Abstract
The tumor microenvironment is comprised of multiple cell types arranged in a three-dimensional structure. Interactions amongst the various cell components play an important role in neoplasia, including the inflammatory reaction that occurs as part of the host response. In this study, the regional lymphocyte subpopulations and cytokine profiles associated with prostate cancer were examined using a quantitative imaging approach and expression microarray analysis. Lymphocytes were measured in four different epithelial phenotypes in prostate cancer specimens: carcinoma; prostatic intraepithelial neoplasia (PIN); benign prostate hyperplasia (BPH); and normal epithelium. The data indicate that CD8 positive, cytotoxic T lymphocytes are significantly decreased in regions adjacent to hyperplasia and carcinoma as compared to normal epithelium and PIN. In contrast the relative number of CD4 positive and CD20 positive lymphocytes did not change markedly. Parallel mRNA expression array analysis of the normal and tumor microenvironments identified a distinct cytokine profile in cancer, with 24 dysregulated genes in tumor epithelium and nine altered in tumor-associated stroma. Overall, these data indicate that the spatial distribution of CD8 positive, cytotoxic T lymphocytes is dysregulated in human prostate glands that contain cancer, and cytokine profiles are altered at the mRNA level.
Keywords: Prostate cancer, lymphocytes, cytokines, histomathematics, histopathology
Introduction
The host inflammatory response in the tumor microenvironment plays an important role in the development of neoplasms and is believed to include a competition between cancer cells and the immune system [1-4]. For example, CD4 lymphocytes secrete cytokines that serve to limit the growth of tumor cells via two separate mechanisms; activation of CD8-positive, cytotoxic T lymphocytes (CTLs) that effect tumor destruction by direct lysis of malignant cells through the Fas or perforin/Granzyme pathways [5-8]; and, differentiation of B lymphocytes into plasma cells that subsequently secrete antibodies against the invading cancer [5]. However, the precise interplay that occurs between tumor and non-tumor cells is not fully understood, thus there is a need to characterize and measure the involved components in detail in clinical samples that harbor tumors. Moreover, since the immune response has spatial as well as temporal characteristics it is useful to examine the immune system status in the context of an entire organ in order to facilitate a comprehensive characterization of the microenvironment, allowing investigators to measure cellular and molecular components in a native tissue setting and gain an appreciation of the relationship amongst the various components. In prostate cancer specimens for example, an ex vivo organ analysis permits study of the wide range of normal biology and pathology present in the samples, including; normal host cells, hyperplasia, pre-malignant lesions, inflammatory cells, and tumors.
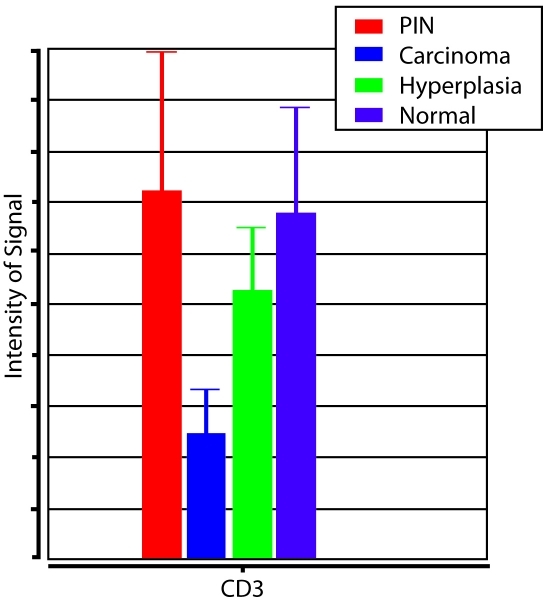
In a previous study, we employed a histomathematical analysis strategy to measure protein levels in the prostate cancer microenvironment and observed a decrease in the number of T lymphocytes in the vicinity of tumor epithelium relative to normal epithelium and prostatic intraepithelial neoplasia (PIN), while benign prostate hyperplasia (BPH) showed an intermediate value between that of the cancer and non-cancer cells (Figure 1) [9]. These results were generated using CD3 antigen as a pan-T cell marker, thus we could not determine the identity of the tumor-dysregulated lymphocytes at that time. The aim of the present study was to further investigate this phenomenon by measuring the lymphocyte sub-types associated with the epithelial phenotypes in prostate cancer specimens, and to investigate the cytokines that mediate the process. Both quantitative immuno-histochemistry and expression microarray analysis of microdissected cell populations were utilized.
