Abstract
Cancer cells exhibit the ability to proliferate indefinitely, but paradoxically, overexpression of cellular oncogenes in primary cells can result in a rapid and irreversible cell cycle arrest known as oncogene-induced senescence (OIS). However, we have shown that constitutive overexpression of the oncogene c-MYC in primary human foreskin fibroblasts results in a population of cells with unlimited lifespan; these immortalized cells are henceforth referred to as iMYC. Here, in order to further elucidate the mechanisms underlying the immortalization process, a gene expression signature of three independently established iMYC cell lines compared with matched early passage c-MYC overexpressing cells was derived. Network analysis of this “iMYC signature” indicated that a large fraction of the downregulated genes were functionally connected and major nodes centered around the TGFβ, IL-6 and IGF-1 signaling pathways. Here, we focused on the functional validation of the alteration of TGFβ response during c-MYC-mediated immortalization. The results demonstrate loss of sensitivity of iMYC cells to activation of TGFβ signaling upon ligand addition. Furthermore, we show that aberrant regulation of the p27 tumor suppressor protein in iMYC cells is a key event that contributes to loss of response to TGFβ. These findings highlight the potential to reveal key pathways contributing to the self-renewal of cancer cells through functional mining of the unique gene expression signature of cells immortalized by c-MYC.
Keywords: c-MYC, gene expression signature, immortalization, TGFβ, p27
Introduction
Replicative senescence is a state of irreversible cell cycle arrest that cultured human fibroblasts enter after approximately 80 population doublings.1,2 Bypass of senescence can result in immortalization, an extension of lifespan that is critical for transformation of normal cells.3 Tumor cells have acquired the ability to proliferate indefinitely, either by disabling senescence pathways or by activating genes that override senescence checkpoints.3 Unsurprisingly, regulators of cellular lifespan and senescence are known tumor suppressors that are often disrupted in tumor cells.
Cancer cells characteristically lose expression of the tumor suppressor genes p16 and ARF encoded by the INK4A/ARF locus,4 which have been shown to directly induce cell cycle arrest or cell death through the Rb- and p53-dependent tumor suppressor pathways,5,6 respectively. Additionally, other cell cycle regulators including p217 and p278,9 are known to have tumor suppressive functions through multiple mechanisms, including limitation of cellular lifespan. Inactivation of the aforementioned tumor suppressors results in extension of lifespan but an additional event required for immortalization is the expression of hTERT, the catalytic subunit of telomerase that is inactive in most somatic cells and is responsible for maintenance of telomere length.10 Indeed, hTERT expression successfully immortalizes human cells,11,12 and has been shown to have a key role in tumorigenesis (reviewed in ref. 13).
Cell immortalization has not been clearly linked to activation of oncogenes, as cells in culture that overexpress the known oncogene RAS soon arrest.14 This phenomenon is known as oncogene-induced senescence (OIS) and is dependent on functional p1615,16 and ARF17,18 as well as mTOR, which has been shown to be required for induction of senescence during oncogene-induced cell cycle arrest.19–22 In contrast, we have shown that primary human foreskin fibroblasts (HFFs) do not undergo OIS.23 HFFs overexpressing RAS not only continue to proliferate but also exhibit properties of transformation including anchorage-independent growth. However, RAS does not extend the lifespan of HFFs.15
In contrast to RAS overexpression, we have shown that overexpression of the oncogene c-MYC in HFFs resulted in the establishment of immortalized cell populations.17 These cells, which we refer to here as iMYC, have continued to proliferate with greater than 220 population doublings to date. Immortalization by c-MYC was shown to be a reproducible event in HFFs isolated from different foreskin donors, providing several independently established iMYC lines. It has been shown that iMYC cells are oligo-clonal and proliferate despite retention of functional p53 and p16 response pathways.17 These observations suggest that additional changes have occurred to enable bypass of cellular lifespan limitations and achieve immortalization. We previously demonstrated that loss of ARF expression due to promoter methylation is one such change,17 but this was not sufficient for immortalization.
In this study, we utilized genome-wide microarray analysis of iMYC cells in comparison with their matched early passage c-MYC overexpressing cells, referred to as eMYC, to elucidate gene expression changes that occur during immortalization by c-MYC. Expression profiles obtained from three independently established iMYC cell lines were analyzed. An iMYC characteristic signature was obtained by identification of genes that were commonly regulated in all three lines relative to their genetically matched eMYC cells. In this iMYC signature, several candidate genes and regulatory pathways were altered that affect cellular proliferation and lifespan. We focused on the TGFβ signaling pathway, which has been shown previously to have a tumor suppressive effect on untransformed cells.24 Indeed, iMYC cells did not show growth inhibition in response to treatment with a TGFβ ligand, while eMYC cells did. Sensitivity to TGFβ ligand in eMYC cells was dependent on increased levels of the tumor suppressor p27 protein. These data reveal that the tumor suppressor function of the TGFβ pathway has been inactivated in iMYC cells, thus eliminating an essential roadblock to c-MYC-induced immortalization.
Results
Establishment of the “iMYC signature” gene expression profile.
