Abstract
Influenza is a human pathogen that continues to pose a public health threat. The use of small mammalian models has become indispensable for understanding the virulence of influenza viruses. Among numerous species used in the laboratory setting, only the ferret model is equally well suited for studying both the pathogenicity and transmissibility of human and avian influenza viruses. Here, we compare the advantages and limitations of the mouse, ferret and guinea pig models for research with influenza A viruses, emphasizing the multifarious uses of the ferret in the assessment of influenza viruses with pandemic potential. Research performed in the ferret model has provided information, support and guidance for the public health response to influenza viruses in humans. We highlight the recent and emerging uses of this species in influenza virus research that are advancing our understanding of virus-host interactions.
Introduction
Influenza viruses are responsible for seasonal epidemics and sporadic pandemics of varying severity in humans. In a typical year, seasonal influenza viruses cause 250,000 to 500,000 deaths globally (approximately 36,000 in the United States), with >90% of these deaths occurring in the elderly (Thompson et al., 2004). Rates of infection and mortality can increase greatly during a pandemic. The influenza pandemic of 1918–1919 was the worst pandemic of modern times, which eventually infected about one-third of the world’s population, killing 20–50 million people (Johnson and Mueller, 2002). Although the recent 2009 H1N1 pandemic virus did not achieve similar levels of mortality (probably owing to both immunological reasons and advances in modern medicine over the past century), both pandemic viruses targeted young, healthy adults – a demographic not usually associated with seasonal influenza-related mortality (Dawood et al., 2009). Highly pathogenic avian influenza (HPAI) H5N1 viruses have caused over 560 human infections, with a 60% mortality rate; among many of these fatalities were young, previously healthy, adults (WHO, 2011). The heterogeneity of influenza viruses in their natural reservoir of wild waterfowl, and the growing diversity of influenza virus lineages affecting mammalian species (including swine, equine, canine and others), suggests that future human infection with viruses of pandemic potential will continue to occur. For these reasons, the availability of small mammalian models to study the virulence of emerging influenza viruses, in addition to understanding the molecular determinants that confer heightened pathogenicity and transmissibility among selected strains, is essential.
Ferret models (Mustela putorius furo) have been established for numerous viruses that cause respiratory infections, including human and avian influenza viruses, coronavirus, nipah virus, morbillivirus and others (Bossart et al., 2009; Martina et al., 2003; Smith and Sweet, 1988; Svitek and von Messling, 2007; Zitzow et al., 2002). Ferrets are an attractive mammalian model for these studies owing to their relatively small size and the fact that they emulate numerous clinical features associated with human disease; this is especially the case with regard to influenza. Ferrets and humans share similar lung physiology, and human and avian influenza viruses exhibit similar patterns of binding to sialic acids (the receptor for influenza viruses), which are distributed throughout the respiratory tract in both species (Maher and DeStefano, 2004; van Riel et al., 2007). The high susceptibility of ferrets to influenza virus, and the ability of sick ferrets to transmit virus to healthy ferrets, was first demonstrated in 1933 (Smith et al., 1933). Since this time, the use of the ferret model has been indispensable in experimental studies to elucidate virus-host interactions following influenza virus infection (Belser et al., 2009; Smith and Sweet, 1988). In this Primer article, we discuss the advantages of using the ferret for research with influenza A viruses, emphasizing the effectiveness of working with a model that can present both the pathogenic and transmissible features of influenza virus infection. We further highlight recent studies that are expanding the utility of the ferret to address novel research questions in this model.