Figure 1.
CD3 Lymphocyte Measurements. Relative number of CD3-positive lymphocytes in histopathological regions of prostatic intraepithelial neoplasia (PIN), carcinoma, hyperplasia, and normal epithelium.
Materials and methods
Ten radical prostatectomy specimens were obtained from the National Institutes of Health and the National Naval Medical center under an IRB-approved protocol. Whole-mount prostate cancer cases were ethanol-fixed and paraffin-embedded as described previously [10].
Five whole-mount prostate tissues were collected anonymously from patients undergoing radical prostatectomy at the Catholic University Hospital in Santiago Chile. A complete transverse section of each excised prostate was embedded in O.C.T. medium, and then flash frozen at -80°C. The frozen tissue blocks were anonymized and shipped to the National Cancer Institute as part of an IRB-approved protocol. Five localized primary prostate cancer cases with low to moderate Gleason grades were selected on the basis of both adequate tumor size and the ability to laser capture microdissect tumor-associated stroma without contamination by infiltrating tumor or inflammatory cells.
Five mm thick tissue sections were immunostained with three antibodies according to a standard immunohistochemistry (IHC) protocol (DAKO Cytomation EnVision system-HRP. Carpinteria, CA). Sections were incubated for 1 hour at RT with one of the following primary antibodies: CD4, mouse anti-human (Zymed labs, San-Francisco CA, cat # 08-0101), pre-diluted; CD8, mouse anti-human (DAKO Cytomation, Carpinteria, CA, cat #M0716), 1:20; CD20, mouse anti-human (DAKO Cytomation, Carpinteria, CA, cat #M0755), 1:200. Slides were counterstained with hematoxylin and positively stained cells were identified by the presence of a brown precipitate. Negative cells were identified by the absence of a brown precipitate and the presence of only blue counterstain.
A detailed experimental protocol appears elsewhere [11]. In short: Recut histological sections from five frozen whole-mounted cases were laser capture microdissected (PixCell lle, Arcturus Engineering, Inc., Mountain View, CA) in four separate histopathologically-defined regions: tumor epithelium; tumor-associated stroma; normal epithelium; and normal stroma. Linearly amplified RNA samples were hybridized to Affymetrix U133 plus 2.0 GeneChips (Affymetrix Inc., Santa Clara, CA). Raw data signal intensities were quantified using the Gene Chip Operating Software (GCOS) provided by Affymetrix. Intensities were normalized to remove background signal and filtered such that only perfectly matched probe sets were retained. The data were then analyzed by comparing tumor epithelium to normal epithelium and tumor stroma to normal stroma. Data sets were uploaded into the National Cancer Institute's microarray analysis program (mAdb) to perform molecular ontology and pathway analysis. Differentially expressed transcripts specifically related to immune function were then identified.