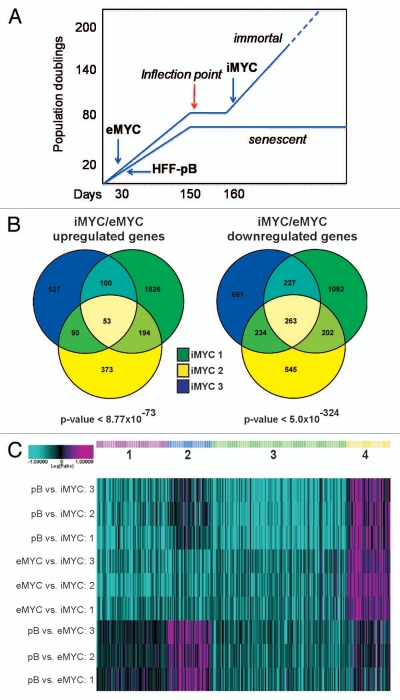
We previously showed that gain of hTERT function and loss of ARF expression were necessary but not sufficient for immortalization of human foreskin fibroblasts (HFFs) by c-MYC.17 In order to identify additional changes that cooperate with c-MYC to immortalize HFFs, microarray analysis was performed on samples from three independently established c-MYC immortalized HFF lines, which we have identified as iMYC lines. These iMYC lines were isolated after the slow-growth “inflection point” that reproducibly occurred prior to establishment of an immortalized cell population (Fig. 1A). Relative gene expression levels from iMYC cells were compared with expression profiles obtained from HFFs transduced with empty control vector (pB) as well as early passage number HFFs transduced with c-MYC retroviral vector (eMYC). These early passage samples were collected within 10 passages after transduction (Fig. 1A). All comparisons were isogenic; each iMYC line was compared with its matched parental HFF-pB line and eMYC line. In order to identify candidate genes involved in the immortalization process, and not merely due to c-MYC overexpression, we focused on gene expression changes in iMYC cells relative to those in eMYC cells. Among all three cell lines analyzed, there were 53 genes upregulated and 263 genes downregulated (Fig. 1B), and we will refer to this gene set as the “iMYC signature” (see Table S1 for gene lists). The statistical significance of the overlap indicated that the common gene expression changes in the iMYC lines were extremely unlikely to be due to chance. For upregulated genes the p-value of the overlap was <8.77E-73, and for downregulated genes the p-value was even more significant at <5E-324. Calculation of the p-values for pairwise overlaps of all possible combinations is also shown in Table S2.
Figure 1.
Gene expression signature of c-MYC-immortalized human fibroblasts, “iMYC.” (A) Schematic of the derivation of c-MYC-immortalized cell lines, iMYC, from three independent human foreskin fibroblast (HFF) isolates. HFFs were tranduced with a c-MYC expressing retroviral vector, pBabe-puro, as described previously in reference 17. Samples of iMYC lines used in the described experiments were collected past the “inflection point” (an event that occurred at >80 population doublings) when cells were uniformly growing as an immortalized population. For comparison, empty vector cells, referred to as HFF-pB and c-MYC expressing HFFs, referred to as eMYC, were collected prior to the “inflection point” within the first 10–30 d in culture (thus “e” for early passage) where cells have undergone fewer than 20 population doublings. (B) Venn diagram indicating the overlap of significantly upregulated (53) and downregulated (263) genes in three independently derived iMYC cells in comparison with eMYC. The p-value of each overlap is shown. (C) K-means clustering analysis identified four groups of genes within the iMYC signature that correctly separated the three cell lines: (1) genes unchanged in eMYC but downregulated in iMYC, (2) genes upregulated in eMYC but downregulated in iMYC to pB levels, (3) genes downregulated in both eMYC and iMYC but more strongly down in iMYC, (4) genes unchanged in eMYC but up in iMYC.
To further investigate more complex patterns of gene expression changes during the immortalization process, microarray data for the iMYC signature gene set was also analyzed by considering the expression levels of genes in the HFF-pB cells as the “basal” level of expression in normal human fibroblasts. Regulation of each gene in the iMYC signature was then evaluated by measuring expression levels in iMYC cells compared with eMYC cells and pB cells. K-means clustering analysis of differentially expressed genes indicated that there were four groups of genes that clustered the three different types of cells, HFF-pB, eMYC and iMYC, correctly (Fig. 1C). One set of genes was unchanged in eMYC cells but downregulated in iMYC cells (Group 1); another set was upregulated in eMYC cells but not in iMYC cells where the levels were comparable to pB (Group 2); a third set was downregulated in eMYC cells, but even further downregulated in iMYC cells (Group 3). Finally, the fourth group of genes identified by K-means clustering included all 53 upregulated genes, which were unchanged in eMYC cells but upregulated in iMYC cells (Group 4) (Fig. 1C). In summary, this analysis supports the conclusion that the gene expression profile of iMYC cell lines identifies these cells as separate entities whether their expression profiles were compared with empty vector control or to eMYC cells. These results also indicate that the gene expression changes detected in the iMYC cell lines did not represent an amplification of a c-MYC overexpression signature, but instead constitute a distinct profile that consistently arises in c-MYC-mediated cell immortalization.
Disruption of the TGFβ signaling pathway in iMYC cells.
