Abstract
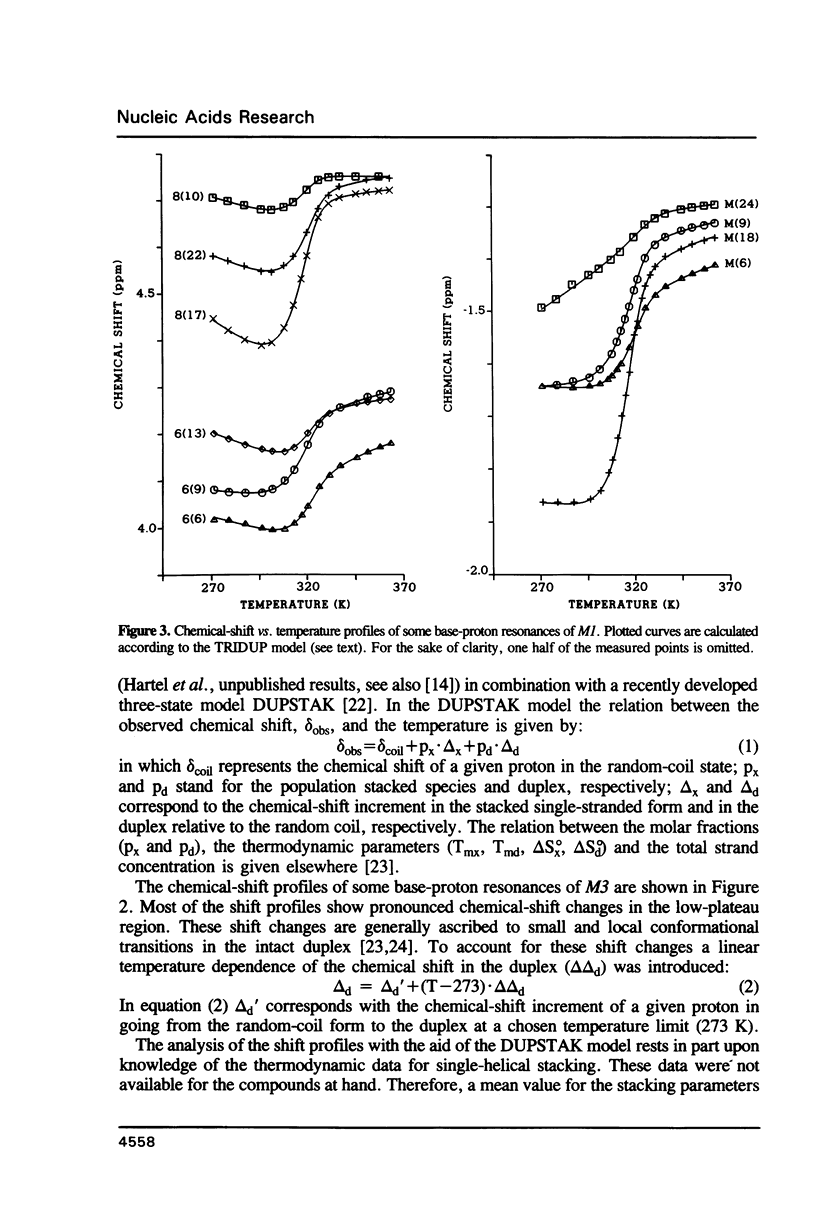
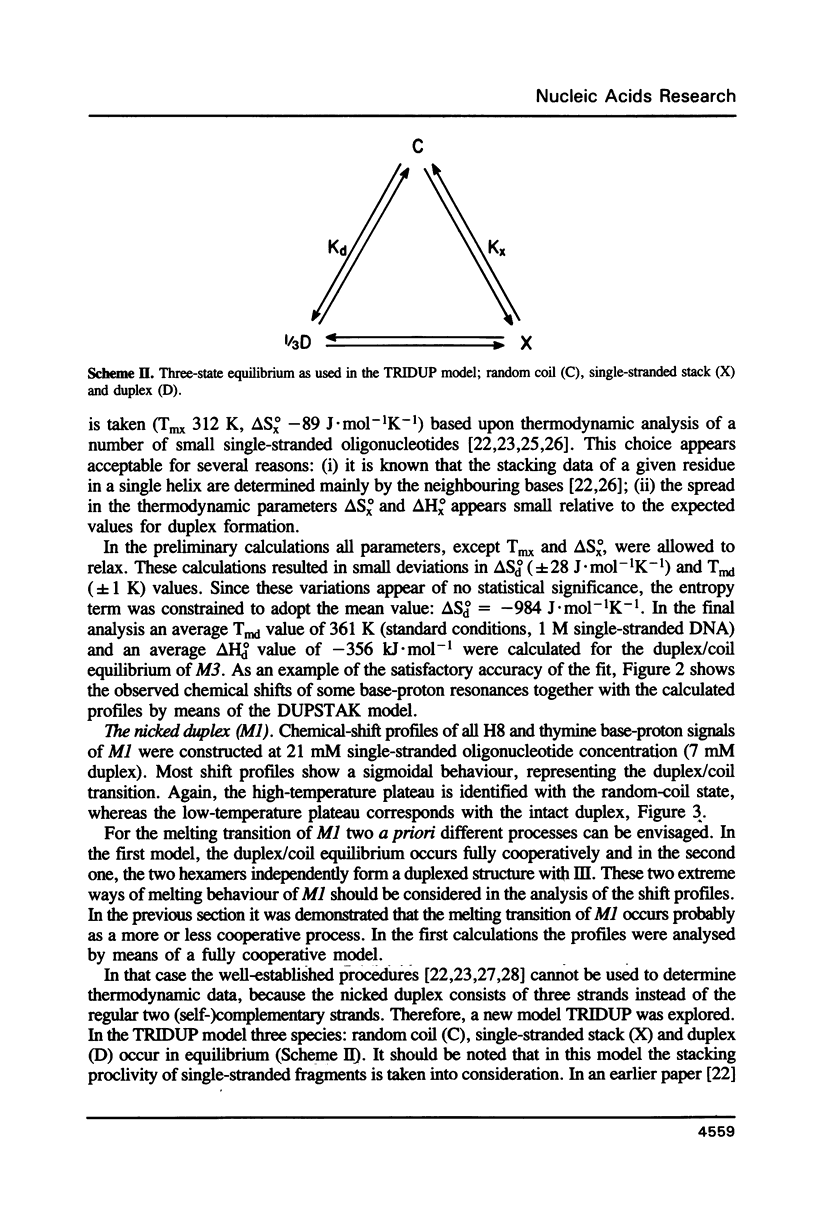
NMR studies were carried out on various equimolar mixtures consisting of a combination of oligomers: d(ACGGCT) (I). d(pACGGCT) (Ia), d(TGCAGT) (II), d(AGCCGTACTGCA) (III), d(TGCAGTACGGCT) (IV). It is shown that I + II + III (MI) and Ia + II + III (M2) form stable duplexes with nicks in the centre of the respective double helices. A close analysis of the NOESY experiments of M1 and M2 revealed that these fragments form B-DNA type duplex structures. A comparison of the chemical-shift data of the nicked duplexes with those of the intact duplex of III + IV (M3) demonstrated that only small local distortions occur when a nick is introduced. The chemical-shift profiles of M1 and M3 were used to obtain the thermodynamic data for the duplex/coil transitions. The profiles of M1 were analysed by means of a new thermodynamic model (TRIDUP). From the calculated thermodynamic data of M1 and M3 it is concluded that the melting behaviour of M1 occurs cooperatively. A ligation experiment demonstrated that the relatively small substrate (M2) was almost completely joined after an overnight incubation at 14 degrees C.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hartel A. J., Lankhorst P. P., Altona C. Thermodynamics of stacking and of self-association of the dinucleoside monophosphate m2(6)A-U from proton NMR chemical shifts: differential concentration temperature profile method. Eur J Biochem. 1982 Dec 15;129(2):343–357. doi: 10.1111/j.1432-1033.1982.tb07057.x. [DOI] [PubMed] [Google Scholar]