In general, the same protocol that was used in our previous histomathematical report was utilized [9]. Briefly, a pathologist (JWG) analyzed the IHC stained tissue sections from all ten cases in the study and photographed four representative epithelial regions: a) Normal epithelium; b) Benign prostate hyperplasia (BPH); c) Prostatic intraepithelial neoplasia (PIN); c) Carcinoma
The stained sections were imaged (constant tissue area of 0.1 mm2) using a magnification of 200x with an Olympus microscope and a charged-coupled device (CCD) camera with a resolution of 2080 × 1542 pixels (CCD color bayer mosaic, Q-Color-3 by Olympus, USA). Sixteen images were taken per slide generating 960 pictures (four pictures per each histological area) × (six antibodies per case) × (10 cases). All 960 images were analyzed for the number of positively and negatively stained cells using the ImagePro Analysis System (ImagePro 4.5, Cybernetics, Chevy-Chase, MD) as previously described [9]. In short: the staining was evaluated per total area of the picture taken (including the epithelial and associated stromal areas) and the percent positive cells was calculated by dividing the positive signal by the sum of the positive and negative signal for every picture. The mean in each histological area was established by averaging the percent positive signal for the 4 pictures taken for the area and was termed the intensity of staining. For the intra-epithelial count, areas of interest (AOI) were annotated around the epithelium in every picture and the percent positive count was calculated by dividing the positive signal in the epithelium by the sum of the positive and negative signal in the epithelium. Pair wise comparison analysis of variance (ANOVA) and a bar graph representation with calculated error bars were then applied to the data sets using PartekPro (Partek, Inc. St. Charles, MO).
Results
Ten whole-mount prostate cancer cases were evaluated for the levels of CD4, CD8, and CD20 positive lymphocytes using a quantitative analysis strategy. Regions of the prostate containing normal epithelium, BPH, PIN, and cancer were individually studied and for each measurement both the epithelial component and immediately surrounding stroma were included.
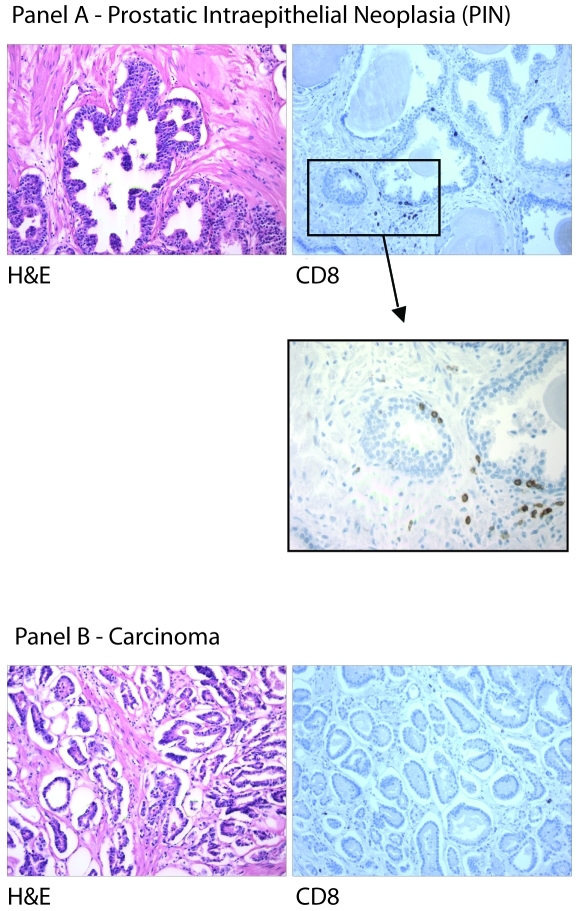
Qualitatively, IHC for CD4 revealed occasional to abundant immuno-positive lymphocytes in the epithelial compartment and the surrounding stroma of all four components analyzed. In contrast, IHC for CD8 cells showed only a few scattered immuno-positive lymphocytes in the epithelium and stroma of normal, BPH, PIN, and tumor (Figure 2). CD20-positive cells (B lymphocytes) were rarely observed in the stroma overall and were not seen associated with any of the epithelial components except for cancer where rare immuno-positive cells were identified.
Figure 2.
CD8 Lymphocytes and Histology. Histological images of CD8-positive cells in prostatic intraepithelial neoplasia (Panel A) and carcinoma (Panel B).