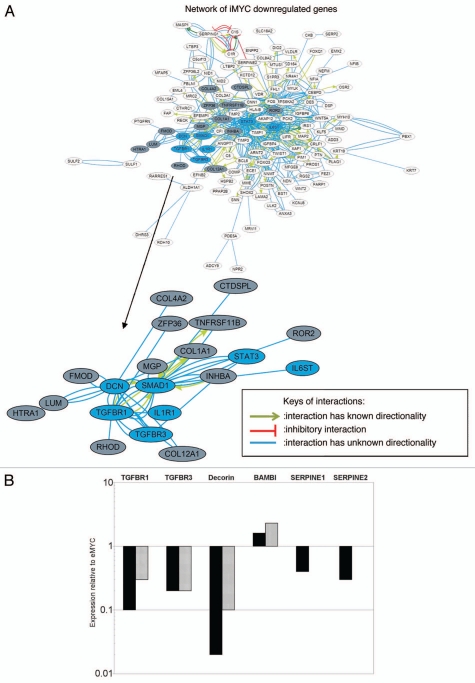
Pathway analysis of the iMYC signature was performed for both the 263 downregulated gene set as well as for the 53 upregulated genes. A broad overview of the known connections among the 263 downregulated genes based on an Ingenuity curated literature database (Fig. 2A) indicated that the majority of the genes are functionally connected. Inspection of the network indicated that several genes belong to the TGFβ signaling pathway, forming a subnetwork (enlarged inset, Fig. 2A). The enrichment for genes belonging to the TGFβ pathway was also verified utilizing Ingenuity Pathway Analysis canonical pathways (Fig. S1A; p-value < 3.56E-03). Furthermore, the majority of these genes were classified in Group 1 and Group 2 in the K-means clustering analysis, indicating that downregulation of genes in the TGFβ pathway occurred only in the iMYC cells.
Figure 2.
Downregulation of TGFβ pathway genes in iMYC cells. (A) Network analysis of the iMYC signature showed that a large number of the 263 commonly downregulated genes form a broad functional network as indicated by the connecting lines, each representing a known literature-based link between the genes (Ingenuity Pathway Analysis database). A color key defining the types of interactions between the genes is shown. The network centered around the TGFβ pathway is enlarged for better visualization and the genes are highlighted in blue to show direct involvement in TGFβ signaling or in gray to show indirect association with the pathway. Other major nodes are centered around IL-6 and IGF-1 (schematics of these pathways and the genes that are altered in iMYC cells are shown in Fig. S1). (B) Real-time quantitative PCR (qPCR) analysis of genes in the iMYC signature that are involved in TGFβ signaling. Two of the three iMYC cell lines (iMYC 1: black bars; iMYC 2: gray bars) that were used in microarray experiments were used for qPCR validation. Expression levels are normalized to GAPDH and shown relative to expression levels of eMYC cells set as “1.”
In addition, other canonical pathways were significantly enriched in the set of downregulated genes: IL-6 (Fig. S1B; p-value < 2.55E-04), and IGF-1 receptor signaling (Fig. S1C; p-value < 8.27E-03). In contrast, pathway analysis of the 53 upregulated genes indicated a single network group of functionally connected transcription factors, including KLF2, MEF2C and several homeobox genes (see Table S1 for complete gene list). These findings, combined with the statistical significance of the gene overlap, suggest roles for these genes as possible co-factors involved in the immortalization of cells by c-MYC.
Because TGFβ signaling serves a tumor suppressive function in early stages of transformation by inducing growth arrest in premalignant cells,25 and, notably, the TGFβ pathway is a well-defined repressor of c-MYC expression,26,27 we decided to focus on this pathway and to validate the microarray results. The genes downregulated in the iMYC signature are involved in both canonical (reviewed in ref. 28) and noncanonical TGFβ signaling.29 These included TGFβ receptor 1 (TGFBR1), which is involved in canonical signaling,28 and betaglycan (TGFBR3), which is involved in non-canonical signaling;29 both receptors have previously been shown to play potential roles in cancer (reviewed in ref. 30). Other genes involved in TGFβ signaling included the accessory protein decorin,31 the signaling protein Smad1,32 and the pseudo-receptor BAMBI.33 BAMBI was identified in the group of upregulated genes and therefore did not appear in the network analysis (Fig. 2A). Altered expression of these genes suggested that iMYC cells had selected for disruption of the TGFβ signaling pathway.
Several of the genes in the TGFβ pathway identified in the iMYC signature were validated by real-time quantitative PCR (qPCR) (Fig. 2B). The qPCR validated downregulation of TGFβ receptors I and III and decorin, as well as upregulation of the TGFβ pathway pseudo-receptor BAMBI in two iMYC cell lines relative to their matched eMYC cells, confirming the findings in the iMYC signature. In addition, qPCR analysis showed downregulation of the TGFβ responsive gene PAI-1 (SERPINE1),34 and its related gene protease nexin-1 (SERPINE2),35 which had been identified in the iMYC signature. In order to determine whether these gene changes were specific to iMYC cells and not observed in other immortalized populations, we assayed gene expression levels in HFFs immortalized by hTERT. Compared with expression levels in eMYC cells, both normal HFFs and immortalized hTERT HFFs do not show the downregulation of TGFβ pathway genes that is observed in iMYC cells (Fig. S2).