Advantages of the ferret as a small animal model for influenza virus pathogenesis and transmission
Several mammalian models are available for influenza virus research, with their use depending on the specific research questions being addressed. Mice have traditionally been used as a model for influenza virus pathogenesis and offer many advantages over other laboratory species, including low cost, broad availability of reagents and the availability of transgenic mice with targeted gene disruptions (Belser et al., 2009). However, most human influenza strains do not replicate efficiently in mice without prior virus adaptation, limiting the utility of mice as models in which to study this group of viruses. Furthermore, many clinical signs of influenza virus infection in humans are not present in mice, and they are a poor model for virus transmission (Lowen et al., 2006; Schulman and Kilbourne, 1963). Guinea pigs have emerged as an alternative small mammalian model for this virus. Unlike mice, they support replication of human influenza viruses without prior adaptation and can be used to model virus transmission to susceptible contact animals (Lowen et al., 2006). Despite this, guinea pigs are a poor model for viral pathogenesis and also lack many clinical signs of infection that are observed in humans (Van Hoeven et al., 2009a). Although not widely used for the study of influenza A viruses, the cotton rat model supports the replication of human influenza viruses without prior adaptation and is suitable for the study of both innate and adaptive immune responses (Eichelberger, 2007; Sadowski et al., 1987). However, similar to the mouse model, cotton rats lack a sneeze reflex and are not well suited to study virus transmissibility (Eichelberger, 2007). In contrast to these small mammalian models, non-human primates offer the ability to study virus infection in an animal that is similarly complex to humans, but size, cost and ethical considerations limit the use of this species in most settings (Kobasa et al., 2007; Rimmelzwaan et al., 2003). Each of the above models offers unique advantages, but none possesses the ability to study both the pathogenesis and transmission of influenza A viruses in most laboratory settings.
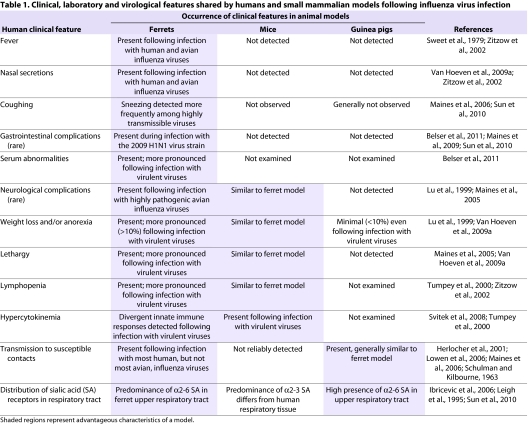
The ferret is currently the only small mammalian model available that is equally well suited to studying both of these aspects of influenza pathobiology. Human and avian influenza viruses replicate efficiently in the respiratory tract of ferrets without prior adaptation, and extra-pulmonary spread of virus results following infection with some HPAI viruses. Unlike other small mammalian models, numerous clinical signs found in humans following seasonal or avian influenza virus infection are present in the ferret following experimental inoculation with these strains by the intranasal route (Table 1). Elevated body temperatures are detected as early as 1 day post-inoculation, and more virulent viruses generally elicit high fevers that can persist for several days (Sweet et al., 1979; Zitzow et al., 2002). Nasal discharge and sneezing are also frequently observed in infected ferrets up to 1 week post-inoculation, with sneezing detected more frequently following seasonal influenza virus infection (Maines et al., 2006). Transmission models have been established using the ferret to measure the capacity of influenza viruses to spread to naïve contact ferrets in the presence of direct contact (i.e. pair-housing an infected and a naïve ferret) or by the spread of respiratory droplets in the absence of direct contact (i.e. separating infected and naïve ferrets with a perforated wall to allow air exchange only) (Herlocher et al., 2001; Maines et al., 2006; Yen et al., 2007). Successful virus transmission is detected by testing contact ferrets for the presence of virus in nasal secretions and seroconversion at the end of the observation period. Daily clinical assessment of inoculated and contact ferrets provides an additional means to evaluate the effects of virus infection and transmission effectiveness.
Table 1.
