Abstract

A major goal in drug design is the improvement of computational methods for docking and scoring. The Community Structure Activity Resource (CSAR) aims to collect available data from industry and academia which may be used for this purpose (www.csardock.org). Also, CSAR is charged with organizing community-wide exercises based on the collected data. The first of these exercises was aimed to gauge the overall state of docking and scoring, using a large and diverse data set of protein–ligand complexes. Participants were asked to calculate the affinity of the complexes as provided and then recalculate with changes which may improve their specific method. This first data set was selected from existing PDB entries which had binding data (Kd or Ki) in Binding MOAD, augmented with entries from PDBbind. The final data set contains 343 diverse protein–ligand complexes and spans 14 pKd. Sixteen proteins have three or more complexes in the data set, from which a user could start an inspection of congeneric series. Inherent experimental error limits the possible correlation between scores and measured affinity; R2 is limited to ∼0.9 when fitting to the data set without over parametrizing. R2 is limited to ∼0.8 when scoring the data set with a method trained on outside data. The details of how the data set was initially selected, and the process by which it matured to better fit the needs of the community are presented. Many groups generously participated in improving the data set, and this underscores the value of a supportive, collaborative effort in moving our field forward.
Introduction
The requests for applications that lead to the creation of CSAR had the goal “to increase the amount of high-quality data publicly available for development, validation, and benchmarking of ligand docking and screening software” (http://grants.nih.gov/grants/guide/rfa-files/RFA-GM-08-008.html). CSAR aims to provide large, complete data sets—like those produced in the pharmaceutical industry —through in-house experiments and donations of data from the wider community. For our first benchmark exercise, we were limited to the data for protein–ligand complexes available in the public domain: PDB,(1) Binding MOAD,(2) and PDBbind.(3) Other published evaluations of docking and scoring have all used different data sets, making comparisons difficult. For this issue of the Journal of Chemical and Information Modeling, all participants have used the CSAR data set, allowing for consistency and an even comparison between different participants’ insights. Participants were asked to score the structures using a standard and an alternative approach, allowing them to gauge the impact of different parameters on the performance of their method.
The benchmark exercise culminated in a symposium at the Fall 2010 National Meeting of the American Chemical Society in Boston, with 14 speakers and open discussion sessions. The goals of the exercise were:
-
(1)
to bring the community together to discuss the issues behind improving docking and scoring.
-
(2)
to establish appropriate metrics for evaluating the performance of scoring functions.
-
(3)
to provide a baseline assessment of current scoring functions using diverse proteins and ligands.
-
(4)
to evaluate participants’ improvements of their parameters, particularly emphasizing what changes were the most universal or had the greatest impact.
-
(5)
to document which complexes are most difficult and require new approaches.
-
(6)
to identify the most important priorities for CSAR to address with custom data sets of specific ligands and targets.
Our second paper, later in this issue, outlines our insights from comparing scores of all participants and how that information impacts our priorities for additional data sets. Here, we describe our process for creating the 2010 benchmark exercise data set that was used in the studies to follow.
The selection of protein–ligand data sets for evaluating docking/scoring has progressed from being only a handful of crystal structures with suitable biological data to now having the luxury of paring down thousands of structures to identify hundreds of exceptional complexes. The initial evaluation set for DOCK4,5 was 2 protein–ligand complexes in 1982. As time progressed, FLEXX(6) was presented in 1996 with an evaluation set of 19 complexes. GOLD(7) followed the next year with an evaluation set of 100 complexes. By 2002, the group at Astex had selected an evaluation set of 305 complexes, forming the CCDC/Astex(8) data set. Their “clean” set had 224 but only 180 with resolution ≤2.5 Å. In 2007, CCDC and Astex(9) further refined the set with more stringent quality assessments, reducing the data set to 85 protein–ligand complexes. The PDBbind refined data set (2007 version) is much larger at 1300, but the quality is not as high.(10) Currently, Binding MOAD has almost 5000 structures of complexes with binding data,(11) but being comprehensive in nature, it contains many that are not of suitable quality for this purpose. Furthermore, the structures have not been setup with hydrogens and a richer set of atom types, which greatly limits their utility in docking and scoring. Table 1 provides additional examples of sets of protein–ligand complexes with associated affinity data.
Table 1. The Evolution of Sets of Protein–Ligand Complexes with Affinities.
| database | year | no. complexes |
|---|---|---|
| Score1(12) | 1994 | 54 |
| Jain(13) | 1996 | 34 |
| VALIDATE(14) | 1996 | 65 |
| ChemScore(15) | 1997 | 112 |
| Score2(16) | 1998 | 94 |
| SCORE(17) | 1998 | 181 |
| PMF(18) | 1999 | 225 |
| BLEEP19,20 | 1999 | 90 |
| DrugScore21,22 | 2000 | 83 |
| LPDB(23) | 2001 | 195 |
| SMoG2001(24) | 2002 | 119 |
| HINT(25) | 2002 | 53 |
| X-Score(26) | 2002 | 230 |
| PLD(27) | 2003 | 485 |
| AffinDB(28) | 2006 | 474 |
| PDBbind – refined set | 2007 | 1300 |
| MOAD with affinities | 2008 | 2948 |
The perfect data set, like all things associated with any form of perfection, is heavily dependent on the viewpoint of the particular person evaluating said perfection. However, there are some common characteristics most would desire. The data set should include only high-resolution protein–ligand structures with well-resolved density and high occupancies in the binding site with no unexplained density. Data should be available for a wide range of protein targets. The binding data should be Ka, Kd, or Ki and include the thermodynamic measurements of entropy, enthalpy, and free energy of binding. The data set should be comprised of several congeneric series of small molecules for each target with at least four orders of magnitude in binding affinity for each series. The compounds should be drug-like with good water solubility and should encompass a wide range of functional groups. In addition to the active molecules in each series, very similar molecules that are inactive are needed. Consistency across, and within, the experiments for targets is a must. As an example, the protein sequence used for crystallography should be the same as in the affinity measurements. One should have actual, measured values for the physical properties of the small molecules such as the pKa’s, log P’s, log D’s, solubility, etc. The CSAR center aims to fulfill as many of these criteria as possible for the data sets it is generating in house and gathering from the wider community of structure-based, drug discovery scientists.
The exercises that are part of CSAR’s goals are a special case. One would ideally like to have unpublished data sets with the above characteristic. “Blinded” data sets would circumvent many of the limitations of studies in the literature, namely the back-and-forth between results and known binding data. It is human nature to track down the causes of discrepancies, but this often leads to a change in the approach that skews the scientist’s results in an immeasurable but tangible way. Future exercises will be based on blinded data sets, preferably those contributed by our colleagues in the pharmaceutical industry.