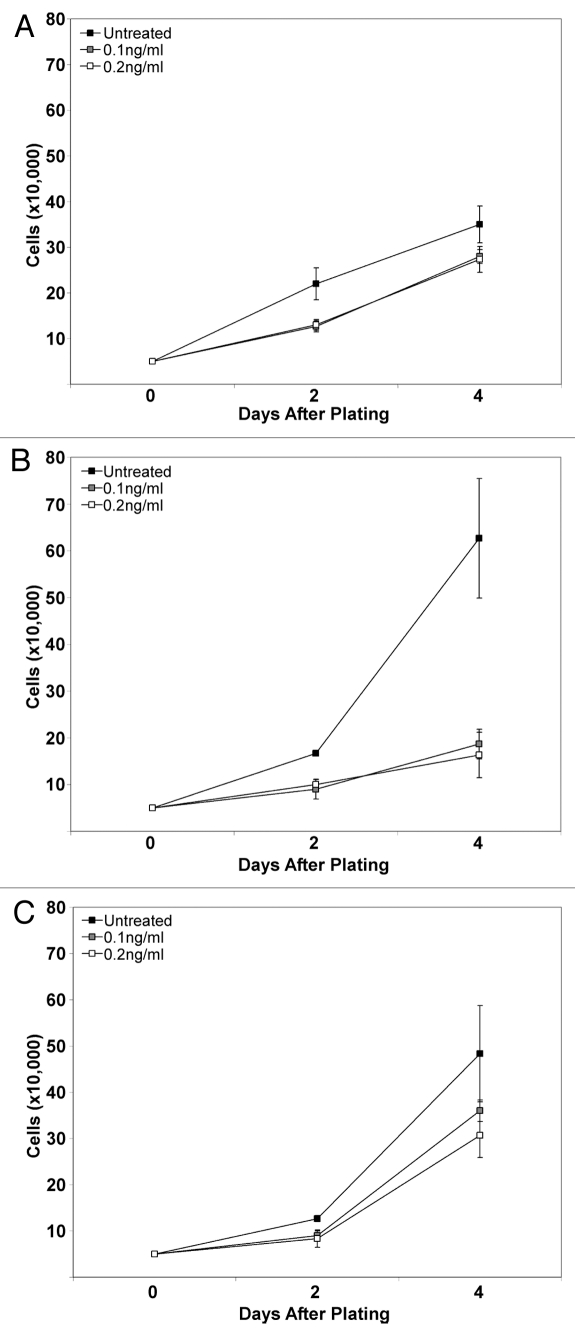
In order to confirm that downregulation of TGFβ pathway genes resulted in functional disruption of TGFβ-induced signaling, specifically whether TGFβ would inhibit cell growth, cells were treated with TGFβ1 ligand and assayed for proliferation as a measure of responsiveness. Cells were counted 2 and 4 d after addition of ligand. As a cell type control, empty vector pB HFFs were also treated and assayed similarly; these cells showed little growth inhibition in the presence of TGFβ1 ligand, even at the higher concentration (Fig. 3A). Strikingly, eMYC cells showed decreased proliferation in the presence of ligand (Fig. 3B), demonstrating the growth inhibition by functional TGFβ signaling in this cell population. In contrast, iMYC cells showed a growth trend similar to control HFFs in the presence of ligand (Fig. 3C), signifying a loss of responsiveness to TGFβ signaling in comparison to eMYC cells. These data indicate a functional disruption of the TGFβ signaling pathway in iMYC cell lines and that resistance to TGFβ was acquired during immortalization by c-MYC.
Figure 3.
Disruption of TGFβ signaling in iMYC cells. (A) Empty vector control (pB) cells were plated, treated with 0.1 ng/ml or 0.2 ng/ml TGFβ1 ligand and counted at 2 d and at 4 d. A growth curve showing cell number on the indicated days is shown. (B) eMYC cells were plated and treated as in (A). (C) iMYC cells were plated and treated as in (A).
The tumor suppressor p27 contributes to inhibitory effects of TGFβ in early c-MYC cells.
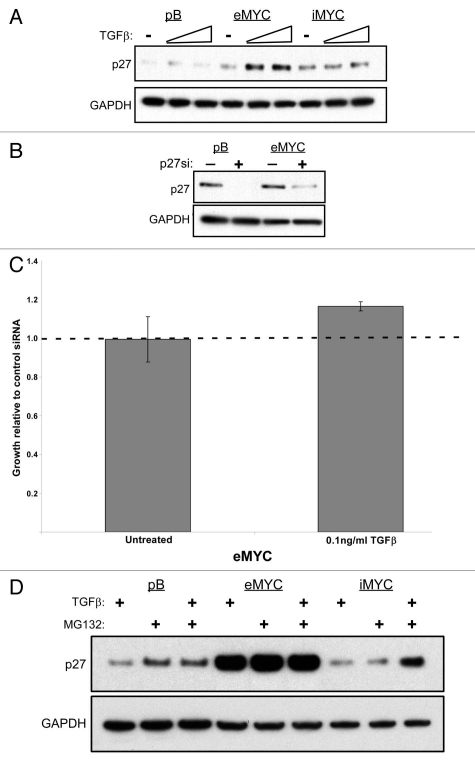
The TGFβ pathway inhibits cell growth through several known tumor suppressors including p15,36–38 p21,39,40 and p27.39,41 We previously showed that p15 and p21 expression remained intact in iMYC cells,17 indicating that loss of sensitivity to TGFβ signaling was not dependent on these proteins. Interestingly, we observed that upon treatment with TGFβ1 ligand, p27 protein levels increased in eMYC cells but remained at basal levels in iMYC cells (Fig. 4A). These data suggest that TGFβ-induced growth inhibition in eMYC cells is at least partly dependent on p27 and that the lack of p27 elevation in iMYC cells could explain the lack of growth arrest.
