Abstract
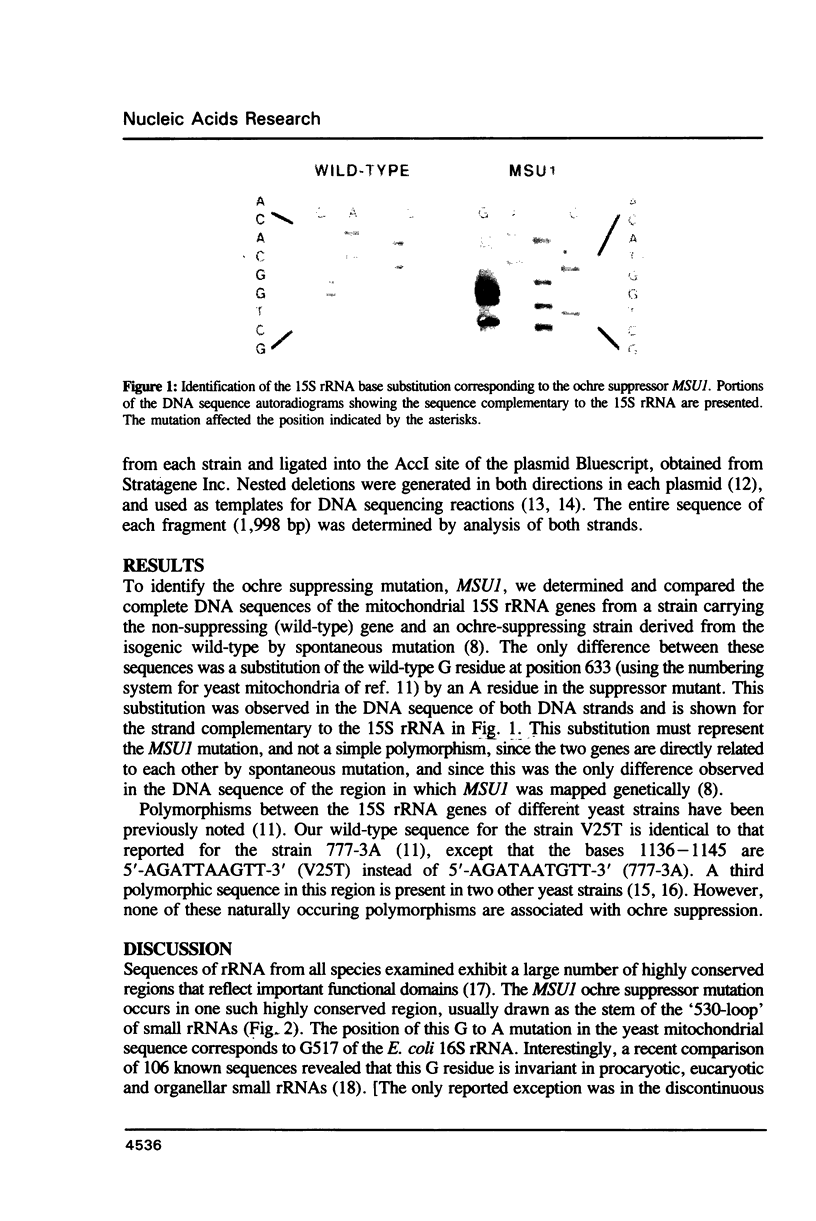
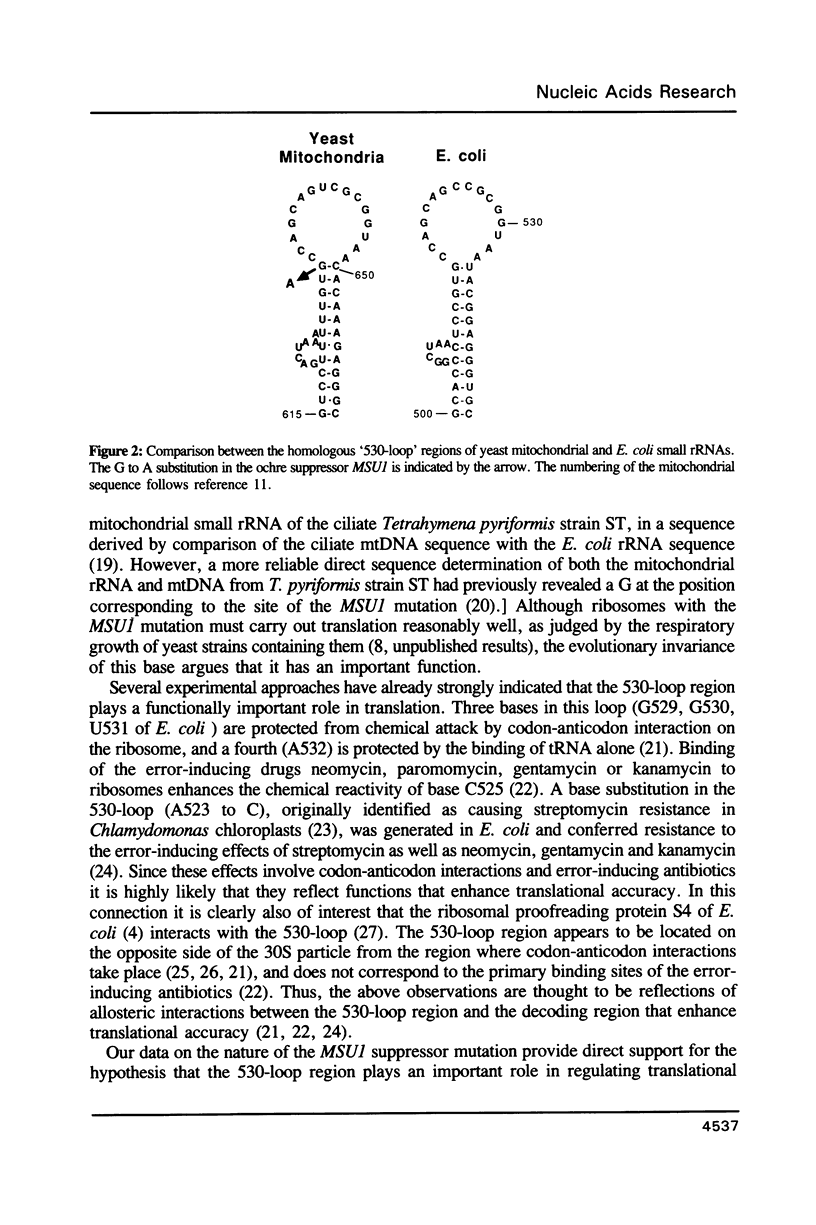
We have determined the nucleotide sequence alteration in the 15S rRNA gene of a Saccharomyces cerevisiae strain carrying the previously described mitochondrial ochre suppressor, MSUI. The suppressor contains an A residue at position 633 of the yeast mitochondrial sequence, in place of the wild-type G. This position, located in the highly conserved region forming the stem of the '530-loop', corresponds to G517 of the Escherichia coli 16S rRNA and is occupied by G in all other known small rRNA sequences. This finding strongly supports the previous conclusions of others that the 530-loop region plays an important role in enhancing translational accuracy.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson D. I., Kurland C. G. Ram ribosomes are defective proofreaders. Mol Gen Genet. 1983;191(3):378–381. doi: 10.1007/BF00425749. [DOI] [PubMed] [Google Scholar]
- Blanc H., Wright C. T., Bibb M. J., Wallace D. C., Clayton D. A. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3' end of the large ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3789–3793. doi: 10.1073/pnas.78.6.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. Mitochondrial and nuclear mutations that affect the biogenesis of the mitochondrial ribosomes of yeast. I. Genetics. Mol Gen Genet. 1979;177(1):39–46. doi: 10.1007/BF00267251. [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B., Bolotin-Fukuhara M. Mutational study of the rRNA in yeast mitochondria: functional importance of T1696 in the large rRNA gene. Nucleic Acids Res. 1988 Oct 11;16(19):9299–9306. doi: 10.1093/nar/16.19.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Eustice D. C., Wakem L. P., Wilhelm J. M., Sherman F. Altered 40 S ribosomal subunits in omnipotent suppressors of yeast. J Mol Biol. 1986 Mar 20;188(2):207–214. doi: 10.1016/0022-2836(86)90305-0. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Genetic and physical analysis of the mitochondrial gene for subunit II of yeast cytochrome c oxidase. J Mol Biol. 1979 May 5;130(1):63–82. doi: 10.1016/0022-2836(79)90552-7. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Staempfli S. Suppressor of yeast mitochondrial ochre mutations that maps in or near the 15S ribosomal RNA gene of mtDNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1583–1587. doi: 10.1073/pnas.79.5.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Weiss-Brummer B. Leaky +1 and -1 frameshift mutations at the same site in a yeast mitochondrial gene. Nature. 1980 Nov 6;288(5786):60–63. doi: 10.1038/288060a0. [DOI] [PubMed] [Google Scholar]
- Fromm H., Edelman M., Aviv D., Galun E. The molecular basis for rRNA-dependent spectinomycin resistance in Nicotiana chloroplasts. EMBO J. 1987 Nov;6(11):3233–3237. doi: 10.1002/j.1460-2075.1987.tb02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A., Turmel M., Lemieux C. Mapping of chloroplast mutations conferring resistance to antibiotics in Chlamydomonas: evidence for a novel site of streptomycin resistance in the small subunit rRNA. Mol Gen Genet. 1988 Oct;214(2):192–197. doi: 10.1007/BF00337710. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Nurse K., Hellmann W., Boublik M., Ofengand J. High resolution localization of the tRNA anticodon interaction site on the Escherichia coli 30 S ribosomal subunit. J Biol Chem. 1984 Aug 25;259(16):10493–10498. [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Sakai H., Weiss-Brummer B. Site-specific AT-cluster insertions in the mitochondrial 15S rRNA gene of the yeast S. cerevisiae. Nucleic Acids Res. 1988 Sep 12;16(17):8665–8674. doi: 10.1093/nar/16.17.8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Isaksson L. A. Involvement of ribosomal protein L7/L12 in control of translational accuracy. Proc Natl Acad Sci U S A. 1985 Feb;82(3):717–721. doi: 10.1073/pnas.82.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labriola J., Weiss I., Zapatero J., Suyama Y. Unexpectedly long 14S ribosomal RNA gene in Tetrahymena mitochondria. Curr Genet. 1987;11(6-7):529–536. doi: 10.1007/BF00384616. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., Underbrink-Lyon K., Martin N. C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982 May 25;257(10):5921–5928. [PubMed] [Google Scholar]