Clinical, laboratory and virological features shared by humans and small mammalian models following influenza virus infection
The ability to use one mammalian model to evaluate both pathogenicity and transmissibility has allowed for a fuller understanding of the capacity of influenza viruses to cause disease, and the ability to place experimental results obtained from this model in the context of human infection. Facilitated by the use of reassortant viruses, research in the ferret model has identified roles for virus gene segments and individual amino acids that contribute towards both virus pathogenicity and transmissibility (Salomon et al., 2006; Tumpey et al., 2007; Van Hoeven et al., 2009b). Furthermore, the efficacy of novel vaccine preparations has been demonstrated in the ferret model: recent studies have demonstrated the ability of vaccination to limit virus spread to susceptible ferret contacts (Ellebedy et al., 2010; Laurie et al., 2010; Pearce et al., 2011). Indeed, ferrets have proved instrumental in the evaluation of vaccine effectiveness and evaluation of vaccine candidates for human use (Bodewes et al., 2010; Robertson et al., 2011; Subbarao and Luke, 2007). Ferret studies have also been instrumental in demonstrating the impact of mutations that confer resistance to antiviral drugs on overall virus fitness and their ability to transmit to uninfected animals (Herlocher et al., 2002; Herlocher et al., 2004; Yen et al., 2005). The polygenic nature of influenza virus pathogenesis and transmission underscores the importance of using a mammalian model capable of assessing both of these parameters.
Recent advances and future uses of the ferret model in influenza research
A greater understanding of influenza viruses and ferret biology as well as the continued development of sophisticated laboratory equipment has led to the ability of scientists to examine numerous properties not previously studied in the ferret model. Below we describe selected areas of investigation that are likely to expand our understanding of influenza virus infection in the coming years.
Aerosol delivery systems
Intranasal administration of virus in a liquid suspension has long been the traditional method for influenza virus inoculation in the ferret model. However, this practice is not truly reflective of ‘natural’ virus infection in humans, which commonly occurs following contact (including direct and indirect contact with contaminated surfaces) with or inhalation of virus-containing respiratory secretions from an infected individual. Exposure of naïve ferrets to aerosolized influenza virus has been performed using a nose-only inhalation exposure system (Lednicky et al., 2010; Tuttle et al., 2010). However, only recently has a rigorous method of ensuring standard doses of virus inoculum during aerosol delivery been reported, and a comprehensive comparison of intranasal and aerosol methods of inoculation been conducted (Gustin et al., 2011). The development of a method to measure the number of viable influenza viruses in droplets and droplet nuclei exhaled from infected animals has furthered the utility of this model (Gustin et al., 2011). The development of influenza virus aerosol delivery systems that rigorously control for virus challenge inoculums, as well as the development of analytical systems for the ferret model, allow a more accurate recreation of natural influenza infection in the laboratory to study influenza virus transmission and perform risk assessments of emerging influenza viruses that pose a threat to public health.
Induction of immune responses
Reports of hypercytokinemia following H5N1 virus infection in humans, resulting in a deleterious ‘cytokine storm’, have underscored the need to study the induction of innate immune responses in severely infected hosts (de Jong et al., 2006). Moreover, knowledge about specific immune responses to vaccines, which correlate with protection against infection, will help to improve the design of novel vaccine candidates. In vivo assessments of immune function (i.e. innate and adaptive responses) following influenza virus infection have been primarily limited to the mouse model owing to a lack of ferret-specific reagents and incomplete genome sequencing of this species. However, the regulation of ferret proinflammatory mediators has been measured by real-time PCR and microarray analysis (Cameron et al., 2008; Svitek et al., 2008). Canine reagents have also been used successfully to assess ferret host responses, owing to the high levels of homology between canine and ferret nucleotide sequences, and numerous mouse monoclonal antibodies have demonstrated cross-reactivity with proteins expressed by ferret lymphocytes (Fang et al., 2010; Rutigliano et al., 2008). Continued immunology-based research in the ferret will result in a valuable outbred mammalian model for the study of innate and adaptive immune responses, leading to an improved understanding of human infection.