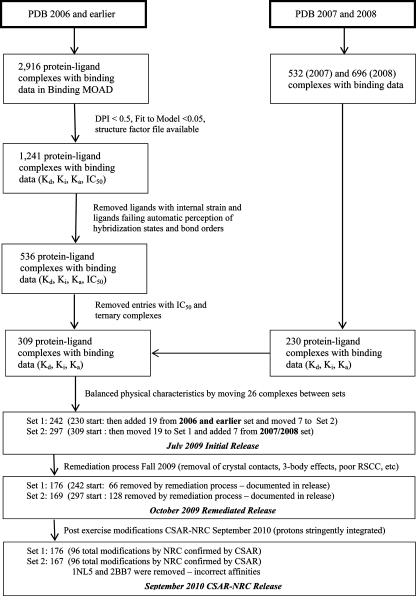
This paper presents the process of developing the data set used in the 2010 CSAR exercise (flowchart in Figure 1). The information is presented in chronological order to show how the process evolved over time. The starting point was a data set constructed from PDB entries up to 2006, which was subsequently augmented with entries from 2007 and 2008. In the paper on PDB_REDO,(29) older entries benefited more from rerefinement than the newer entries, with the break point being in the range of 2004–2006. The full data set was subdivided into two parts, Sets 1 and 2, allowing one to be used as a training set and the other as a test set if desired (we requested participants to train on Set 1 and test on Set 2, then repeat the exercise by training on Set 2 and testing on Set 1 to show robustness of their approach). Setting the break point at 2006 provided almost identical sets in terms of size and calculated physical properties (see Figure 2). Set 2 is primarily comprised of the older entries (up to but not including those deposited on January 1, 2007), and that is where we will begin the discussion of the selection process.
Figure 1.
Flowchart of how the data set was curated.
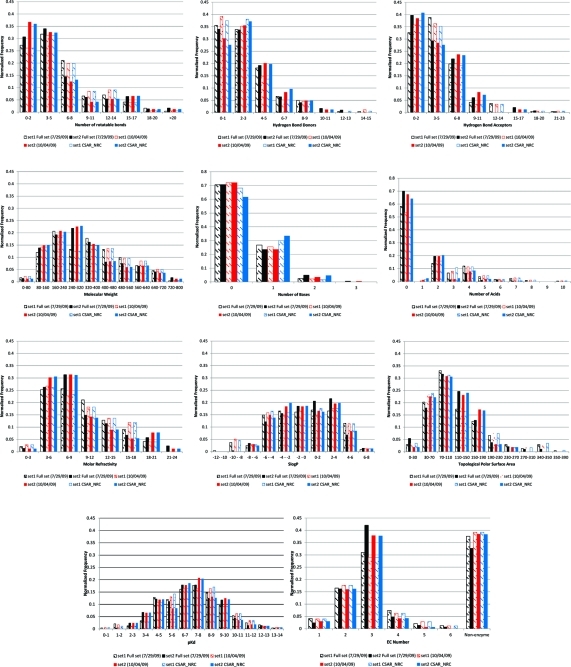
Figure 2.
Distribution analysis of the calculated physical properties of the data sets.
In the Strengths and Weaknesses section at the end of this paper, we discuss the possible influence that the error in the pKd or pKi values might have. Understanding the limitations of integral parts of the data set is essential for using the data properly and making solid conclusions. A description of the experimental error in the biological assay data is just as critical as describing the measures of error used in selecting the crystal structures. The Supporting Information provides a discussion of the 95% and 90% confidence intervals possible for Pearson R, R2, Spearman ρ, and Kendall τ. The intervals are dictated by the size of the data set, and users should know the confidence intervals in the values they obtain when fitting methods to reproduce these affinities. This is essential for interpreting whether improving a method from R2 = 0.6 to 0.7, using the final 343 complexes of the CSAR-NRC set, is statistically significant (it is at the 90% confidence level).
Initial Selection Process (Structures Later Named Set 2)
As part of the update effort for the 2006 version of the Binding MOAD database, we had identified an initial set of 2916 PDB structures deposited by December 31, 2006 with binding data noted in the crystallography paper. This large starting set was filtered using the quality metrics described below. We have also coordinated our efforts with PDBbind and included those structures that were unique to that database. This collection of structures formed the basis of Set 2, the set containing PDB entries up through 2006. A flowchart illustrating the general steps involved is shown in Figure 1. The other set (Set 1), those from 2007 and 2008, followed the same procedure.
For both sets, an initial set of thousands of PDB entries with valid ligands and binding data in Binding MOAD was identified. We then calculated the diffraction-coordinate precision index (DPI)30,31 for each structure using code based on work by Goto et al.(32) and modified to handle unique parsing issues. Hydrogen atoms were ignored in the calculation because they are usually a vestigial result of refinement and inadvertently reported. Only one entry had resolution <1 Å where hydrogens might be resolved (1us0 at 0.66 Å). It had a few hydrogens reported (i.e., Arg63, Trp161, etc.), but we chose not to use them so that all entries were processed in a consistent manner. In addition to the DPI, we also examined the “fit to model” (Rfree – R). We downloaded the structure factor files for each of the 2916 PDB entries, but many were not available. With this information, the data set was filtered for DPI < 0.5, fit to model <0.05, and the presence of a structure factor file. All of the criteria had to have been met simultaneously. Combined with the appropriate unique entries from PDBbind, the resultant list of possible candidates was cut to 1241 entries.
The next phase examined the ligands in those structures to identify those of acceptable quality. This involved a check of the ligands as independent entities. All of the ligands for the 1241 entries were extracted in PDB file format, giving a set of 2494 individual ligand .pdb files. In some cases, there were several copies of the ligand in a given entry.
As part of our assessment of the ligands, the ligand .pdb files were stripped of the connect records, retaining only atom records and coordinate information. By stripping the connect records, we intended to test of the ability of available software to correctly perceive the molecule’s bonds and bond orders correctly, based on just the coordinates and general atom type. If software recognized a part of a molecule as a phenyl ring, when it was actually a cyclohexyl ring, then the ligand may require hand processing or may not be of high local quality. If independent software packages from different sources correctly perceived the given ligand’s bond orders and bond types, then the ligand was most likely of acceptable local quality, as it conformed to definitions of standard bond lengths, bond angles, etc.
