentrations of NaCl, and the dependence of the yield of nucleosomes on the concentration of NaCl measured after 30 min incubation in the presence of 1.6 mM CHAPS is shown Figure S3.
Figure 3.
Time dependent yields of nucleosomes in 0.4 nM solutions containing 1.6 mM CHAPS and different amount of NaCl: a – 0 mM NaCl (triangles); b – 150 mM NaCl (spheres); c – 300 mM NaCl (crosses).
Figure S3.
The dependence of the yields of nucleosomes (red line) and free DNA (black line) on the concentration of NaCl in the presence of 1.6 mM CHAPS (0.1%).
Effect of CHAPS on the sequence specificity of nucleosomes
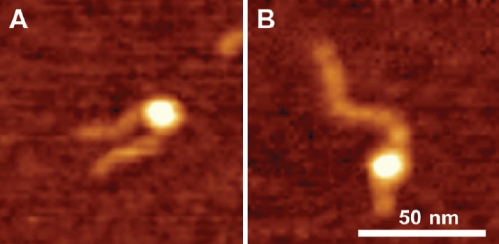
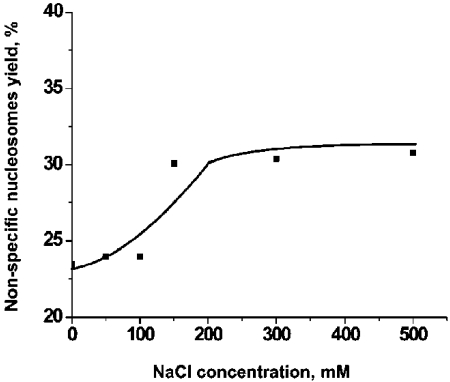
Throughout this work we used DNA substrate containing 147 bp segment (601 sequence) with a high specificity for the nucleosome formation. Indeed, the nucleosome position on the AFM images (Figure 1) is consistent with the location of this segment. However, rarely we found nucleosomes located far off this motif that we called non-specific nucleosomes. Figure 4 shows the pair of AFM images for specific (A) and non-specific (B) nucleosomes. The lengths of the arms for image B are 6 nm and 65 nm compared to 25 nm and 46 nm for image (A). According to Table 2, the yield of such nonspecific complex in the initial sample (no CHAPS) was 3-4%. However, in the presence of 1.6 mM CHAPS the number of non-specific nucleosomes after incubation for 2 hours reaches the value as high as 20-22%. 3-fold increase CHAPS concentration (4.8 mM) leads to the higher yield of non-specific nucleosome (25-27%). Results in Figure 5 show that the yield of non-specific nucleosomes increases at high ionic strength. Non-specific nucleosomes are formed from specific ones that require translocation of histone core to a new position. High ionic strength facilitates the disassemblyassembly process of the nucleosome, and the data obtained confirm this synergetic effect of ionic strength.
Figure 4.
AFM images of specific (A) and non-specific (B) nucleosomes. The sizes of small and large arms are 25 nm and 46 nm for image (A) and 6 nm and 65 nm for image (B) respectively.
Table 2.
Efect of CHAPS on the yield of non-specific nucleosomes
| 0.4. nM NCP no CHAPS | 0.4. nM NCP 1. 6 mM CHAPS | 0.4. nM NCP 4.8 mM CHAPS | |
|---|---|---|---|
| Yield of non-specific nucleosomes (%) | 3-4 | 20-22 | 25-27 |
Figure 5.
The dependence of the yield on non-specific nucleosomes on the concentration of NaCl. The solution of nucleosomes (0.4 nM) was incubated in the buffer solution (10 mM HEPES, 4 mM MgCl2) in the presence of 1.6 mM of CHAPS and various concentrations of NaCl for 2 hours. The nonspecific and specific complexes were identified by the measurements of the lengths of the arms on the AFM images. Nucleosomes with a short arm less than 10 nm were assigned to the class of non-specific nucleosomes.