Figure 4.
p27 protein turnover contributes to the differential growth response of eMYC and iMYC cells to TGFβ. (A) Western analysis of protein lysates from empty vector control (pB), eMYC (MYC) and iMYC (iMYC) cells treated with TGFβ1 ligand. Levels of p27 and GAPDH are shown. (B) Western analysis of protein lysates from empty vector control (pB) and eMYC (MYC) cells transfected with either non-targeting siRNA (−) or against p27 (+). Levels of p27 and GAPDH are shown. (C) eMYC cells transfected with siRNA against p27 were plated 24 h after transfection, treated with 0.1 ng/ml TGFβ1 ligand and counted at 3 d after treatment. Levels of growth in TGFβ1-untreated and treated cells are shown relative to levels of growth in eMYC cells transfected with control siRNA and treated with ligand. (D) Western analysis of protein lysates from empty vector control (pB), eMYC (eMYC) and iMYC (iMYC) cells treated with TGFβ1 ligand, MG132, or both 2 h prior to protein harvest. Levels of p27 and GAPDH are shown.
In order to determine whether p27 upregulation was responsible for growth inhibition by TGFβ in eMYC cells, we knocked down p27 by siRNA transfection prior to treatment with TGFβ1 ligand (Fig. 4B). Upon knockdown of p27, the TGFβ-induced growth inhibition was partially relieved in eMYC cells compared with cells that were transfected with a non-targeting siRNA control (Fig. 4C). These data support a key role of p27 in the proliferative inhibition of eMYC cells by TGFβ; furthermore, the disruption of this response in iMYC cells is likely mediated by the failure to upregulate p27 protein levels in response to TGFβ. It has been shown that TGFβ signaling prevents proteasome-dependent degradation of p27,42 a finding that is consistent with our observation that TGFβ-induced increase of p27 protein in iMYC cells is only observed in the presence of proteasomal inhibitor (Fig. 4D). Although the toxicity of the inhibitor prevented determination of whether increased p27 sensitized iMYC cells to TGFβ dependent growth inhibition, these results indicate that stabilization of p27 protein in response to TGFβ is disabled in iMYC cells. Overall, our observations that p27 protein increased in response to TGFβ in eMYC cells and that this response is lost in iMYC cells strongly suggest a mechanism by which iMYC cells have bypassed the TGFβ growth inhibitory response.
Discussion
We have previously shown that constitutive overexpression of c-MYC consistently immortalizes human fibroblasts. Furthermore, this reproducible phenomenon results in a cell population that shows no gross chromosomal changes that would indicate crisis events.17 The ability of c-MYC to immortalize human fibroblasts isolated from different donors demonstrates that this is a reproducible phenomenon attributable to c-MYC. Additionally, iMYC cells are distinguishable from their HFF donor-matched eMYC cells in several aspects, including tolerance to intact p16 and p53 response pathways and epigenetic downregulation of ARF.17 These properties strongly suggest that overexpression of c-MYC may favor and selects for additional epigenetic changes to bypass replicative lifespan limitations and oncogene-induced senescence, resulting in immortalization.
In this study, we compared gene expression changes in three independent iMYC cell lines from their matched eMYC cells and generated an expression profile common to iMYC cells, which we referred to as the iMYC signature. The iMYC signature represents genes that are candidate co-factors for immortalization by c-MYC. These co-factors likely cooperate to bypass regulators of lifespan, including oncogene-induced senescence and apoptosis pathways, in immortalization of human cells by c-MYC.17 K-means clustering analysis revealed that over half of the downregulated genes in the iMYC signature were already downregulated in eMYC cells to some degree (Fig. 1C), suggesting that regulation of these genes occurred during early stages of c-MYC overexpression but iMYC cells selected for lower levels of expression. Perhaps more intriguing are genes downregulated in iMYC cells that were unchanged or even upregulated in eMYC cells, as the regulation suggests a selection for decreased expression of these genes in iMYC cells.
Analysis of individual genes in the iMYC signature showed that several genes in the TGFβ pathway were downregulated, suggesting that iMYC cells selected for disruption of TGFβ signaling. These genes were mostly classified in Groups 1 and 2 of the K-means clustering (Fig. 1C). The primary receptor TGFβ receptor I and TGFβ receptor III were downregulated in iMYC cells, as were the genes decorin and Smad1. Downregulation of these genes associated with the pathway and upregulation of the pseudo-receptor BAMBI suggested additional means through which TGFβ signaling was disabled. Paradoxically, TGFβ inhibits cellular proliferation of pre-malignant cells but promotes growth and metastasis of established tumors (reviewed in ref. 25). Indeed, transformed cells often lose sensitivity to TGFβ-induced growth inhibition,43,44 and many cancers have downregulated key genes in the pathway.45–49 Overexpression of c-MYC alone does not bypass growth inhibition by TGFβ, as eMYC cells are sensitive to growth inhibitory effects of activated TGFβ signaling (Fig. 3B), similarly to what has been previously observed.44 In contrast, iMYC cells have lost sensitivity to TGFβ signaling and continue to proliferate despite activation of the TGFβ pathway (Fig. 3C), suggesting that downregulation of TGFβ signaling is key to the immortalization process. Bypass of TGFβ-induced senescence has previously been shown in mouse keratinocytes overexpressing activated v-ras,50 and we showed here that the case may also be true for human fibroblasts immortalized by c-MYC.