The ligand files were converted into Tripos(33) .mol2 format, a format with a relatively rich set of possible atom types, using two software packages Omega2 from Openeye(34) and MOE from CCG.(35) Omega2 was used to construct a low-energy conformation, while MOE used the exact conformation as entered (we exploited the difference in the two methods to check for high-energy ligand conformations in the complexes). The .mol2 files from Omega2 were then loaded into MOE, and both sets of files were washed and had hydrogens added in an internally consistent fashion for the MOE energy calculations. Various descriptors were then calculated for the two sets: atom count (a_count), number of hydrogens (nH), number of aromatic bonds (b_ar), number of double bonds (b_double), number of single bonds (b_single), number of triple bonds (b_triple), number of heavy atoms (a_heavy), and the energy (E) using the MMF94x forcefield. This was output from MOE as a .csv file and moved to JMP(36) for additional analysis. If both programs perceive the ligands with the same bonds and bond orders, then the difference between these descriptors should be zero. The difference of the descriptors for each set of ligands was calculated, and those with no difference for all descriptors except E were moved on in the process, while the remainder were flagged for by-hand processing. The filtering provided a set of ligands with very standard bond lengths and angles, minimizing any unusual structure characteristics in the ligands in the data set.
The trimmed set of 1283 ligands was examined by a distribution analysis to look for potential outliers based on the difference in energy between the conformations from MOE and Omega2, which should identify ligands in high-energy conformations in the crystal structures. Based on this analysis, 73 ligands fell outside 1.5 times the interquartile range. Removal of these structures resulted in a final set of 1210 ligands.
The final phase was the manual (visual) inspection of each ligand in the context of the protein with any associated cofactors. Here, we looked for conditions, such as alternate densities, obvious strain in the ligand, ambiguity in identification of the ligand(s), and obvious strain in the cofactors or other associated nonprotein units. If there were multiple ligands per PDB entry, each ligand was visually examined to ensure all were acceptable. After the manual curation phase, we arrived at the 536 PDB entries that passed all of the above selection criteria. The list was further reduced by dropping all entries with IC50 data, keeping only those with Ki, Kd, or Ka data. Structures with any cofactor or ternary molecule within 4 Å of the ligand were also removed. This resulted in a final number of 309 entries to be included as Set 2. The entries, both protein and ligand, were then processed using Sybyl(33) in exactly the same manner as the PDBbind refined set,(37) including the readdition of hydrogens and Gasteiger–Huckel charges for the ligands. Each entry was successfully scored in GOLD,(7) Drugscore,21,22,38 X-Score,(26) M-Score,(39) and FRED(33) as a further check for robustness of the data sets.
Addition of PDB Structures from 2007 and 2008 (Set 1)
The initial selection of Set 1 followed the same procedure as that for Set 2. As part of the Binding MOAD 2007 and 2008 updates, we had an additional 1228 PDB files with binding data to process. There were 113 PDB entries from the 2007 update and 117 entries from the 2008 update that met our criteria as defined above. The distributions of several calculated properties of the ligands of Sets 1 and 2 were compared. The properties considered were: number of rotatable bonds, number of hydrogen-bond donors, number of hydrogen acceptors, molecular weight, number of acidic atoms, number of basic atoms, SLog P, molar refractivity, and topological surface area. In order to obtain a more similar distribution of these properties between the two sets, 19 PDB entries were moved from Set 1 to Set 2 and 7 PDB entries from Set 2 were moved to Set 1. The final numbers associated with the two initial sets was then at 242 PDB entries for Set 1 and 297 PDB entries for Set 2. The distributions for the ligand properties for the two sets are shown in Figure 2.
Initial Release of the CSAR-HIQ Set (July 2009)
The initial sets were released as file archives for the community to download. Within each set, each entry consisted of a high-level directory named with the PDB code. Within each directory the following files were included: the ligand, in both a .mol2 (Tripos) and a .sdf (MDL) format, the protein in the .pdb format, and the binding data in a kd.dat file. If the structure also contained a valid small molecule besides the ligand, according to Binding MOAD’s definition, then the coordinates for these molecules were also included in appropriate .mol2 and .sdf files within the directory. At this point, we had constructed two sets of high-quality PDB entries to be used as reasonable starting points and put them forward to the community on July 29, 2009.
This initial release reflected our choice of automated processing of the ligands and the proteins to simulate real-world limitations in an industrial setting.(40) A typical, high-throughput, docking-and-scoring approach in industry would start with processing the protein target based on the actual docking method being used and would vary from method to method. This impacts if and how hydrogens are added, how covalently modified side chains are handled, termini, etc.
In real applications in the pharmaceutical industry, hundreds of thousands to millions of ligands would be represented in a generic state chosen automatically when the database was created. Proper ligand conformations and orientations of hydrogens would be unknown. Therefore, we made no attempt to orient hydrogens in the ligand to maximize the interactions (or minimize the clash) with their respective proteins. We did not adjust the protonation state based on the crystallographic or assay conditions (also the setup of choice for BindingDB’s(41) validation sets, www.bindingdb.org/validation_sets/index.jsp). Expert users expect good software to accommodate and correct for hydrogen mismatches.
One of the major goals of this exercise was to find areas that could be targeted for improvement in the prediction of affinity. Known potential difficulties were left in the data set to determine what impact they may have in the bigger picture and learn whether the community at large—using many different methods—could statistically prove these as problems. Most stumbling blocks are known simply through anecdotal examples. Are metalloenzymes really harder to score well versus other targets? Do ligands with poor fits to density really score much worse than well-fit ligands? Do ligands affected by crystal contacts score much worse than those without contacts? Most importantly, which of these issues is the most detrimental to scoring? Indeed, if users wish to assess their method’s susceptibility to these limitations, performance could be compared between this initial release and the final CSAR-NRC set used by authors in this special issue. All of the sets described here are available at www.csardock.org or by request.
Remediated CSAR-HIQ Set (Released October 2009)
Based on immediate feedback from the community, the quality assessments of crystal structures were significantly expanded and the original two sets subjected to more stringent assessment criteria. Those entries not meeting the new requirements were removed, and these remediated sets were used for the symposium at the Boston, MA 2010 ACS National Meeting. The improved criteria for quality assessment and the remediation of several errors in the initial set are presented below.
The global metrics of structural quality were expanded. We eliminated those structures with Rfree >0.29 and those where the recalculated R-factor disagreed with the reported R-factor from the PDB. Additionally, we assessed the fit of the ligand to its density (recall that all entries were required to have structure factors available). Those with a real-space correlation coefficient42,43 (RSCC) of 0.9 or better were accepted, and those with <0.9 were examined by hand by G. Warren at OpenEye. Those of unacceptable quality were removed (removal also verified by the CSAR group). There were a handful of entries where the CSAR group disagreed. For example, ligand P34 in Set 1–149 (2q6m) was well resolved in its cyclic region but had poorer fitting for a solvent-exposed tail. The CSAR team retained this structure, despite its lower RSCC.