Laboratory abnormalities
Severe human infection with influenza viruses can result in substantial abnormalities of the lymphohematopoietic system, including lymphopenia, leukopenia, anemia and quantitative alterations of numerous serum analytes. Although lymphopenia has been previously demonstrated in ferrets, comprehensive analysis of white and red blood cell populations in whole blood from severely infected ferrets has only recently been performed (Belser et al., 2011; Zitzow et al., 2002). Furthermore, comprehensive analyses of serum chemistry profiles have allowed for detailed measurement of numerous parameters in the ferret that are frequently observed in severe human influenza virus infection (Belser et al., 2011). The ability to measure these parameters throughout the duration of an experiment by collecting samples during scheduled anesthesia of ferrets makes this an important contribution towards the standard pathotyping of viruses in this model.
Opportunistic infections
The 1918 H1N1 virus was extraordinarily virulent, but much of the mortality associated with this pandemic was due to bacterial pneumonia that developed post-infection (Morens et al., 2010). Studies in mouse and ferret models corroborate the finding that previous infection with influenza virus can render a host more susceptible to secondary bacterial infection (Peltola et al., 2006). Although research in the ferret model has been limited to date, recent work has further demonstrated that prior infection with influenza virus can influence the severity and transmissibility of Streptococcus pneumoniae in ferrets, a finding that corroborates historical data in humans documenting that the incidence of pneumonia increases during pandemic influenza (McCullers, 2006; McCullers et al., 2010). Although histopathology provides evidence of bacterial involvement following wild-type virus infection, further work is needed to more precisely measure the onset of naturally occurring opportunistic infections following influenza virus infection in this model.
Immunocompromised hosts
Humans with immunocompromised immune systems represent an especially susceptible population to influenza virus infection, and pose numerous challenges with respect to effectively preventing and treating infection. Although ferrets have not been used extensively to research influenza virus infection in the context of immunosuppression, this remains an understudied area that could benefit from such a model. Immunocompromised ferrets (achieved either by treatment with chemotherapy drugs or by depleting CD8+ and/or CD4+ T cells) were found to safely tolerate live, attenuated influenza vaccines (Huber and McCullers, 2006; Min et al., 2010). Although the research in this model is largely limited to drug- or antibody-induced immunosuppression, additional future studies evaluating influenza virus pathogenicity and transmissibility in the context of immunosuppression should allow for a greater understanding of influenza virulence in immunodeficient populations.
Conclusion
Influenza viruses continue to emerge and cause human infection, so there remains a need to characterize the pathogenicity and transmissibility of these viruses in animal models. This information serves a vital role in public health with respect to evaluating the efficacy of novel vaccines and therapeutics against currently circulating strains. As described in this Primer article, the ferret is an excellent small mammalian model that can recapitulate many of the hallmarks of human influenza virus infection. Given the wealth of information that can be generated while studying respiratory virus infection in this model, ferrets offer numerous advantages over other small mammalian models and are an ideal laboratory species to measure the most crucial aspects of influenza virus infection in the context of improving public health.
Advantages of using ferrets compared with other small mammalian models for research of influenza A viruses.
Ferrets are highly susceptible to both human and avian influenza viruses.
Pathogenesis and transmission studies can both be performed in the ferret model.
Numerous clinical signs of human infection with influenza viruses are present in the ferret.
Ferrets are well suited to vaccine efficacy studies.