- Heerschap A., Walters J. A., Hilbers C. W. Interactions of some naturally occurring cations with phenylalanine and initiator tRNA from yeast as reflected by their thermal stability. Biophys Chem. 1985 Aug;22(3):205–217. doi: 10.1016/0301-4622(85)80044-2. [DOI] [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Jellema A. K., Haasnoot C. A., Van Boom J. H., Altona C. Conformational analysis of the single-helical DNA fragment d(T-A-A-T) in aqueous solution. The combined use of NMR proton chemical shifts and coupling constants obtained at 300 MHz and 500 MHz. Eur J Biochem. 1984 May 15;141(1):165–175. doi: 10.1111/j.1432-1033.1984.tb08171.x. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Pieters J. M., van der Marel G. A., van Boom J. H., Haasnoot C. A., Altona C. Sequence-dependent structural variation in single-helical DNA. Proton NMR studies of d(T-A-T-A) and d(A-T-A-T) in aqueous solution. Eur J Biochem. 1984 Sep 3;143(2):285–301. doi: 10.1111/j.1432-1033.1984.tb08371.x. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Gilham P. T. Anomalous hairpin formation in an oligodeoxyribonucleotide. Nucleic Acids Res. 1985 Nov 25;13(22):8259–8274. doi: 10.1093/nar/13.22.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. M., Mellema J. R., van den Elst H., van der Marel G. A., van Boom J. H., Altona C. Thermodynamics of the various forms of the dodecamer d(ATTACCGGTAAT) and of its constituent hexamers from proton nmr chemical shifts and UV melting curves: three-state and four-state thermodynamic models. Biopolymers. 1989 Mar;28(3):717–740. doi: 10.1002/bip.360280304. [DOI] [PubMed] [Google Scholar]