Several entries required corrections to the binding affinity data. Most were nM-level affinities that were misprocessed into μM for the kd.dat files. This only affected seven entries. We had misprocessed ∼30 of the older files and improperly assigned some Rfree and Rfactor information in the original Set 2. These errors were remediated.
Non-natural amino acids were improperly processed in our original automated preparation. The non-natural amino acids were not recognized in Sybyl and simply dropped from the structure in the original sets. Fortunately, they are uncommon, and only ∼20 structures in the original data sets required correction. For the final release, all non-natural amino acids (PCA, LLP, KCX, SEP, etc) appear in the .pdb structure file.
Some crystallographic additives were eliminated very early in our examinations and lead to the release of a few structures where we initially failed to identify significant contact between the ligand and a “third body”. Removing structures with three-body interactions, whether biologically relevant or due to a crystallographic additive, simplifies the problem to a single ligand binding to a single pocket. For the remediation, we also examined crystal-packing contacts that could complicate the bound orientation of the ligands.(10) Any ligand with significant contacts (≤4 Å) to a well-resolved symmetry partner was eliminated. Contact to surface residues modeled in without density to support the conformation was not a cause to eliminate a complex from the data set. All biounits were consulted to differentiate crystal-packing contacts from inherent multimeric structure. Ultimately, 343 structures from Sets 1 and 2 passed the more stringent requirements.
Preparation of the Remediated Structures
Each protein structure was prepared via Sybyl (8.0)(33) to give a standard PDB file format with partial header information. The following curation was performed on each structure: The first and last resolved amino acids of each chain were charged, but any “internal” termini due to missing loops were neutralized by addition of an NME or ACE group. The histidine residues in the binding sites were examined and renamed to HIP or HIE based on visual inspection (all HIS in the current files are the HID tautomer). Histidines associated to a metal ion anywhere in the protein were also examined to determine the proper protonation state and orientation. Histidines outside the binding site and away from a metal have not been examined manually, and all are modeled as HID. Users may choose to alter these in their applications.
All Cys were examined for participation in disulfide bonds (denoted as CYX) and interactions with metals (deprotonated Cys is denoted as CYM in our .pdb files).
When hydrogens were added to the structures, some clashes were inevitably introduced. Some strain between the ligand and the active site was reduced by manually reorienting an occasional hydrogen (almost all were rotation of a hydroxyl group in the ligand) or by removing the hydrogen if ionization/tautomerization was applicable. Heavy atoms were never moved, except in two straightforward cases. If needed, some histidine rings were flipped by 180°, and very rarely, an asparagine or glutamine side chain was flipped to reduce clashes with the ligand.
Missing side chains of flexible surface residues have been added so that the charge of the protein is more appropriate for the system. This was done in an automated fashion using the mutate command in Sybyl. The additions were not minimized, but NACCESS(44) was used to identify added side chains that were more buried, and those were manually inspected and corrected when needed. Newly built heavy atoms within 2 Å of existing heavy atoms in the structures were rebuilt with alternate rotameric states when possible. The 2 Å limit between heavy atoms still permits some slight clash between the added hydrogen atoms. It is likely that strain has been introduced in some structures, especially given the unusual backbone orientations assigned within flexible regions. We stress that none of these side chain additions were in the binding sites, and we simply included them so that the proteins were as complete as possible. (However, we repeat that missing loop regions were capped with NME and ACE rather than built, given the obvious limitations.) Water molecules within 1 Å of a built residue were removed.
Non-natural amino acids have been included, noted as HETATMs in the .pdb files. Metals and any covalent modifications (glycosylation, etc) have also been retained and noted as HETATMs. All unusual modifications (SF4, PLP) are >10 Å from the binding site. A list of all unusual residues was provided for each entry. The protein, its metals, and its covalent modifications do not have any partial charges assigned, and the protein was not energy minimized in any way. Our criteria did not include whether or not any of the modifications were potentially difficult to treat in current scoring methods. The lack of partial charges and minimization was intended to avoid any force field bias.
Several structures have amino acids that were modified in association with the crystallography (MSE, CAS, CSO, CME, CSS, and HIC residues). In these cases, the papers were consulted, and the amino acids changed back to their respective natural amino acids for consistency with the experiments that provided the binding data reported for the structure. We have verified that these residues are not close to the binding sites. Several structures had ABA as a non-natural amino acid, which was changed to ABU in keeping with Sybyl’s amino acid dictionary.
For Set 1 no. 2 (2arb) and no. 3 (2are), we inspected the electron density to conclude that the first amino acid for each chain should be pyroglutamic acid (PCA) as opposed to glutamic acid, as reported in the original PDB file. We have made such changes to both structures through simple atomic replacement, not a rerefinement of the structures. During the inspection of ligand densities, it was observed that in Set 1 no. 148 (2pyy) the side-chain conformation of Arg10 placed the NH1 atom outside the available density. The deposited conformation, in addition to being outside the density, did not form an optimum salt bridge to the carboxylate of the bound ligand (glutamic acid). A side chain rotomer search, using the Richardson rotomer library, was performed. A new conformation was discovered that fit the density and allowed for the formation of a bidentate interaction between Arg10 and the carboxylate of the ligand. The structure was then rerefined using Refmac5 (v5.5.01009) with density generated from the deposited structure factors.
All waters >5 Å from the protein surface were stripped away. All crystallographic additives were removed. To allow users to examine the locations of additives, ions, and other copies of the ligands in multimeric structure, we included the complete, corrected biounits from Binding MOAD.
Each ligand was then prepared via the Sybyl program. AM1-BCC charges assigned to the ligand were calculated via OpenEye QUACPAC(15) 1.3.1. File conversion and postprocessing was performed to add the CSAR project information before the field of “@<TRIPOS>MOLECULE”. Ligand density was evaluated to determine the best resolved and most appropriate ligand to use in each structure. Again, if the user wishes to examine the locations of other copies of the ligands in multimeric structures, the raw biounits should be consulted. There were a few cases where a better resolved ligand was not chosen for the structure, but those were cases where the second ligand was outside of the active site in crystal-packing induced locations.