Effect of cholesteryl sulfate on the stability of nucleosomes
In parallel with CHAPS we studied the effect of cholesteryl sulfate. Compared to CHAPS cholesteryl sulfate is physiologically active derivative of cholesterol and its chemical structure is very close to the one of CHAPS. The data in Figure 6 show that the nucleosome stability increases in the presence of cholesteryl sulfate, although that effect is much less than that of CHAPS.
Figure 6.
Time dependent yields of nucleosomes prepared in 10 mM HEPES, 4 mM MgCl2 with addition of (a) 1.6 mM CHAPS, (b) 1.6 mM cholesteryl sulfate and (c) no additives.
Discussion
Stabilization of nucleosomes in diluted solutions
The data in the paper show directly and unambiguously that zwitter-ionic detergent CHAPS increases the stability of nucleosomes in very dilute sub-nanomolar solutions of nucleosomes. Importantly, the structure of nucleosomes as analyzed by AFM remains unchanged, suggesting that CHAPS methodology is useful for physical chemical studies of nucleosomes in diluted solutions. Recently, NP-40 and NP-40 detergents was used for the stabilization of nucleosomes for single molecule fluorescence study [8, 21]. The results were not reproducible in the absence of NP-40 [8]. The use of the detergent enabled the authors to study the dynamics of nucleosomes with the single molecule Florescence Resonance Energy Transfer (FRET); however, the NP-40 approach did not work at physiological salt concentrations. We showed that CHAPS in similar concentrations stabilizes nucleosomes in a broad range of ionic strengths covering physiological conditions. Moreover, CHAPS in concentration as low as 16 µM prevents nucleosomes from dissociation if they are in the sub-nanomolar concentration that is even lower than traditional requirements for the single molecule FRET.
We also performed control experiment to rule out the possibility that interaction of CHAPS with APS-mica surface contributes to the observed stabilization effect. In these experiments the nucleosome samples were incubated different times in the absence of CHAPS that was added only to the aliquots prior to the sample deposition on APS-mica. The kinetic curve of the nucleosomes decay was practically the same as obtained without adding CHAPS (Figure 1).
The stabilization of nucleosomes is an attractive feature of CHAPS for various single-molecule experiments in which very low concentration of nucleosomes is required. However, the change of the sequence specificity of nucleosomes in the presence of CHAPS should receive a careful consideration. The accumulation of non-specific nucleosomes in the sample initially containing specific nucleosomes predominantly (Figure 5) indicates that CHAPS increases the efficiency of translocation of nucleosomes along the DNA template. Removal of CHAPS after the preparation of the sample for the single molecule experiment can decrease the translocation effect of CHAPS.
Effect of CHAPS on nucleosome dynamics
We used for these experiments DNA substrate containing inside 601 sequence characterized by very high specificity for assembly nucleosomes [31]. Indeed, in these experiments as well as in our prior papers [18, 35, 37] the position of nucleosomes coincides with the location of the 601 segment. Moreover, time-lapse experiments showed that dynamics of nucleosomes occurs by unwrapping of both arms by shortening of the 147 bp regions [18, 35]. In the presence of CHAPS this strong sequence specificity pattern changes. Incubation in 1.6 mM CHAPS leads to the 6-fold increase of the number of nucleosomes with positions of NCP far off the central 601 region (Figure 4B). Importantly, this number grows if the concentration of CHAPS increases suggesting that the stringency of sequence specific position of nucleosomes in the presence of CHAPS is lowered. The role of the DNA sequence in positioning of nucleosomes in chromatin is a problem of great importance (review [38] and references therein). Positioning of nucleosomes is one of the major factors regulating the genes activity, and our knowledge on mechanism regulating the sequence specificity of nucleosomes is rather limited. Here we show that the sequence specific stringency even for 601 sequence can be decreased in the presence of CHAPS. The importance of this finding is supported by the fact that CHAPS is a very close analog of cholesterol, a ubiquitous physiological compound. To test the hypothesis on potential role of cholesterol in the chromatin structure and dynamics we studied the effect of the water-soluble lipid cholesteryl sulfate on stability of nucleosomes. The data in Figure 6 show that the nucleosome stability increases in the presence of cholesteryl sulfate, although that effect is much less than that of CHAPS. Strong negative charge of this type of cholesterol prevents its interaction with DNA of nucleosomes and can contribute to lower effect on the nucleosome stability of cholesteryl sulfate compared to CHAPS. However, regardless of this unfavorable electrostatic property, the stabilization effect of cholesteryl sulfate is observed. Altogether the observation of the change of dynamics of nucleosomes by CHAPS prompts us to hypothesize that small molecules can modulate the dynamics and stability of chromatin and metabolites of cholesterol can be such molecules.