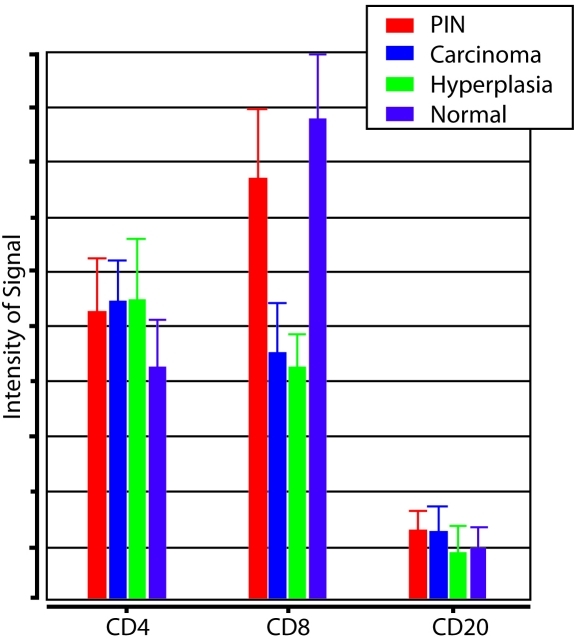
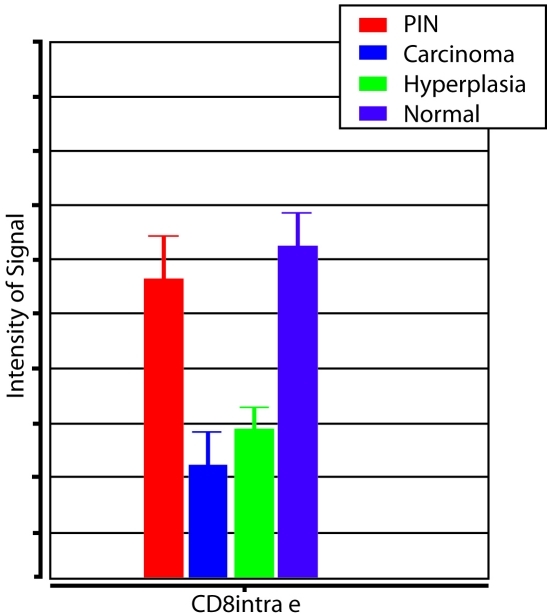
Quantitative analyses of these results are summarized in Figure 3 and Table 1. CD4 positive cells were observed at a similar abundance level in all sub-regions of the prostate. In contrast, CD8 positive cells were significantly less common in regions containing cancer and BPH as opposed to normal epithelium and PIN. Reductions in CD8+ lymphocytes were noted for both the overall histological field (tumor and stroma) as well as the epithelial components of BPH and cancer (Figure 4). CD20 positive cells were generally scarce throughout the prostate and were not significantly associated with any of the epithelial regions (Figure 3).
Figure 3.
CD4, CD8, CD20 Lymphocyte Measurements. Relative number of CD4-, CD8-, and CD20-positive lymphocytes in histopathological regions of prostatic intraepithelial neoplasia (PIN), carcinoma, hyperplasia, and normal epithelium.
Table 1.
Statistical analysis of lymphocyte number comparison amongst PIN, carcinoma, hyperplasia, and normal epithelium. Statistically significant p-values are in blue
| PIN - Carcinoma | PIN - Hyperplasia | PIN – Normal Epithelium | Carcinoma - Hyperplasia | Carcinoma – Normal Epithelium | |
|---|---|---|---|---|---|
| CD3 | 0.05464 | 0.415919 | 0.852019 | 0.250407 | 0.0802136 |
| CD4 | 0.893656 | 0.874042 | 0.443344 | 0.980169 | 0.369018 |
| CD8 | 0.0314564 | 0.0216791 | 0.451762 | 0.872492 | 0.00488537 |
| CD8 Epithelial | 0.000430376 | 0.00376864 | 0.491397 | 0.440662 | 0.0000550 |
| CD20 | 0.94937 | 0.42308 | 0.502703 | 0.460226 | 0.543616 |
Figure 4.