TGFβ signaling effects are enforced through multiple tumor suppressors, including p15,36–38 which is derepressed in the presence of TGFβ.37 Mechanisms of growth suppression by TGFβ independent of p15 have been previously observed, via repression of CDC25A51 and E2F152 and activation of p21 and p27.39,53 However, iMYC cells continue to express p15 and p2117 and therefore the desensitization of iMYC cells to activated TGFβ signaling cannot be attributed to these tumor suppressors. In contrast, p27 appeared necessary for maintenance of TGFβ-induced cell cycle arrest in tumor-derived cell lines.41 Here we demonstrated that levels of p27 protein were increased in response to TGFβ only in eMYC cells and not in iMYC cells and this correlated with the ability of iMYC cells to bypass TGFβ-induced growth inhibition. We also showed that treatment of iMYC cells with a proteasomal inhibitor resulted in accumulation of p27 after TGFβ treatment, indicating a disruption of the TGFβ response that prevents degradation of p27. Therefore, the TGFβ-induced increase in p27 protein levels in eMYC cells compared with iMYC cells can be explained by the inhibition of p27 protein degradation in iMYC cells. Our findings strongly suggest that p27 contributes to the differential response of eMYC and iMYC cells to TGFβ, and that in iMYC cells the lack of response to TGFβ includes, in addition to the transcriptional downregulation of key TGFβ pathway genes, a failure to stabilize p27.
Cross-talk between c-MYC and TGFβ centered around p27 has been previously highlighted by the observation that TGFβ directly represses c-MYC53–56 and this results in downregulation of CKS1, a transcriptional target of c-MYC and a co-factor of the E3 ubiquitin ligase SKP2 that targets p27 for proteasomal degradation.57 In addition, TGFβ has been shown to directly decrease protein levels of both SKP2 and CKS1, thus stabilizing levels of p27 protein, which is essential for the growth inhibition response.42 Finally, the direct connection between TGFβ and c-MYC in regulation of cellular growth is also underscored by the finding that c-MYC overexpression enables cancer cells to bypass the growth arrest induced by TGFβ.46 Our results correlate with these findings and strongly suggest that in order to continuously proliferate, iMYC cells selected for disruption of major components of the TGFβ response, including loss of the accumulation of p27.
Interestingly, tumor suppressors characteristically disabled in immortalized cells remain intact in the iMYC population. Retention of functional p53 and p21 enables iMYC cells to activate the p53-dependent DNA damage response, which may prevent gross genomic abnormalities that would prevent the stable propagation of the iMYC population. However, downregulation of ARF in iMYC cells prevents p53 upregulation in response to c-MYC overexpression, thereby inactivating the growth suppressive response to oncogene deregulation. Therefore, retaining intact tumor suppressors that function in normal cellular maintenance may be necessary to ensure survival of the iMYC cell population and disruption of the TGFβ pathway is likely an alternative mechanism to avoid growth inhibition.
Genes regulated by c-MYC have been well-defined previously in reference 58. Here, we focused on genes that are associated with c-MYC-dependent extension of lifespan, which may include both direct as well as indirect effects of stable overexpression of c-MYC. In addition to the TGFβ pathway, the signature identifies additional candidate genes that may be significant in later stages of tumor progression or that may contribute to c-MYC function in facilitating establishment of pluripotent stem cells.59 In this respect, it is interesting that the upregulated genes in the iMYC signature included several transcription factors, among these Kruppel-like factor 2 (KLF2), myogenic factor 2C (MEF2C) and several homeobox genes (MSX1, DLX1 and 2), which are functionally connected in the myogenic cell lineage. While the potential roles of these transcription factors in the self-renewal capacity of iMYC cells remain to be established, it is intriguing that a transcription factor of the same class, KLF4, is one of the known combination of four genes that enables induction of pluripotency of human fibroblasts.60 It is tempting to speculate that c-MYC overexpression may prevent differentiation, promoting self-renewal and thus immortality of mesenchymal stem cells existing in the initial HFF populations. As self-renewal, immortalization and lack of differentiation are crucial features of tumor formation, elucidating pathways that cooperate with oncogenes to extend cellular lifespan could provide new clues to this process as well pinpoint relevant therapeutic targets.
Materials and Methods
Cell culture.
Human foreskin fibroblast cell lines expressing empty vector pBABE or vector with gene sequence encoding c-MYC were previously described in reference 17. Cells were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin. For cell growth assays, equal number of cells per cell line were plated and counted on the specified days after plating.
Microarray analysis.
Total RNA was purified using an RNeasy kit (Qiagen, Valencia, CA). Genome-scale expression analysis was performed using custom microarrays (Affymetrix, Santa Clara, CA) containing oligonucleotide probes corresponding to approximately 22,000 human genes. Microarray analysis was performed as described in reference 61. Data were analyzed using Rosetta Resolver™ software. We determined a p-value cut-off of 0.01 for genes in all three samples. Upon publication of this manuscript, microarray data will be available in GEO (www.ncbi.nlm.nih.gov/geo/info/linking.html), under accession number GSE31002.
iMYC signature analysis.
Fifty-three upregulated genes and 263 downregulated genes were listed and input into the Ingenuity Pathways Analysis program (Ingenuity, Redwood City, CA) and analyzed for functional categorization and network associations.
Real-time quantitative PCR.
Cellular RNA was isolated with Trizol (Invitrogen) and purified using RNeasy Mini Kit (Qiagen, Valencia, CA). RT reactions were performed using Superscript II (Gibco, Carlsbad, CA) and random hexamers (Invitrogen). Real-time quantitative PCR was performed using Taqman Gene Expression Assays (Applied Biosystems, Foster City, CA) specific for the genes tested. Expression levels were normalized to GAPDH control.
Protein gel blotting.