Mechanisms of CHAPS-nucleosome interaction
As we mentioned above, CHAPS was successfully applied for the stabilization of various bio-molecular systems, including a range of protein-DNA complexes [26]. Moreover, there were reports that CHAPS does not interfere with enzymatic activities of the systems [27–30]. However, a molecular mechanism of the CHAPS stabilization effect remains unclear. There is evidence that CHAPS interacts with proteins and nucleic acids, but how these interactions change the interactions within the systems is not clarified. CHAPS as any surfactant forms micelles, and its Cmc value is 6-10 mM, so at concentrations 3 mM CHAPS exists primarily as a monomer [39]. Substantial increase of oli-gomers in [39] was observed at 10 times higher concentration of CHAPS. Primarily our experiments were performed at 1.6 mM CHAPS suggesting that the concentration of oligomers of CHAPS in our experiments is negligible, so a monomeric form of it needs to be considered. Importantly, CHAPS in 16 µM efficiently stabilizes nucleosomes with concentration 0.4 nM (Figure 2d). Given the size of CHAPS (MW 614.9) comparable with the sizes of the DNA base pairs, it is instructive to compare the concentration of CHAPS with concentrations of DNA nucleotides and histone amino acids which are ~500 nM combined. This value is only ~30 times less than CHAPS concentration, suggesting that direct interactions of the detergents with the nucleosome in addition to the change of the solvent properties need to be taken into account for understanding molecular mechanism of CHAPS stabilization. According to the recent studies [9, 20–22, 40], the depletion of histone dimer (H2A and H2B) from the histone core is one of the factors determining the stability of nucleosome. Therefore, we can speculate that CHAPS increases the interaction of H2A and H2B dimer within the nucleosome to stabilize the dissociation of the entire histone particle.
In our previous time-lapse AFM data [32], we observed that nucleosome stability depends on the number of the DNA nucleosomal turns. A potential explanation of the CHAPS stabilization effect would be the increase of the length of nucleosomal DNA. However, we did not observe nucleosome unwrapping after adding CHAPS. Moreover, our data show that time-course dissociation of nucleosomes leads to the accumulation of free DNA without the formation of stable transient states. Similar to the time-course experiments with no CHAPS added, we did not reveal transient states as well (data not shown). Therefore, CHAPS does not induce the formation of stable nucleosome intermediates with the characteristics different from the original ones.
Acknowledgment
We thank Luda S. Shlyakhtenko for advice in the use of CHAPS and the interpretation of the data, and Alex Portillo for proofreading of the manuscript. The work was supported by grants the EPS-0701892 (NSF), DOE (DE-FG02-08ER64579), Nebraska Research Initiative (NRI) and CBN.NR.NRSFP 983204 (NATO) to YLL.
Abbreviations:
- CHAPS
3-[((3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate
- AFM
Atomic Force Microscopy
- NCP
nucleosome core particles
- APS
1-(3-aminopropyl)silatrane.
Supplemental information
Measurement of DNA length by AFM
Figure S1.