CD8 Lymphocyte Measurements. Relative number of CD8-positive, intraepithelial lymphocytes in histopathological regions of prostatic intraepithelial neoplasia (PIN), carcinoma, hyperplasia, and normal epithelium.
We next assessed expression levels of immune-related transcripts in normal and tumor areas to examine the associated cytokine profile. Data from previous global mRNA measurements from five prostatectomy specimens with cancer were re-analyzed specifically for immune-related features [11]. Both the epithelial and stromal components were individually assessed. Twenty-four genes related to immune function were differentially expressed in the tumor epithelium relative to the normal epithelium, four up-regulated and twenty down-regulated (Table 2). Nine immune-related transcripts were differentially expressed in the tumor-associated stroma (six up-regulated, three down-regulated). The two most highly up-regulated immune-related transcripts in the tumor epithelium were chemokine ligand 14 (CXCL14, 2.55 fold difference), and interleukin 17 receptor B (IL17RB, 2.08 fold difference). CXCL14 was also the most highly dysregulated transcript in the tumor-associated stroma (3.26 fold difference). In contrast, CXCR4 was the third-most negatively regulated transcript (-1.98 fold difference) in the tumor epithelium behind suppressor of cytokine signaling 3 (SOCS3.-2.21 fold difference) and CCAAT/enhancer binding protein (CEBPB.-2.21 fold difference).
Table 2.
Expression microarray comparing tumor versus normal epithelium and comparing tumor stroma versus normal stroma
| Immune Features – Tumor Cells vs Normal Epithelium | ||||
|---|---|---|---|---|
| Gene | Fold Change | OMIM | UniGene ID | Description |
| CXCL14 | 2.55 | 604186 NCBI | 483444 | Chemokine (C-X-C motif) ligand 14 |
| IL17RB | 2.08 | 605458 NCBI | 558512 | Homo sapiens interleukin 17 receptor B |
| PAWR | 1.52 | 601936 NCBI | 406074 | PRKC, apoptosis, WT1, regulator |
| IGSF4 | 1.38 | 605686 NCBI | 370510 | Immunoglobulin superfamily, member 4 |
| C7 | -1.21 | 217070 NCBI; 610102 NCBI | 78065 | Complement component 7 |
| FAS | -1.34 | 134637 NCBI; 601859 NCBI | 244139 | Fas (TNF receptor superfamily, member 6) |
| PRKCA | -1.39 | 176960 NCBI | 531704 | Protein kinase C, alpha |
| NKTR | -1.43 | 161565 NCBI | 529509 | Natural killer-tumor recognition sequence |
| ELF4 | -1.43 | n/a | 271940 | E74-like factor 4 (ets domain transcription factor) |
| HLA-DPA1 | -1.44 | 142880 NCBI | 347270 | Major histocompatibility complex, class II, DP alpha 1 |
| CXCR7 | -1.48 | 610376 NCBI | 471751 | Chemokine (C-X-C motif) receptor 7 |
| IFIT3 | -1.50 | n/a | 47338 | Interferon-induced protein with tetratricopeptide repeats 3 |
| ETS1 | -1.65 | 164720 NCBI | 369438 | v-ets erythroblastosis virus E26 oncogene homolog 1 |
| CD69 | -1.68 | 107273 NCBI | 208854 | CD69 antigen (p60, early T-cell activation antigen) |
| BCL2 | -1.70 | 151430 NCBI | 150749 | B-cell CLL/lymphoma 2 |
| CX3CL1 | -1.73 | 601880 NCBI | 531668 | Chemokine (C-X3-C motif) ligand 1 |
| IFI16 | -1.74 | 147586 NCBI | 380250 | Interferon, gamma-inducible protein 16 |
| CLU | -1.75 | 185430 NCBI | 436657 | Clusterin |
| IL6ST | -1.77 | 600694 NCBI | 532082 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| PTGER4 | -1.89 | 601586 NCBI | 199248 | Prostaglandin E receptor 4 (subtype EP4) |
| IL6ST | -1.91 | 600694 NCBI | 532082 | Interleukin 6 signal transducer (gp130) |
| CXCR4 | -1.98 | 162643 NCBI; 193670 NCBI | 421986 | Chemokine (C-X-C motif) receptor 4 |