- Quignard E., Fazakerley G. V., Teoule R., Guy A., Guschlbauer W. Consequences of methylation on the amino group of adenine. A proton two-dimensional NMR study of d(GGATATCC) and d(GGm6ATATCC). Eur J Biochem. 1985 Oct 1;152(1):99–105. doi: 10.1111/j.1432-1033.1985.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., Sanderson M. R., van der Marel G. A., van Boom J. H., Altona C. Conformational analysis of the octamer d(G-G-C-C-G-G-C-C) in aqueous solution. A one-dimensional and two-dimensional proton NMR study at 300 MHz and 500 MHz. Eur J Biochem. 1986 Aug 15;159(1):85–93. doi: 10.1111/j.1432-1033.1986.tb09836.x. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of the base sequence on the conformational behaviour of DNA polynucleotides in solution. Eur J Biochem. 1987 Jul 1;166(1):87–101. doi: 10.1111/j.1432-1033.1987.tb13487.x. [DOI] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H., Joordens J., De Bruin S. H., Hilbers C. W. Destabilization of secondary structure in 16S ribosomal RNA by dimethylation of two adjacent adenosines. Nucleic Acids Res. 1981 Sep 11;9(17):4413–4422. doi: 10.1093/nar/9.17.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters J. A., Geerdes H. A., Hilbers C. W. On the binding of Mg2+ and Mn2+ to tRNA. Biophys Chem. 1977 Sep;7(2):147–151. doi: 10.1016/0301-4622(77)80007-0. [DOI] [PubMed] [Google Scholar]
- van der Marel G. A., van Boeckel C. A., Wille G., van Boom J. H. A general method for the synthesis of 5'-monophosphates of DNA fragments via phosphotriester intermediates. Nucleic Acids Res. 1982 Apr 10;10(7):2337–2351. doi: 10.1093/nar/10.7.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]