REFERENCES
- Belser J. A., Szretter K. J., Katz J. M., Tumpey T. M. (2009). Use of animal models to understand the pandemic potential of highly pathogenic avian influenza viruses. Adv. Virus Res. 73, 55–97 [DOI] [PubMed] [Google Scholar]
- Belser J. A., Gustin K. M., Maines T. R., Blau D. M., Zaki S. R., Katz J. M., Tumpey T. M. (2011). Pathogenesis and transmission of triple-reassortant swine H1N1 influenza viruses isolated before the 2009 H1N1 pandemic. J. Virol. 85, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Rimmelzwaan G. F., Osterhaus A. D. (2010). Animal models for the preclinical evaluation of candidate influenza vaccines. Expert Rev. Vaccines 9, 59–72 [DOI] [PubMed] [Google Scholar]
- Bossart K. N., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J., McEachern J. A., Green D., Hancock T. J., Chan Y. P., et al. (2009). A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 5, e1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C. M., Cameron M. J., Bermejo-Martin J. F., Ran L., Xu L., Turner P. V., Ran R., Danesh A., Fang Y., Chan P. K., et al. (2008). Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J. Virol. 82, 11308–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawood F. S., Jain S., Finelli L., Shaw M. W., Lindstrom S., Garten R. J., Gubareva L. V., Xu X., Bridges C. B., Uyeki T. M. (2009). Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615 [DOI] [PubMed] [Google Scholar]
- de Jong M. D., Simmons C. P., Thanh T. T., Hien V. M., Smith G. J., Chau T. N., Hoang D. M., Chau N. V., Khanh T. H., Dong V. C., et al. (2006). Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12, 1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger M. C. (2007). The cotton rat as a model to study influenza pathogenesis and immunity. Viral Immunol. 20, 243–249 [DOI] [PubMed] [Google Scholar]
- Ellebedy A. H., Ducatez M. F., Duan S., Stigger-Rosser E., Rubrum A. M., Govorkova E. A., Webster R. G., Webby R. J. (2010). Impact of prior seasonal influenza vaccination and infection on pandemic A(H1N1) influenza virus replication in ferrets. Vaccine 29, 3335–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Rowe T., Leon A. J., Banner D., Danesh A., Xu L., Ran L., Bosinger S. E., Guan Y., Chen H., et al. (2010). Molecular characterization of in vivo adjuvant activity in ferrets vaccinated against influenza virus. J. Virol. 84, 8369–8388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K. M., Belser J. A., Wadford D. A., Pearce M. B., Katz J. M., Tumpey T. M., Maines T. R. (2011). Influenza virus aerosol exposure and analytical system for ferrets. Proc. Natl. Acad. Sci. USA 108, 8432–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlocher M. L., Elias S., Truscon R., Harrison S., Mindell D., Simon C., Monto A. S. (2001). Ferrets as a transmission model for influenza: sequence changes in HA1 of type A (H3N2) virus. J. Infect. Dis. 184, 542–546 [DOI] [PubMed] [Google Scholar]
- Herlocher M. L., Carr J., Ives J., Elias S., Truscon R., Roberts N., Monto A. S. (2002). Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54, 99–111 [DOI] [PubMed] [Google Scholar]
- Herlocher M. L., Truscon R., Elias S., Yen H. L., Roberts N. A., Ohmit S. E., Monto A. S. (2004). Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190, 1627–1630 [DOI] [PubMed] [Google Scholar]
- Huber V. C., McCullers J. A. (2006). Live attenuated influenza vaccine is safe and immunogenic in immunocompromised ferrets. J. Infect. Dis. 193, 677–684 [DOI] [PubMed] [Google Scholar]
- Ibricevic A., Pekosz A., Walter M. J., Newby C., Battaile J. T., Brown E. G., Holtzman M. J., Brody S. L. (2006). Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 80, 7469–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. P., Mueller J. (2002). Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76, 105–115 [DOI] [PubMed] [Google Scholar]
- Kobasa D., Jones S. M., Shinya K., Kash J. C., Copps J., Ebihara H., Hatta Y., Kim J. H., Halfmann P., Hatta M., et al. (2007). Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323 [DOI] [PubMed] [Google Scholar]
- Laurie K. L., Carolan L. A., Middleton D., Lowther S., Kelso A., Barr I. G. (2010). Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J. Infect. Dis. 202, 1011–1020 [DOI] [PubMed] [Google Scholar]