- Melançon P., Lemieux C., Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988 Oct 25;16(20):9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Montandon P. E., Nicolas P., Schürmann P., Stutz E. Streptomycin-resistance of Euglena gracilis chloroplasts: identification of a point mutation in the 16S rRNA gene in an invariant position. Nucleic Acids Res. 1985 Jun 25;13(12):4299–4310. doi: 10.1093/nar/13.12.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montandon P. E., Wagner R., Stutz E. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 1986 Dec 20;5(13):3705–3708. doi: 10.1002/j.1460-2075.1986.tb04703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Hijazi K. A., Göringer H. U., Dahlberg A. E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Oakes M. I., Clark M. W., Henderson E., Lake J. A. DNA hybridization electron microscopy: ribosomal RNA nucleotides 1392-1407 are exposed in the cleft of the small subunit. Proc Natl Acad Sci U S A. 1986 Jan;83(2):275–279. doi: 10.1073/pnas.83.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset R., Gorini L. A ribosomal ambiguity mutation. J Mol Biol. 1969 Jan 14;39(1):95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Heinonen T. Y., Young P. G., Gray M. W. A discontinuous small subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J Biol Chem. 1986 Apr 15;261(11):5187–5193. [PubMed] [Google Scholar]
- Singh A., Mason T. L., Zimmermann R. A. A cold-sensitive cytoplasmic mutation of Saccharomyces cerevisiae affecting assembly of the mitochondrial 50S ribosomal subunit. Mol Gen Genet. 1978 May 3;161(2):143–151. doi: 10.1007/BF00274184. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Hüttenhofer A., Kaudewitz F. Leakiness of termination codons in mitochondrial mutants of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;198(2):62–68. doi: 10.1007/BF00328702. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Sakai H., Kaudewitz F. A mitochondrial frameshift-suppressor (+1) [corrected] of the yeast S. cerevisiae maps in the mitochondrial 15S rRNA locus. Curr Genet. 1987;11(4):295–301. doi: 10.1007/BF00355403. [DOI] [PubMed] [Google Scholar]