A tar file of both sets (Sets 1 and 2) with all the accompanying informational files described below was made available on the CSAR Web site (www.csardock.org) in the download area. Within the tar file, there is a main directory for each set (Sets 1 and 2) which consists of a series of subdirectories for each complex. The numbered subdirectories each include three files: the ligand structure (#.mol2), the protein (#.pdb), and the binding affinity (kd.dat). The file “kd.dat” provides information about the structure in the following format: number, PDB id, and affinity [given as −log(Kd or Ki)]. The following summary files (located in the SUMMARY_FILES directory) are given to describe the data set, again subdivided into Sets 1 and 2. “DROP_LIST.txt” describes the complexes from the original release that have been excluded from this release. Reasons are given for each complex. “KEEP_LIST.txt” describes the complexes that have been retained. (Note: Set 2 no. 258 was dropped as an entry but inadvertently retained an entry in the KEEP_LIST.txt, and there was a typographical error in Set 2 no. 213, the binding data should have read 5.08 and not 2.08.) The structures have been grouped by 100% and 90% sequence identity, and the resulting protein families can be found in “100idFamilies.txt” and “90idFamilies.txt”. These lists are provided to facilitate users examination of relative binding trends or rankings of a series of compounds bound to the same protein.
Comma separated files, “set1.csv” and “set2.csv”, detail the contents of the sets in the following format: structure number, its PDB id, the binding affinity (in pKd), and the ligand name. In the #.pdb, “set1_unusual-molecules.txt” and “set2_unusual-molecules.txt” list any molecules files that differ from the typical amino acids and may require additional parameters. Non-natural amino acids (ABU, PCA, LLP, KCX, SEP, TRQ, etc.), metal ions, and covalent modifications of the protein are listed. While metalloenzymes may be difficult for some methods, we stress that many metals only play a structural role and are well outside the binding sites. We discouraged users from shying away from entries that contain metals since many are quite tractable. Lastly, there are two cases, Set 2 no. 178 (2fxu) and no. 202 (1bky), where a cofactor (ATP and SAH, respectively) is located well outside the binding site and was retained for completeness.
The release of the remediated set contained 343 entries, organized into: Set 1 (176 complexes, most deposited to the PDB in 2007 and 2008) and Set 2 (167 complexes, most deposited in 2006 or earlier). In order to avoid confusion, the numbering of the structures is the same as the original July 29, 2009 release. Thus, removing structures has resulted in missing numbered directories in each data set. These remediated sets were used for the scoring exercise starting on October 4, 2009. Participants scores were accepted well into February 2010, and the results of that exercise were discussed in three sessions held at the August 2010 meeting (240th) of the ACS in Boston, MA.
Modifications to the Dataset, Post-Meeting Exercise: CSAR-NRC HIQ Dataset (September 24, 2010 Release)
In the course of working through the exercise, Traian Sulea (National Research Council of Canada, NRC) had identified necessary modifications to both the ligands and the proteins in the October 4 release of the data set. The vast majority were changes in the protonation state of either the protein or the ligand and also included changes in tautomerism or reorientations of a polar hydrogen. Here, we will present a very brief description of those modifications and our collaborative effort to improve the data set based on his work. For any additional details on his work, the reader is directed to Sulea, Cui, and Purisma’s paper in this issue of Journal of Chemical Information and Modeling.
Examples of the modifications include: deprotonation of the sulfonamide ligands of carbonic anhydrase complexes (9 instances), deprotonation of Cys215 in the protein tyrosine phosphatase complexes (9 instances), and protonation of Asp25/A in the HIV protease complexes (26 instances, though Set 1 no. 164 and Set 2 no. 7 are still doubly deprotonated as is appropriate for the bound ligands). In all, we made protonation changes on 41 ligand entries and 69 proteins. There were polar hydrogen reorientations in 72 entries. Our NRC colleagues provided the CSAR group with the full data set of 343 complexes with all of his modifications, including a minimized version of each entry. There were only five instances where the CSAR group disagreed, and we worked together to resolve the differences.
During the final discussion sessions of the CSAR symposium at the ACS meeting, it was decided that most of the participants would repeat their scoring using the NRC modifications to the CSAR sets. These changes should improve performance of the various methods and allow users to assess the impact of protonation states if desired. The major difference in the overall formatting of the CSAR-NRC release is in the structures directory. It consists of two directories (Sets 1 and 2) with each containing subdirectories based on the initial numbering system. Each of the subdirectories has the kd.dat file, the unminimized complex in .mol2 format, and the minimized complex in .mol2 format. The waters were stripped out of the .mol2 files for the NRC’s methods, but we have included them in a .pdb formatted file for others to add back in if they so choose. We stress that users will need to use the unminimized coordinate set if they add the water in their application. This tar file set was made available late in September 2010 on the CSAR Web site as the CSAR_NRC_HiQ_Set_24Sept2010.tar.
Strengths and Weaknesses
Here, we present an assessment of the experimental error and outline the limits they impose upon use of the data set. The greatest asset for the final CSAR-NRC set is the high level of curation, generously contributed by many experts. It likely represents the “best of the PDB” for structures deposited before January 1, 2009, augmented with binding data from Binding MOAD and PDBbind. It also represents the full diversity in targets and ligands for publically available complex structures, given the stringent constraints applied in the curation process. It provides a relatively unrestricted landscape that good docking and scoring methods should be able to process. Part of the reason for seeking this diversity was to document the issues that limit docking and scoring. Metalloenzymes are thought to be difficult targets because of anecdotal experience. Are they truly more difficult than other targets, if one examines a larger spectrum of available targets? Without a data set like the CSAR-NRC set, one cannot hope to answer this question or to prioritize the issues that most limit our field.
However, that diversity can also be considered a weakness. Data are only available for 16 small, congeneric series (see Table 2), and data on known inactive compounds are not available. Though it is limited, a user could easily incorporate more ligand data if needed for their purpose. Each protein has 3–11 structures, completely setup and ready for docking, which is a very good starting point for docking and scoring studies on series of compounds. Furthermore, the data being collected by the CSAR center from industry and our in-house efforts are specifically intended to expand data on congeneric series for a variety of drug targets.