- Lednicky J. A., Hamilton S. B., Tuttle R. S., Sosna W. A., Daniels D. E., Swayne D. E. (2010). Ferrets develop fatal influenza after inhaling small particle aerosols of highly pathogenic avian influenza virus A/Vietnam/1203/2004 (H5N1). Virol. J. 7, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh M. W., Connor R. J., Kelm S., Baum L. G., Paulson J. C. (1995). Receptor specificity of influenza virus influences severity of illness in ferrets. Vaccine 13, 1468–1473 [DOI] [PubMed] [Google Scholar]
- Lowen A. C., Mubareka S., Tumpey T. M., Garcia-Sastre A., Palese P. (2006). The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. USA 103, 9988–9992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Tumpey T. M., Morken T., Zaki S. R., Cox N. J., Katz J. M. (1999). A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73, 5903–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J. A., DeStefano J. (2004). The ferret: an animal model to study influenza virus. Lab. Anim. (NY) 33, 50–53 [DOI] [PubMed] [Google Scholar]
- Maines T. R., Chen L. M., Matsuoka Y., Chen H., Rowe T., Ortin J., Falcon A., Nguyen T. H., Mai L. Q., Sedyaningsih E. R., et al. (2006). Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. USA 103, 12121–12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines T. R., Lu X. H., Erb S. M., Edwards L., Guarner J., Greer P. W., Nguyen D. C., Szretter K. J., Chen L. M., Thawatsupha P., et al. (2005). Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79, 11788–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines T. R., Jayaraman A., Belser J. A., Wadford D. A., Pappas C., Zeng H., Gustin K. M., Pearce M. B., Viswanathan K., Shriver Z. H., et al. (2009). Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325, 484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B. E., Haagmans B. L., Kuiken T., Fouchier R. A., Rimmelzwaan G. F., Van Amerongen G., Peiris J. S., Lim W., Osterhaus A. D. (2003). Virology: SARS virus infection of cats and ferrets. Nature 425, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A. (2006). Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A., McAuley J. L., Browall S., Iverson A. R., Boyd K. L., Henriques Normark B. (2010). Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J. Infect. Dis. 202, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J. Y., Vogel L., Matsuoka Y., Lu B., Swayne D., Jin H., Kemble G., Subbarao K. (2010). A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J. Virol. 84, 11950–11960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M., Taubenberger J. K., Harvey H. A., Memoli M. J. (2010). The 1918 influenza pandemic: lessons for 2009 and the future. Crit. Care Med. 38, e10–e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. B., Belser J. A., Houser K. V., Katz J. M., Tumpey T. M. (2011). Efficacy of seasonal live attenuated influenza vaccine against virus replication and transmission of a pandemic 2009 H1N1 virus in ferrets. Vaccine 29, 2887–2894 [DOI] [PubMed] [Google Scholar]
- Peltola V. T., Boyd K. L., McAuley J. L., Rehg J. E., McCullers J. A. (2006). Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect. Immun. 74, 2562–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan G. F., Kuiken T., van Amerongen G., Bestebroer T. M., Fouchier R. A., Osterhaus A. D. (2003). A primate model to study the pathogenesis of influenza A (H5N1) virus infection. Avian Dis. 47, 931–933 [DOI] [PubMed] [Google Scholar]
- Robertson J. S., Nicolson C., Harvey R., Johnson R., Major D., Guilfoyle K., Roseby S., Newman R., Collin R., Wallis C., et al. (2011). The development of vaccine viruses against pandemic A(H1N1) influenza. Vaccine 29, 1836–1843 [DOI] [PubMed] [Google Scholar]
- Rutigliano J. A., Doherty P. C., Franks J., Morris M. Y., Reynolds C., Thomas P. G. (2008). Screening monoclonal antibodies for cross-reactivity in the ferret model of influenza infection. J. Immunol. Methods 336, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski W., Wilczynski J., Semkow R., Tulimowska M., Krus S., Kantoch M. (1987). [The cotton rat (Sigmodon hispidus) as an experimental model for studying viruses in respiratory tract infections. II. Influenza viruses types A and B]. Med. Dosw. Mikrobiol. 39, 43–55 [PubMed] [Google Scholar]