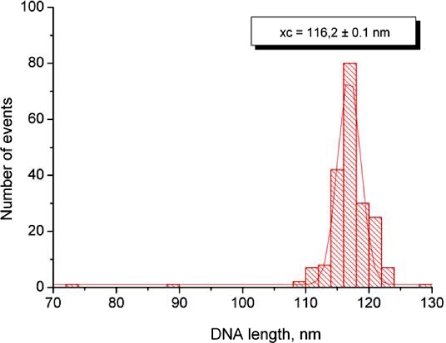
The length distribution of the DNA substrate measured with FemtoScan Online software using parameter “curve”. The data were plotted as statistical histogram and fitted with Gaussian distribution (Origin software). The most probable value of 116 nm was taken as the total length of DNA molecule. The DNA substrate consists of 353 bp; thus the linear density of the DNA filaments on the AFM images is 0.33 nm/bp that is very close to cristallographic value 0.34 nm/bp.
The number of DNA turns around histone octamer
Figure S2 shows the histograms for the number of DNA turns calculated as described above. Data were plotted as statistical histograms fitted by Gaussian distributions using Origin software.
References
- [1].Widom J. Chromatin: the nucleosome unwrapped. Curr Biol. 1997;7:R653–655. doi: 10.1016/s0960-9822(06)00327-7. [DOI] [PubMed] [Google Scholar]
- [2].Sana A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- [3].Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- [4].Lovullo D, Daniel D, Yodh J, Lohr D, Woodbury NW. A fluorescence resonance energy transfer-based probe to monitor nucleosome structure. Anal Biochem. 2005;341:165–172. doi: 10.1016/j.ab.2005.03.022. [DOI] [PubMed] [Google Scholar]
- [5].Leuba SH, Anand SP, Harp JM, Khan SA. Expedient placement of two fluorescent dyes for investigating dynamic DNA protein interactions in real time. Chromosome Res. 2008;16:451–467. doi: 10.1007/s10577-008-1235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gurunathan K, Levitus M. Single-molecule fluorescence studies of nucleosome dynamics. Curr Pharm Biotechnol. 2009;10:559–568. doi: 10.2174/138920109788922074. [DOI] [PubMed] [Google Scholar]
- [8].Koopmans WJ, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kelbauskas L, Chan N, Bash R, DeBartolo P, Sun J, Woodbury N, Lohr D. Sequence-dependent variations associated with H2A/H2B depletion of nucleosomes. Biophys J. 2008;94:147–158. doi: 10.1529/biophysj.107.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koopmans WJ, Buning R, Schmidt T, van Noort J. spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophys J. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zlatanova J, Leuba SH. Chromatin fibers, one-at-a-time. J Mol Biol. 2003;331:1–19. doi: 10.1016/s0022-2836(03)00691-0. [DOI] [PubMed] [Google Scholar]
- [12].Brower-Toland BD, Smith CL, Yen RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Nati Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bussiek M, Toth K, Brun N, Langowski J. DNA-loop formation on nucleosomes shown by in situ scanning force microscopy of supercoiled DNA. J Mol Biol. 2005;345:695–706. doi: 10.1016/j.jmb.2004.11.016. [DOI] [PubMed] [Google Scholar]
- [14].Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bussiek M, Muller G, Waldeck W, Diekmann S, Langowski J. Organisation of nucleosomal arrays reconstituted with repetitive African green monkey alpha-satellite DNA as analysed by atomic force microscopy. Eur Biophys J. 2007;37:81–93. doi: 10.1007/s00249-007-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Montel F, Menoni H, Castelnovo M, Bednar J, Dimitrov S, Angelov D, Faivre-Moskalenko C. The dynamics of individual nucleosomes controls the chromatin condensation pathway: direct atomic force microscopy visualization of variant chromatin. Biophys J. 2009;97:544–553. doi: 10.1016/j.bpj.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol Biol. 2009;543:337–351. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- [18].Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Bio/chemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suzuki Y, Higuchi Y, Hizume K, Yokokawa M, Yoshimura SH, Yoshikawa K, Takeyasu K. Molecular dynamics of DNA and nucleosomes in solution studied by fast-scanning atomic force microscopy. Ultramicroscopy. 2010;110:682–688. doi: 10.1016/j.ultramic.2010.02.032. [DOI] [PubMed] [Google Scholar]