Table 2. Congeneric Series Present in the Data Set.
| protein | count | ligand series type | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id | pdb id |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 protease (L63P) | 11 | hydroxyethylamine | 2cem | 2cen | 2qnq | 1g2k | 1d4i | 1d4j | 1ebz | 1ec0 | 1ec1 | 1ec2 | 1xl5 |
| HIV-1 protease (WT) | 11 | hydroxyethylamine | 2psv | 2q54 | 2qhy | 2qhz | 2qi1 | 2qi3 | 2qi4 | 2qi5 | 2qi6 | 2i0a | 2i0d |
| tyrosine-protein phosphatase | 8 | thiophene-dicarboxylic acid | 2hb1 | 2qbq | 2qbr | 2qbs | 2zmm | 2zn7 | 2azr | 2b07 | |||
| coagulation factor X | 6 | pyrrolidin-2-one | 2uwl | 2uwp | 2cji | 2j2u | 2j34 | 2j4i | |||||
| carbonic anhydrase 2 | 6 | sulfamide analogues | 2h15 | 2pov | 2pow | 3bl0 | 2hd6 | 2pou | |||||
| tRNA guanine transglycosylase | 5 | quinazolin-4-one | 2bbf | 1r5y | 1s38 | 1s39 | 1q4w | ||||||
| HMGCoA reductase | 5 | atorvastatin analogues | 3ccw | 3ccz | 3cd7 | 3cd0 | 3cda | ||||||
| acetylcholinesterase | 4 | huperzine/hupyridone | 1vot | 1gpk | 1h22 | 1h23 | |||||||
| estrogen receptor alpha | 4 | phenol | 2pog | 2r6w | 2ayr | 1qkt | |||||||
| urokinase | 4 | benzamidine analogues | 1gja | 1gj8 | 1gjd | 1gi9 | |||||||
| β-1,4-xylanase | 3 | aza-sugar analogues | 1fh8 | 1fh9 | 1fhd | ||||||||
| glutamate [NMDA] receptor ζ1 | 3 | small ring amino carboxylic acid | 1y1m | 1y1z | 1y20 | ||||||||
| lectin | 3 | saccharides | 1ax1 | 1ax0 | 1ax2 | ||||||||
| membrane lipoprotein tmpC | 3 | purine neucloside analogues | 2fqw | 2fqx | 2fqy | ||||||||
| retropepsin | 3 | macrocyclic peptidomimetic | 1b6j | 1b6l | 1b6m | ||||||||
| transporter (LeuT) | 3 | small amino acid | 3f3d | 3f48 | 3f4j |
The range of affinities in the CSAR-NRC set is quite large, 14 orders of magnitude in pKd. Though a large range of affinity is the property most important for obtaining a reasonably good fit from linear regression,(45) it is not the range that is most useful in practical applications. Instead, the ability to score ligands with nM level affinities over μM affinities is the critical range for identifying leads over mere hits. The bulk of the data in the CSAR-NRC set have pKd in that key range between 5 and 10, and a user could choose to limit his/her studies to those complexes. Brown et al.(45) have shown that 3 orders of magnitude or more with about 50 data points per target protein is appropriate for obtaining good fit to experimental data.
Another concern is that the diversity of proteins requires users to try to calculate absolute free energies of binding. With series of compounds against a single target, one can focus on relative ranking. Some thought that an exercise based on absolute free energies was impossible, but the papers in this issue show that the correlations for absolute free energies are no worse than the correlations seen for studies based on congeneric series.(46) The ability to properly calculate absolute free energies of binding is the Holy Grail of computational drug design, and it appears to be nearly as tractable as relative ranking. If we were to make significant progress in this aim, then all of our problems would be solved simultaneously. If absolute free energies can be calculated, then relative ranking is trivial. Furthermore, enrichment in database searching would be de facto. Even determining selectivity for ligands against homologues or possible side effects from binding to alternate targets would be possible.
Lastly, one could argue that the uncertainty inherent to comparing all proteins, all experimental techniques, and Kd vs Ki creates more “noise” than “signal” in the data set. Indeed, this does raise an important point that is frequently misunderstood by computational chemists. The error given for experiments in almost all publications is underestimated! Reported error is typically for multiple measurements on the same day, under the same conditions, by the same scientist. Experimentalists understand that independent laboratories can measure Kd (or Ki) of the same complex and easily differ by 3-fold (0.5 log Kd or 0.7 kcal/mol). Occasionally, two laboratories differ by an order of magnitude (1.0 log Kd or 1.4 kcal/mol), and this is acceptable. Yet, we often require the best calculations to match experimental values within 1 kcal/mol. Depending on the system, we can often require accuracy for our calculations that is not possible in the experiments we benchmark against.
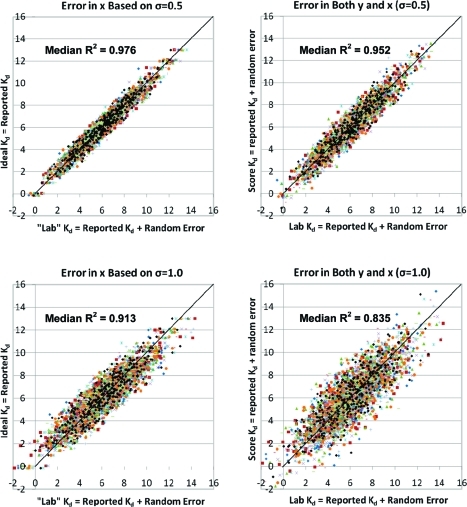
Returning to the question of signal over noise, the values above can be used as estimates for appropriate standard deviations (σ) in the experimental affinities reported in the CSAR-NRC set. With a normal distribution of experimental error, measurements within ±1σ will occur ∼68.3% of the time (±2σ = 95.4%, ± 3σ = 99.7%). A σ of 0.5 log K would make differences of up to 3-fold common (2 in 3 measures) and errors >1.0 log K would be seen only on rare occasion (1 in 22 observations). Let us assume that each measured value should be normally distributed around its “ideal” Kd (or Ki), the value obtained under the exact same conditions with no error. To approximate this, we can use the reported affinities in the CSAR-NRC set as ideal measures, generate random error around those points, and assess the potential influence on the analyses (Figure 3). When 100 independent generations of random error are used, there is a minimal addition of “noise” to the affinity data; the R2 range from 0.970 to 0.980 with a median of 0.976 (ave |error| = 0.40 log K, median |error| = 0.34 log K, RMSE = 0.50 log K). If error is added to both x and y directions to simulate the same variation in scores and experimental values, then the R2 range from 0.943 to 0.962 with a median of 0.952 (ave |error| = 0.56 log K, median |error| =0 .47 log K, RMSE = 0.71 log K).
Figure 3.
The addition of random error with standard deviations of 0.5 log K (top) or 1.0 log K (bottom) do not significantly degrade the “signal to noise” in the CSAR-NRC data set. Only 10 of the 100 randomly generated sets are shown for clarity, and a line with a slope of 1.0 is given as a guideline in all the graphs. (Left) Correlations based on the model that the reported affinities are ideal (y-axis) and random, normally distributed error can be added to generate possible measurements found in another lab (x-axis). (Right) Correlations based on the model that both the reported value and another measured value could have the same, random error. These plots also approximate the variation between scores and measured affinity values.