| CEBPB | -2.11 | 189965 NCBI | 517106 | CCAAT/enhancer binding protein (Interleukin 6-dependent DNA-binding protein) |
| SOCS3 | -2.21 | 604176 NCBI | 527973 | Suppressor of cytokine signaling 3 |
| Immune Features – Tumor Stroma vs Normal Stroma | ||||
| Gene | Fold Change | OMIM | UniGene ID | Description |
| CXCL14 | 3.26 | 604186 NCBI | 483444 | Chemokine (C-X-C motif) ligand 14 |
| IGSF4D | 1.97 | 609938 NCBI | 164578 | Immunoglobulin superfamily, member 4D |
| CD24 | 1.78 | 126200 NCBI; 600074 NCBI | 632285 | CD24 antigen |
| TNFSF10 | 1.62 | 603598 NCBI | 478275 | Tumor necrosis factor (ligand) superfamily, member 10 |
| AZGP1 | 1.34 | 194460 NCBI | 546239 | Alpha-2-glycoprotein 1, zinc-binding |
| ARTS-1 | 1.20 | 606832 NCBI | 436186 | Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator |
| C3 | -1.14 | 120700 NCBI | 529053 | Complement component 3 |
| IGKC | -1.48 | 147200 NCBI | 552522 | Immunoglobulin kappa constant |
| HLA-C | -1.80 | 134580 NCBI; 147200 NCBI | 552522 | Major histocompatibility complex, class I, C |
Discussion
The results of the present study indicate that decreased numbers of CD8+ lymphocytes are associated with prostate tumors, and that there is a significant dysregulation of cytokines in histological regions that contain cancer. T-cell lymphocytes are an important element in the immune response and are regulated through a cascade of cellular and cytokine-based interactions. CD4+ lymphocytes, or T-helper cells, recognize antigens in conjunction with antigen presenting cells (APC) and produce cytokines that induce a wide response against foreign proteins on viruses or those on tumor cells [12-14]. In contrast, CD8+ cytotoxic T lymphocytes directly attack and kill infected cells or cancers via binding of the CD8 glycoprotein and T-cell receptor to Class I MHC molecules on the target cells. CD8+ cells release cytotoxic perforin and granzymes that initiate a series of cell killing events that include formation of aqueous channels, activation of cellular cysteine proteases and caspases, and ultimately apoptosis of the infected or cancerous cell [13]. A second way to induce apoptosis is via cell-surface interactions between the CD8+ cells and the surface protein FAS ligand (FasL)(Apo1L)(CD95L) that binds to Fas (Apo1)(CD95) molecules expressed on the target cell [13]. Engagement of Fas with FasL allows for recruitment of the death-induced silencing complex (DISC). The Fas-associated death domain (FADD) translocates with the DISC, allowing recruitment of procaspases 8 and 10 which then activate the effector caspases 3, 6, and 7, leading to cleavage of death substrates such as lamin A, lamin B1, lamin B2, PARP, and DNAPK. The final result is apoptosis of the target cell that expresses Fas [13].
With respect to the current study, the observed decrease in CD8+ lymphocytes may facilitate tumor development by removing this important tumor-cytotoxic aspect of the immune response. This result is consistent with the previous work of Naoe and colleagues [14] who assessed MHC class I expression and lymphocytes in both benign and malignant prostate tissue and found significantly fewer CD8+ T lymphocytes associated with prostate tumor cells than in benign prostate, including a significant correlation between the number of CD8+ T lymphocytes and MHC class I expression. The authors concluded this relationship might be important in cancer growth. Our study extends this observation further by analyzing CD8+ lymphocytes in the context of a detailed histopathological examination, and further analyzing cytokine profiles at the transcript level.