- [20].Godde JS, Wolffe AP. Disruption of Reconstituted Nucleosomes. Journal of Biological Chemistry. 1995;270:27399–27402. doi: 10.1074/jbc.270.46.27399. [DOI] [PubMed] [Google Scholar]
- [21].Claudet C, Angelov D, Bouvet P, Dimitrov S, Bednar J. Histone Octamer Instability under Single Molecule Experiment Conditions. Journal of Biological Chemistry. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- [22].Adkins NL, Hall JA, Georgel PT. The use of Quantitative Agarose Gel Electrophoresis for rapid analysis of the integrity of protein-DNA complexes. Journal of Biochemical and Biophysical Methods. 2007;70:721–726. doi: 10.1016/j.jbbm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- [23].Wu H-M, Dattagupta N, Hogan M, Crothers DM. Structural changes of nucleosomes in low-salt concentrations. Biochemistry. 1979;18:3960–3965. doi: 10.1021/bi00585a018. [DOI] [PubMed] [Google Scholar]
- [24].Watkins JF, Smerdon MJ. Nucleosome rearrangement in vitro. 1. Two phases of salt-induced nucleosome migration in nuclei. Biochemistry. 1985;24:7279–7287. doi: 10.1021/bi00346a039. [DOI] [PubMed] [Google Scholar]
- [25].Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, TTith K, Langowski Jr, Seidel CAM. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proceedings of the National Academy of Sciences. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hassanain HH, Dai W, Gupta SL. Enhanced Gel Mobility Shift Assay for DNA-Binding Factors. Analytical Biochemistry. 1993;213:162–167. doi: 10.1006/abio.1993.1400. [DOI] [PubMed] [Google Scholar]
- [27].Ezgimen MD, Mueller NH, Teramoto T, Padmanabhan R. Effects of detergents on the West Nile virus protease activity. Bioorganic & Medicinal Chemistry. 2009;17:3278–3282. doi: 10.1016/j.bmc.2009.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang H, Kiserow DJ, McGown LB. Effects of bile salts on the solubility and activity of yeast alcohol dehydrogenase in AOT reversed micelles. Journal of Molecular Catalysis B: Enzymatic. 2001;14:7–14. [Google Scholar]
- [29].Bernhard G, Hugo S. Stabilization of photosystem II reaction centers: influence of bile salt detergents and low pH. FEBS Letters. 1998;431:161–166. doi: 10.1016/s0014-5793(98)00739-x. [DOI] [PubMed] [Google Scholar]
- [30].Culbert AA, Brown MJ, Frame S, Hagen T, Cross DAE, Bax B, Reith AD. GSK-3 inhibition by adenoviral FRAT1 overexpression is neuroprotective and induces Tau dephosphorylation and [beta]-catenin stabilisation without elevation of glycogen synthase activity. FEBS Letters. 2001;507:288–294. doi: 10.1016/s0014-5793(01)02990-8. [DOI] [PubMed] [Google Scholar]
- [31].Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- [32].Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL. Dynamics of Nucleosomes Revealed by Time-Lapse Atomic Force Microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Luger K, Rechsteiner TJ, Richmond TJ, Paul M, Wassarman APW. Methods in Enzymology. Academic Press: 1999. Preparation of nucleosome core particle from recombinant histones; pp. 3–19. [DOI] [PubMed] [Google Scholar]
- [34].Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- [35].Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luger K, Mader A, Richmond R, Sargent D, Richmond T. Crystal structure of the nucleosome core particle at 2.8BT)%or... resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [37].Filenko NA, Kolar C, West JT, Smith SA, Hassan Yl, Borgstahl GEO, Zempleni J, Lyubchenko YL. The Role of Histone H4 Biotinylation in the Structure of Nucleosomes. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0016299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Funasaki N, Fukuba M, Hattori T, Ishikawa S, Okuno T, Hirota S. Micelle formation of bile salts and zwitterionic derivative as studied by two-dimensional NMR spectroscopy. Chem Phys Lipids. 2006;142:43–57. doi: 10.1016/j.chemphyslip.2006.02.025. [DOI] [PubMed] [Google Scholar]
- [40].Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]