Of course, differences between laboratories come from both standard error and differences in experimental conditions, which exist even when trying to replicate the exact same experiment. If experimental conditions change, then no one would be surprised at differences in Kd of an order of magnitude or more. A σ of 1.0 log K would make differences of up to an order of magnitude common (2 in 3 measures) and beyond 2 orders of magnitude seen on rare occasion (1 in 22 observations). Still the random error gives small noise to affinity data; the R2 range from 0.892 to 0.931 with a median of 0.913 (ave |error| = 0.80 log K, median |error| = 0.68 log K, RMSE = 1.00 log K). Based on this, we can set an upper limit to the performance of scoring methods. If a method has many tunable parameters all trained to specifically reproduce the data in Sets 1 + 2 of the CSAR-NRC data set, the uncertainty in the underlying experimental data should make it impossible to achieve a perfect correlation. We expect the limit to be R2 ≈ 0.9. In fact, the limit to a scoring function trained on outside data and tested against the CSAR-NRC set should be R2 ≈ 0.8 because adding random error with σ = 1.0 log K to both scores and experimental values produced R2 between 0.798 and 0.864 with a median of 0.835 (ave |error| = 1.13 log K, median |error| = 0.95 log K, RMSE = 1.41 log K).
Pushing the limits beyond what is likely: σ = 2.0 log K leads to median R2 of 0.744 for correlations to ideal values (ave |error| = 1.59 log K, median |error| = 1.34 log K, RMSE = 1.99 log K) and 0.553 for correlations between scores and experimental values with the same range of error (ave |error| = 2.27 log K, median |error| = 1.91 log K, RMSE = 2.85 log K); σ = 3.0 log K gives median R2 of 0.590 (ave |error| = 2.41 log K, median |error| = 2.04 log K, RMSE = 3.01 log K) and 0.355 (ave |error| = 3.39 log K, median |error| = 2.88 log K, RMSE = 4.24 log K) for correlations to ideal values and between scores and experimental values, respectively.
Summary
This data set is the product of many different contributions from the modeling and crystallographic communities and has benefitted greatly from users’ input, comments, and constructive critiques over the course of approximately a year. The participants have used this single data set in many different scoring approaches (knowledge based, force-field based, grid based, etc), with expert hands in each case, to a common goal of calculating absolute free energies. This impressive array of information has been collected, collated, and analyzed with regard to what is the current state of the art and what trends (both global and specific) can be gleaned from this exercise. The knowledge gained from this exercise is reflected in the papers in this issue. While based on the final CSAR-NRC data set of September 24, 2010, it is certainly grounded in the thoughts and critiques from the larger community on its various incarnations; the journey, if you will, on the path to this data set.
Acknowledgments
We wish to thank Traian Sulea (NRC), Gregory Warren and Matt Geballe (OpenEye), and Matt Jacobson and C. Kalyanaraman (UCSF) for their contributions to remediating the data set. The CSAR Center is funded by the National Institute of General Medical Sciences (U01 GM086873). K.W.L. thanks Rackham Graduate School, the Pharmacological Sciences Training Program (GM07767), and the American Foundation for Pharmaceutical Education for funding. N.A.K. thanks the Bioinformatics Training Grant (GM070449) for support. We also thank the Chemical Computing Group and OpenEye Scientific Software for generously donating the use of their software.
Supporting Information Available
Confidence intervals possible for R, R2, ρ, and τ are provided. This information is available free of charge via the Internet at http://pubs.acs.org
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Berman H. M.; Westbrook J; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Benson M. L.; Smith R. D.; Lerner M. G.; Carlson H. A. Binding MOAD (Mother of All Databases). Proteins: Struct., Funct., Bioinf. 2005, 60, 333–340. [DOI] [PubMed] [Google Scholar]
- Wang R.; Fang X.; Lu Y.; Wang S. The PDBbind Database: Collection of Binding Affinities for Protein-Ligand Complexes with Known Three-Dimensional Structures. J. Med. Chem. 2004, 47, 2977–2980. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D.; Blaney J. M.; Oatley S. J.; Langridge R.; FerrinT. E. A Geometric Approach to Macromolecule-Ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [DOI] [PubMed] [Google Scholar]
- Meng E. C.; Shoichet B. K.; Kuntz I. D. Automated docking with grid-based energy evaluation. J. Comput. Chem. 1992, 13, 505–524. [Google Scholar]
- Rarey M.; Kramer B.; Lengauer T.; Klebe G. A fast flexible docking method using and incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [DOI] [PubMed] [Google Scholar]
- Jones G.; Willett P.; Glen R. C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995, 245, 43–53. [DOI] [PubMed] [Google Scholar]
- Nissink J. W. M.; Murray C. W.; Hartshorn M. J.; Verdonk M. L.; Cole J. C.; Taylor R. A. A new test set for validating predictions of protein-ligand interaction. Proteins: Struct., Funct., Genet. 2002, 49, 457–471. [DOI] [PubMed] [Google Scholar]
- Hartshorn M. J.; Verdonk M. L.; Chessari G.; Brewerton S. C.; Mooij W. T. M.; Mortenson P. N.; Murray C. W. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007, 50, 726–741. [DOI] [PubMed] [Google Scholar]
- Sondergard C. R.; Garrett A. E.; Carstensen T.; Pollastri G.; Nielsen J. E. Structural artifacts in protein-ligand x-ray structures: implications for the development of docking scoring functions. J. Med. Chem. 2009, 52, 5673–5684. [DOI] [PubMed] [Google Scholar]
- Benson M. L; Smith R. A.; Khazanov N. A.; Dimcheff B.; Beaver J.; Dresslar P.; Nerothin J; Carlson H. A. Binding MOAD, A high-quality protein-ligand database. Nucleic Acids Res. 2008, 36, D674–D678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm H. J. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput.-Aided Mol. Des. 1994, 8, 243–256. [DOI] [PubMed] [Google Scholar]