Preparation of whole-cell lysates and Protein gel blotting were performed as previously described in reference 62. Equal amounts of protein (10–30 µg) per sample were loaded onto pre-cast NuPAGE gels (Invitrogen, Carlsbad, CA), transferred to PVDF membrane and probed with antibodies to p27Kip1 (Clone 57, BD Biosciences, San Jose, CA), actin (I-19, Santa Cruz Biotechnology, Santa Cruz, CA) and GAPDH (ab8245, Abcam, Cambridge, MA). For samples treated with MG132, MG132 was administered to a final concentration of 10 ng/ml 2 h prior to harvest.
siRNA transfection.
ONTARGETplus non-targeting siRNA or ONTARGETplus SMARTpool siRNA against CDKN1B/p27 (Dharmacon, Lafayette, CO) were transfected using the Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA).
TGFβ treatment.
Recombinant human TGFβ1 (R&D Systems, Minneapolis, MN) was resuspended in HCl/BSA as directed and added to cells for a final concentration of 0.1 ng/ml or 0.2 ng/ml.
Acknowledgments
We wish to thank the members of the Galloway, Katzenellenbogen and Grandori laboratories for technical advice and valuable feedback. This work was supported in part by PHS NRSA T32 GM07270 from NIGMS (to M.L. Wang), RO1 CA64795 from NCI (to D.A. Galloway) and RO1 AGO26661 from NIA (to C. Grandori).
Supplementary Material
References
- 1.Hayflick L. THE LIMITED IN VITRO LIFETIME OF HUMAN DIPLOID CELL STRAINS. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9.. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9.. [DOI] [PubMed] [Google Scholar]
- 4.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003.. [DOI] [PubMed] [Google Scholar]
- 5.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021.. [DOI] [PubMed] [Google Scholar]
- 6.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061.. [DOI] [PubMed] [Google Scholar]
- 7.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belletti B, Nicoloso MS, Schiappacassi M, Chimienti E, Berton S, Lovat F, et al. p27(kip1) functional regulation in human cancer: a potential target for therapeutic designs. Curr Med Chem. 2005;12:1589–1605. doi: 10.2174/0929867054367149.. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501.. [DOI] [PubMed] [Google Scholar]
- 10.Günes C, Lichtsteiner S, Vasserot AP, Englert C. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60:2116–2121. [PubMed] [Google Scholar]
- 11.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349.. [DOI] [PubMed] [Google Scholar]
- 12.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962.. [DOI] [PubMed] [Google Scholar]
- 13.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9.. [DOI] [PubMed] [Google Scholar]
- 15.Benanti JA, Galloway DA. Normal human fibroblasts are resistant to RAS-induced senescence. Mol Cell Biol. 2004;24:2842–2852. doi: 10.1128/MCB.24.7.2842-52.2004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drayton S, Rowe J, Jones R, Vatcheva R, Cuthbert-Heavens D, Marshall J, et al. Tumor suppressor p16INK4a determines sensitivity of human cells to transformation by cooperating cellular oncogenes. Cancer Cell. 2003;4:301–310. doi: 10.1016/S1535-6108(03)00242-3.. [DOI] [PubMed] [Google Scholar]
- 17.Benanti JA, Wang ML, Myers HE, Robinson KL, Grandori C, Galloway DA. Epigenetic downregulation of ARF expression is a selection step in immortalization of human fibroblasts by c-Myc. Mol Cancer Res. 2007;5:1181–1189. doi: 10.1158/1541-7786.MCR-06-0372.. [DOI] [PubMed] [Google Scholar]
- 18.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blagosklonny MV. Cell cycle arrest is not senescence. Aging (Albany NY) 2011;3:94–101. doi: 10.18632/aging.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606.. [DOI] [PubMed] [Google Scholar]
- 21.Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010;2:344–352. doi: 10.18632/aging.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leontieva OV, Blagosklonny MV. DNA damaging agents and p53 do not cause senescence in quiescent cells, while consecutive re-activation of mTOR is associated with conversion to senescence. Aging (Albany NY) 2010;2:924–935. doi: 10.18632/aging.100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benanti JA, Galloway DA. The normal response to RAS: senescence or transformation? Cell Cycle. 2004;3:715–717. doi: 10.4161/cc.3.6.948.. [DOI] [PubMed] [Google Scholar]
- 24.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/S0092-8674(00)00121-5.. [DOI] [PubMed] [Google Scholar]
- 25.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietenpol JA, Holt JT, Stein RW, Moses HL. Transforming growth factor beta1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc Natl Acad Sci USA. 1990;87:3758–3762. doi: 10.1073/pnas.87.10.3758.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu S, Hultquist A, Hydbring P, Cetinkaya C, Oberg F, Larsson LG. TGFbeta enforces senescence in Myc-transformed hematopoietic tumor cells through induction of Mad1 and repression of Myc activity. Exp Cell Res. 2009;315:3099–3111. doi: 10.1016/j.yexcr.2009.09.009.. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Massague J. Mechanisms of TGFbeta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YE. Non-Smad pathways in TGFbeta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon KJ, Blobe GC. Role of transforming growth factor beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Ständer M, Naumann U, Wick W, Weller M. Transforming growth factor beta and p-21: multiple molecular targets of decorin-mediated suppression of neoplastic growth. Cell Tissue Res. 1999;296:221–227. doi: 10.1007/s004410051283.. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Hata A, Baker JC, Doody J, Carcamo J, Harland RM, et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0.. [DOI] [PubMed] [Google Scholar]