Overall, the cytokine mRNA pattern showed mostly down-regulation of genes in tumor cells and up-regulation in the tumor stroma. The directionality of immune-related mRNA changes is the same as we have previously observed in microdissection-based global gene expression studies of both prostate cancer and esophageal squamous cell carcinoma; more down-regulated transcripts in tumor cells as compared to corresponding normal epithelium, and more up-regulated mRNAs in tumor associated stroma versus normal quiescent stroma [11]. Changes in genes encoding inflammatory proteins included up-regulation of PAWR and IGSF4 and down regulation of SOCS3, CEBPB, IL6ST, CLU and IFI16 in the tumor cells. In the tumor-associated stroma, up-regulation of IGSF4D, CD24, and TNFSF10 was seen, and down-regulation of HLA-C, IGKC, and C3 was observed .
The data on CXCR4 were among the most interesting findings in the study as these results differed from some reports in the literature and there is extensive evidence that CXCR4 is involved in development or progression of multiple different tumor types [15-19]. CXCR4 is a seven transmembrane G protein-coupled receptor that plays a role in many physiological processes involving cell migration and cell fate, such as vascular formation in the gastrointestinal tract and other sites of angiogenesis, stem cell homing, neural development, and immune cell trafficking [15, 16]. CXCR4's ligand, CXCL12/ SDF-1, is constitutively expressed in many cell types, including endothelium, keratinocytes, sweat glands, and sites of chronic inflammation [17]. The signaling mechanism by which CXCR4 promotes cell migration through CXCL12 is mediated through the Ga13-Rho pathway [16]. Apart from leukocytes, in which CXCR4 is ubiquitously present, expression of CXCR4 is low or absent in many normal tissues, including breast, ovary, and colon. However, CXCR4 is implicated in various pathological conditions, including metastatic spread and human immunodeficiency virus infection, and CXCL12 is expressed in common sites of metastasis (lung, liver, bone). CXCR4 is also frequently expressed by human cancer cells and to date more than 20 different tumor types are known to up-regulate CXCR4 protein [18]. In contrast, our data showed decreased levels of CXCR4 mRNA in microdissected tumor cells. A recent report by Singh et al studying CXCR4 in prostate cancer provides a potential mechanistic explanation for this finding [19]. These investigators found that CXCR4 induces caspase-mediated apoptosis in prostate tumor cells when stimulated by gp-120-IIIB, thus diminished levels of CXCR4 could have a protective effect. Based on these findings, additional study of CXCR4 and its ligands are warranted in prostate cancer.