- Jain A. N. Scoring noncovalent protein-ligand interactions: A continuous differentiable function tuned to compute binding affinities. J. Comput.-Aided Mol. Des. 1996, 10, 427–440. [DOI] [PubMed] [Google Scholar]
- Head R. D.; Smythe M. L.; Oprea T. L.; Waller C. L.; Green S. M.; Marshall G. R. (1996) VALIDATE: A new method for the receptor-based prediction of binding affinities of novel ligands. J. Am. Chem. Soc. 1996, 118, 3959–3969. [Google Scholar]
- Eldridge M. D.; Murray C. W.; Auton T. R.; Paolini G. V.; Mee R. P. (1997) Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des. 1997, 11, 425–445. [DOI] [PubMed] [Google Scholar]
- Bohm H. J. Prediction of binding constants of protein ligands: A fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput.-Aided Mol. Des. 1998, 12, 309–323. [DOI] [PubMed] [Google Scholar]
- Wang R.; Gao Y.; Lai L. SCORE: A new empirical method for estimating the binding affinity of a protein-ligand complex. J. Mol. Model. 1998, 4, 379–394. [Google Scholar]
- Muegge I.; Martin Y. C. A general and fast scoring function for protein-ligand interactions: A simplified approach. J. Med. Chem. 1999, 52, 791–804. [DOI] [PubMed] [Google Scholar]
- Mitchell J. B. O.; Laskowski R. A.; Alex A.; Forster M. J.; Thornton J. M. BLEEP: Potential of mean force describing protein-ligand interactions. I. Generating potential. J. Comput. Chem. 1999, 20, 1165–1176. [Google Scholar]
- Mitchell J. B. O; Laskowski R. A.; Alex A.; Forster M. J.; Thornton J. M. BLEEP: Potential of mean force describing protein-ligand interactions. II. Calculation of binding energies and comparison with experimental data. J. Comput. Chem. 1999, 20, 1177–1185. [Google Scholar]
- Gohlke H.; Hendlich M.; Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000, 295, 337–356; http://pc1664.pharmazie.uni-marburg.de/drugscore/introduction.php. Accessed June 14, 2011. [DOI] [PubMed] [Google Scholar]
- Gohlke H.; Hendlich M.; Klebe G. Predicting binding modes, binding affinities and “hot spots” for protein-ligand complexes using a knowledge-based scoring function. Perspect. Drug Discovery Des. 2000, 20, 115–144. [Google Scholar]
- Roche O.; Kiyama R.; Brooks C. L. III Ligand-Protein DataBase: Linking protein-ligand complex structures to binding data. J. Med. Chem. 2001, 44, 3592–3598. [DOI] [PubMed] [Google Scholar]
- Ishchenko A. V.; Shakhnovich E. I. Small Molecule Growth 2001 (SMoG2001): An improved knowledge-based scoring function for protein-ligand interactions. J. Med. Chem. 2002, 45, 2770–2780. [DOI] [PubMed] [Google Scholar]
- Cozzini P.; Fornabaio M.; Marabotti A.; Abraham D. J.; Kellogg G. E.; Mozzarelli A. A simple, intuitive calculation of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J. Med. Chem. 2002, 45, 2469–2483. [DOI] [PubMed] [Google Scholar]
- Wang R.; Lai L.; Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002, 16, 11–26. [DOI] [PubMed] [Google Scholar]
- Puvanendrampillai D.; Mitchell J. B. O. Protein Ligand Database (PLD): Additional understanding of the nature and specificity of protein-ligand complexes. Bioinformatics 2003, 19, 1856–1857. [DOI] [PubMed] [Google Scholar]
- Block P.; Sotriffer C. A.; Dramburg I.; Klebe G. AffinDB: a freely accessible database of affinities for protein–ligand complexes from the PDB. Nucleic Acids Res. 2006, 34, D522–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten P; Salzemann J.; Bloch V.; Stockinger H.; Berglund A.-C.; Blanchet C.; Bongcam-Rudloff E.; Combet C.; Da Costa A. L.; Deleage G.; Diarena M.; Fabbretti R.; Fettahi G.; Flegel V.; Gisel A.; Kasam V.; Kervinen T.; Korpelainen E.; Mattila K.; Pagni M.; Reichstadt M.; Breton V.; Tickle I. J.; Vreind G. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009, 42, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. C. D.; Skillman A. G.; Warren G. L.; Ellingson B. A.; Stahl M. T. Conformer generation with OMEGA: Algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins P. C. D.; Warren G. L.; Skillman A. G.; Nicholls A. How to do an evaluation: pitfalls and traps. J. Comput.-Aided Mol. Des. 2008, 22, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto J.; Katakoa R.; Hirayama N. Ph4Dock: Pharmacophore-based protein-ligand docking. J. Med. Chem. 2004, 47, 6804–6813. [DOI] [PubMed] [Google Scholar]
- Sybyl, version 8.0; Tripos International: St. Louis, MO, 2008.
- Omega2; OpenEye Scientific Software: Santa Fe, NM, 2010.
- MOE; Chemical Computing Group: Montreal, Quebec, Canada, 2009.
- JMP, version 8; SAS Institute Inc.: Cary, NC, 2009.
- Wang R.; Fang X.; Lu L.; Yang C.-Y.; Wang S. The PDBbind Database: Methodologies and updates. J. Med. Chem. 2005, 48, 4111–4119. [DOI] [PubMed] [Google Scholar]
- Velec H. F.; Gohlke H.; Klebe G. DrugScore(CSD)-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J. Med. Chem. 2005, 48 (20), 6296–6303. [DOI] [PubMed] [Google Scholar]
- Yang C.-Y.; Wang R.; Wang S. M-score: a knowledge-based potential scoring function accounting for protein atom mobility. J. Med. Chem. 2006, 49, 5903–5911. [DOI] [PubMed] [Google Scholar]
- Jain A. N.; Nicholls A. Recommendations for evaluations of computational methods for docking and ligand-based modeling. J. Comput.-Aided Mol. Des. 2008, 22, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Lin Y.; Wen X.; Jorrisen R. N.; Gilson M. K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden C. I.; Jones T. A. Between objectivity and subjectivity. Nature 1990, 343, 687–689. [Google Scholar]
- Rupp B. Real-space solution to the problem of full disclosure. Nature 2006, 444, 817. [DOI] [PubMed] [Google Scholar]
- Hubbard S. J.; Thornton M. J.. NACCESS; Department of Biochemistry and Molecular Biology, University College: London, 1993; http://www.bioinf.manchester.ac.uk/naccess/. Accessed June 9, 2011.
- Brown S. P.; Muchmore S. W.; Hajduk P. J. Healthy skepticism: assessing realistic model performance. Drug Discovery Today 2009, 14, 420–427. [DOI] [PubMed] [Google Scholar]
- Warren G. L.; Andrews C. W.; Capelli A. M.; Clarke B.; LaLonde J.; Lambert M. H.; Lindvall M.; Nevins N.; Semus S. F.; Senger S.; Tedesco G.; Wall I. D.; Woolven J. M.; Peishoff C. E.; Head M. S. A critical assessment of docking programs and scoring. J. Med. Chem. 2006, 49, 5912–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.