- Salomon R., Franks J., Govorkova E. A., Ilyushina N. A., Yen H. L., Hulse-Post D. J., Humberd J., Trichet M., Rehg J. E., Webby R. J., et al. (2006). The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman J. L., Kilbourne E. D. (1963). Experimental transmission of influenza virus infection in mice. Ii. Some factors affecting the incidence of transmitted infection. J. Exp. Med. 118, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H., Sweet C. (1988). Lessons for human influenza from pathogenicity studies with ferrets. Rev. Infect. Dis. 10, 56–75 [DOI] [PubMed] [Google Scholar]
- Smith W., Andrewes C. H., Laidlaw P. P. (1933). A virus obtained from influenza patients. Lancet 222, 66–68 [Google Scholar]
- Subbarao K., Luke C. (2007). H5N1 viruses and vaccines. PLoS Pathog. 3, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Bi Y., Pu J., Hu Y., Wang J., Gao H., Liu L., Xu Q., Tan Y., Liu M., et al. (2010). Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One 5, e15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitek N., von Messling V. (2007). Early cytokine mRNA expression profiles predict Morbillivirus disease outcome in ferrets. Virology 362, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitek N., Rudd P. A., Obojes K., Pillet S., von Messling V. (2008). Severe seasonal influenza in ferrets correlates with reduced interferon and increased IL-6 induction. Virology 376, 53–59 [DOI] [PubMed] [Google Scholar]
- Sweet C., Bird R. A., Cavanagh D., Toms G. L., Collie M. H., Smith H. (1979). The local origin of the febrile response induced in ferrets during respiratory infection with a virulent influenza virus. Br. J. Exp. Pathol. 60, 300–308 [PMC free article] [PubMed] [Google Scholar]
- Thompson W. W., Shay D. K., Weintraub E., Brammer L., Bridges C. B., Cox N. J., Fukuda K. (2004). Influenza-associated hospitalizations in the United States. JAMA 292, 1333–1340 [DOI] [PubMed] [Google Scholar]
- Tumpey T. M., Lu X., Morken T., Zaki S. R., Katz J. M. (2000). Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J. Virol. 74, 6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey T. M., Maines T. R., Van Hoeven N., Glaser L., Solorzano A., Pappas C., Cox N. J., Swayne D. E., Palese P., Katz J. M., et al. (2007). A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315, 655–659 [DOI] [PubMed] [Google Scholar]
- Tuttle R. S., Sosna W. A., Daniels D. E., Hamilton S. B., Lednicky J. A. (2010). Design, assembly, and validation of a nose-only inhalation exposure system for studies of aerosolized viable influenza H5N1 virus in ferrets. Virol. J. 7, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoeven N., Belser J. A., Szretter K. J., Zeng H., Staeheli P., Swayne D. E., Katz J. M., Tumpey T. M. (2009a). Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J. Virol. 83, 2851–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoeven N., Pappas C., Belser J. A., Maines T. R., Zeng H., Garcia-Sastre A., Sasisekharan R., Katz J. M., Tumpey T. M. (2009b). Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl. Acad. Sci. USA 106, 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Munster V. J., de Wit E., Rimmelzwaan G. F., Fouchier R. A., Osterhaus A. D., Kuiken T. (2007). Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171, 1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2011). Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. Geneva: http://www.who.int/csr/disease/avian_influenza/en/ [Google Scholar]
- Yen H. L., Herlocher L. M., Hoffmann E., Matrosovich M. N., Monto A. S., Webster R. G., Govorkova E. A. (2005). Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 49, 4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H. L., Lipatov A. S., Ilyushina N. A., Govorkova E. A., Franks J., Yilmaz N., Douglas A., Hay A., Krauss S., Rehg J. E., et al. (2007). Inefficient transmission of H5N1 influenza viruses in a ferret contact model. J. Virol. 81, 6890–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzow L. A., Rowe T., Morken T., Shieh W. J., Zaki S., Katz J. M. (2002). Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 76, 4420–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]