- 33.Sekiya T, Adachi S, Kohu K, Yamada T, Higuchi O, Furukawa Y, et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279:6840–6846. doi: 10.1074/jbc.M310876200.. [DOI] [PubMed] [Google Scholar]
- 34.Kortlever RM, Nijwening JH, Bernards R. Transforming growth factor beta requires its target plasminogen activator inhibitor-1 for cytostatic activity. J Biol Chem. 2008;283:24308–24313. doi: 10.1074/jbc.M803341200.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.GR-1478R.. [DOI] [PubMed] [Google Scholar]
- 36.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan CH, Yaswen P, et al. Transforming growth factor beta stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997;17:2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086.. [DOI] [PubMed] [Google Scholar]
- 38.Hannon GJ, Beach D. p15INK4B is a potential effector of TGFbeta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0.. [DOI] [PubMed] [Google Scholar]
- 39.Flørenes VA, Bhattacharya N, Bani MR, Ben-David Y, Kerbel RS, Slingerland JM. TGFbeta mediated G1 arrest in a human melanoma cell line lacking p15INK4B: evidence for cooperation between p21Cip1/WAF1 and p27Kip1. Oncogene. 1996;13:2447–2457. [PubMed] [Google Scholar]
- 40.Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donovan JC, Rothenstein JM, Slingerland JM. Non-malignant and tumor-derived cells differ in their requirement for p27Kip1 in transforming growth factor-beta-mediated G1 arrest. J Biol Chem. 2002;277:41686–41692. doi: 10.1074/jbc.M204307200.. [DOI] [PubMed] [Google Scholar]
- 42.Lecanda J, Ganapathy V, D'Aquino-Ardalan C, Evans B, Cadacio C, Ayala A, et al. TGFbeta prevents proteasomal degradation of the cyclin-dependent kinase inhibitor p27kip1 for cell cycle arrest. Cell Cycle. 2009;8:742–756. doi: 10.4161/cc.8.5.7871.. [DOI] [PubMed] [Google Scholar]
- 43.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letterio J, Rudikoff E, Voong N, Bauer SR. Transforming growth factor-beta1 sensitivity is altered in Abl-Myc- and Raf-Myc-induced mouse pre-Bcell tumors. Stem Cells. 2006;24:2611–2617. doi: 10.1634/stemcells.2005-0623.. [DOI] [PubMed] [Google Scholar]
- 45.Bernabeu C, Lopez-Novoa JM, Quintanilla M. The emerging role of TGFbeta superfamily coreceptors in cancer. Biochim Biophys Acta. 2009;1792:954–973. doi: 10.1016/j.bbadis.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen CR, Kang Y, Massague J. Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor beta growth arrest program. Proc Natl Acad Sci USA. 2001;98:992–999. doi: 10.1073/pnas.98.3.992.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen G, Ghosh P, Osawa H, Sasaki CY, Rezanka L, Yang J, et al. Resistance to TGFbeta 1 correlates with aberrant expression of TGFbeta receptor II in human B-cell lymphoma cell lines. Blood. 2007;109:5301–5307. doi: 10.1182/blood-2006-06-032128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parekh TV, Gama P, Wen X, Demopoulos R, Munger JS, Carcangiu ML, et al. Transforming growth factor beta signaling is disabled early in human endometrial carcinogenesis concomitant with loss of growth inhibition. Cancer Res. 2002;62:2778–2790. [PubMed] [Google Scholar]
- 49.Diaz-Chavez J, Hernandez-Pando R, Lambert PF, Gariglio P. Downregulation of transforming growth factor-beta type II receptor (TGFbetaRII) protein and mRNA expression in cervical cancer. Mol Cancer. 2008;7:3. doi: 10.1186/1476-4598-7-3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tremain R, Marko M, Kinnimulki V, Ueno H, Bottinger E, Glick A. Defects in TGFbeta signaling overcome senescence of mouse keratinocytes expressing v-Ha-ras. Oncogene. 2000;19:1698–1709. doi: 10.1038/sj.onc.1203471.. [DOI] [PubMed] [Google Scholar]
- 51.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell cycle arrest by cytokine TGFbeta in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0.. [DOI] [PubMed] [Google Scholar]
- 52.Spender LC, Inman GJ. TGFbeta induces growth arrest in Burkitt lymphoma cells via transcriptional repression of E2F-1. J Biol Chem. 2009;284:1435–1442. doi: 10.1074/jbc.M808080200.. [DOI] [PubMed] [Google Scholar]
- 53.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, et al. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9.. [DOI] [PubMed] [Google Scholar]
- 54.Reynisdóttir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492.. [DOI] [PubMed] [Google Scholar]
- 55.Slingerland JM, Hengst L, Pan CH, Alexander D, Stampfer MR, Reed SI. A novel inhibitor of cyclin-Cdk activity detected in transforming growth factor beta-arrested epithelial cells. Mol Cell Biol. 1994;14:3683–3694. doi: 10.1128/mcb.14.6.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warner BJ, Blain SW, Seoane J, Massague J. Myc downregulation by transforming growth factor beta required for activation of the p15(Ink4b) G(1) arrest pathway. Mol Cell Biol. 1999;19:5913–5922. doi: 10.1128/mcb.19.9.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller UB, Old JB, Dorsey FC, Nilsson JA, Nilsson L, MacLean KH, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 2007;26:2562–2574. doi: 10.1038/sj.emboj.7601691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014.. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024.. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019.. [DOI] [PubMed] [Google Scholar]
- 61.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249.. [DOI] [PubMed] [Google Scholar]
- 62.Benanti JA, Williams DK, Robinson KL, Ozer HL, Galloway DA. Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol Cell Biol. 2002;22:7385–7397. doi: 10.1128/MCB.22.21.7385-97.2002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.