In summary, by using an open, non-directed histomathematics analysis technique, we were able to identify differential expression in the inflammatory infiltrate in human prostate cancer. This report highlights the importance of quantitative, high throughput approaches in pathological specimens, efforts that can lead to new insight into disease pathogenesis.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Kim R, Emi M, Tanabe K, Arihiro K. Tumor-driven evolution of immunosuppressive networks during malignant progression. Cancer Res. 2006;66:5527–36. doi: 10.1158/0008-5472.CAN-05-4128. [DOI] [PubMed] [Google Scholar]
- 2.Nanda NK, Birch L, Greenberg NM, Prins GS. MHC class I and class II molecules are expressed in both human and mouse prostate tumor microenvironment. Prostate. 2006;66:1275–84. doi: 10.1002/pros.20432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrulio CA, Kim-Schulze S, Kaufman HL. The tumour microenvironment and implications for cancer immunotherapy. Expert Opin Biol Ther. 2006;6:671–84. doi: 10.1517/14712598.6.7.671. [DOI] [PubMed] [Google Scholar]
- 4.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264:169–84. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 5.Pardoll D. Does the immune system see tumors as foreign or self? Annu Rev Immunol. 2003;21:807–39. doi: 10.1146/annurev.immunol.21.120601.141135. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnett CT, Palena C, Chakraborty M, Tsang KY, Schlom J, Hodge JW. Sublethal irradiation of human tumor cells modulates phenotype resulting in enhanced killing by cytotoxic T lymphocytes. Cancer Res. 2004;64:7985–94. doi: 10.1158/0008-5472.CAN-04-1525. [DOI] [PubMed] [Google Scholar]
- 8.Karja V, Aaltomaa S, Lipponen P, Isotalo T, Talja M, Mokka R. Tumour-infiltrating lymphocytes: A prognostic factor of PSA-free survival in patients with local prostate carcinoma treated by radical prostatectomy. Anticancer Res. 2005;25:4435–8. [PubMed] [Google Scholar]
- 9.Gannot G, Gillespie JW, Chuaqui RF, Tangrea MA, Linehan WM, Emmert-Buck MR. His-tomathematical analysis of clinical specimens: challenges and progress. J Histochem Cytochem. 2005;53:177–85. doi: 10.1369/jhc.4A6457.2005. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie JW, Best CJ, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JI, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan WM, Duray PH, Bova GS, Emmert-Buck MR. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160:449–57. doi: 10.1016/S0002-9440(10)64864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson AM, Woodson K, Wang Y, Rodriguez-Canales J, Erickson HS, Tangrea MA, Novakovic K, Gonzalez S, Velasco A, Kawasaki ES, Emmert-Buck MR, Chuaqui RF, Player A. Global expression analysis of prostate cancer associated stroma and epithelia. Diagn Mol Pathol. 2007;16:189–97. doi: 10.1097/PDM.0b013e3180de20ac. [DOI] [PubMed] [Google Scholar]
- 12.Gannot G, Gannot I, Vered H, Buchner A, Keisari Y. Increase in immune cell infiltration with progression of oral epithelium from hyperkeratosis to dysplasia and carcinoma. Br J Cancer. 2002;86:1444–8. doi: 10.1038/sj.bjc.6600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez-Galan L, Arenas-Del Angel MC, Zenteno E, Chavez R, Lascurain R. Cell death mechanisms induced by cytotoxic lymphocytes. Cell Mol Immunol. 2009;6:15–25. doi: 10.1038/cmi.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naoe M, Marumoto Y, Ishizaki R, Ogawa Y, Nakagami Y, Yoshida H. Correlation between major histocompatibility complex class I molecules and CD8+ T lymphocytes in prostate, and quantification of CD8 and interferon-gamma mRNA in prostate tissue specimens. BJU Int. 2002;90:748–53. doi: 10.1046/j.1464-410x.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 15.lyengar S, Schwartz DH, Hildreth JE. T cell-tropic HIV gpl20 mediates CD4 and CD8 cell chemo-taxis through CXCR4 independent of CD4: Implications for HIV pathogenesis. J Immunol. 1999;162:6263. [PubMed] [Google Scholar]
- 16.Tan W, Martin D, Gutkind JS. The Galphal3-Rho signaling axis is required for SDF-1-induced migration through CXCR4. J Biol Chem. 2006;281:39542–9. doi: 10.1074/jbc.M609062200. [DOI] [PubMed] [Google Scholar]
- 17.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–31. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Baruch A. The multifaceted roles of chemokines in malignancy. Cancer Metastasis Rev. 2006;25:357–71. doi: 10.1007/s10555-006-9003-5. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Bond VC, Powell M, Singh UP, Bumpers HL, Grizzle WE, Lillard JW Jr. CXCR4-gpl20-IIIB interactions induce caspase-mediated apoptosis of prostate cancer cells and inhibit tumor growth. Mol Cancer Ther. 2009;8:178–84. doi: 10.1158/1535-7163.MCT-08-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]