Abstract
The retinoblastoma (RB) tumor suppressor belongs to a cellular pathway that plays a crucial role in restricting the G1-S transition of the cell cycle in response to a large number of extracellular and intracellular cues. Research in the last decade has highlighted the complexity of regulatory networks that ensure proper cell cycle progression, and has also identified multiple cellular functions beyond cell cycle regulation for RB and its two family members, p107 and p130. Here we review some of the recent evidence pointing to a role of RB as a molecular adaptor at the crossroads of multiple pathways, ensuring cellular homeostasis in different contexts. In particular, we discuss the pro- and anti-tumorigenic roles of RB during the early stages of cancer, as well as the importance of the RB pathway in stem cells and cell fate decisions.
Introduction
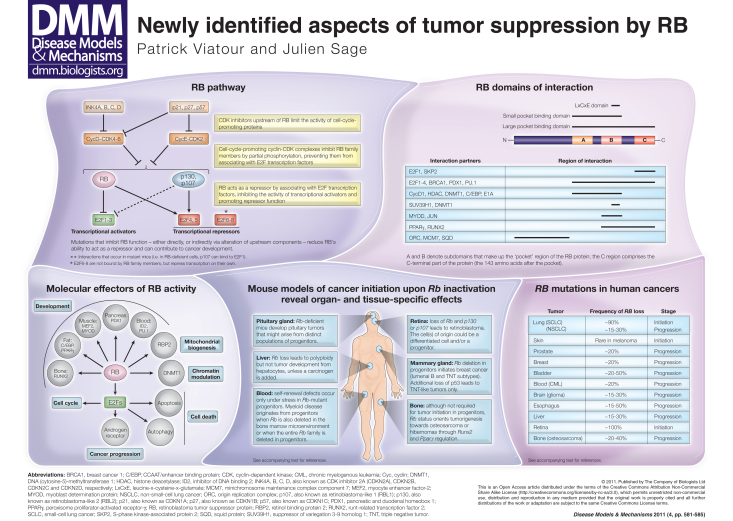
Since it was cloned 25 years ago, there has been much emphasis on determining the role of the retinoblastoma tumor suppressor gene (RB) in controlling the G1-S transition during cell cycle progression. The RB protein and its family members, p107 and p130, are components of a pathway that mediates cellular responses to a variety of signals by controlling the activity of E2F transcription factors and the expression of their target genes during cell cycle progression. In normal conditions, RB family members are inhibited by cyclin-dependent kinase (CDK) complexes in response to growth factors. Cyclin-CDK complexes are themselves inhibited by small cell cycle inhibitors of the p16INK4a and p21CIP1 families under cytostatic conditions. Events (e.g. mutation, promoter methylation, overexpression) that inhibit the function of RB family members (directly, or indirectly via alteration of upstream components) reduce their ability to regulate E2F transcription factors and can contribute to cancer development (see ‘RB pathway’ section of the poster) (Burkhart and Sage, 2008). Additional lines of research have described multiple roles for RB family proteins in tumor initiation and progression, beyond cell cycle regulation and interactions with E2F transcription factors (Chinnam and Goodrich, 2011; Knudsen and Knudsen, 2008). In this article and the accompanying poster, we review recent evidence indicating that RB acts as a molecular adaptor at the crossroads of multiple pathways, depending on the cellular context. We also discuss the idea that intact RB function might in some cases promote the early steps of tumorigenesis, a provocative possibility for the first-identified tumor suppressor.
The RB pathway in cancer
Mutations targeting the RB pathway are almost universal in cancer, but different components of this pathway are selectively affected in distinct cancer types (see ‘RB mutations in human cancers’ section of the poster). Events that affect upstream members of the pathway (e.g. methylation of the INK4a promoter or amplification of CDK genes) lead to the concomitant inactivation of the three RB family members and are often sufficient to induce uncontrolled cellular proliferation. In contrast to p107 and p130, which are seldom mutated in human cancers, the RB gene is inactivated in most cases of retinoblastoma, osteosarcoma and small cell lung cancer (SCLC), as well as at a lower frequency in other types of cancer. This discrepancy among RB family members indicates that RB possesses tumor suppressor functions that are not shared with p130 and p107 (Burkhart and Sage, 2008; Knudsen and Knudsen, 2008) (see ‘RB pathway’ section of the poster).
Recent insights into the molecular mechanisms of tumor suppression by RB
The RB protein as an adaptor
RB can suppress cancer development through its interactions with more than 100 partners, including the E2F family of transcription factors, lineage-specific transcription factors, DNA-modifying enzymes and members of chromatin remodeling complexes (Chinnam and Goodrich, 2011). The vast majority of these interactions involve the C-terminal portion of RB (which also serves as the docking site for E2F), but the N-terminal part of RB might also be involved in inhibiting E2F transactivation ability (Burke et al., 2010), and recent data in Drosophila have shown that binding partners that interact with the N-terminal portion of RB are involved in the control of DNA replication (Ahlander et al., 2008) (see ‘RB domains of interaction’ section of the poster). However, the role of the N-terminal part of RB in tumor suppression is still unclear (Riley et al., 1997; Yang et al., 2002).
Preserving RB functions to promote tumorigenesis
RB mutations often occur late during tumor progression (with some major exceptions, such as retinoblastoma and SCLC), raising the possibility that tumor cells initially preserve RB function to gain a competitive advantage. Indeed, loss of RB function can lead to an increase in cell death, which is mediated by the transcriptional activity of E2F1. The fact that E2F1 activates ARF, which acts upstream of p53 to suppress tumor development, connects the RB and p53 pathways (Aslanian et al., 2004; Chen, D. et al., 2009) and might partially explain why loss of p53 function is frequent in RB-deficient tumors. Another recently uncovered potential mechanism by which RB might prevent cell death is the increased production of reactive oxygen species (ROS) in RB-deficient cells, which leads to autophagy in cells that are also deficient for the tumor suppressor tuberous sclerosis 2 (TSC2) (Chicas et al., 2010; Ciavarra and Zacksenhaus, 2010; Ciavarra and Zacksenhaus, 2011). Why some cell types respond to loss of RB by triggering cell death programs whereas others do not is still unknown. Nevertheless, cells in which loss of RB function induces death might benefit from an active RB protein at least until they have mutated other components of the cell death machinery (see ‘RB mutations in human cancers’ and ‘Molecular effectors of RB activity’ sections of the poster). Interestingly, RB function is nearly always preserved in colorectal cancer and the RB locus is even sometimes amplified in this cancer type. It is thought that, in colorectal cancer cells, RB promotes tumor development by preventing the inhibition of β-catenin transcription by E2F1 (Firestein et al., 2008; Morris et al., 2008). This idea that RB might be oncogenic in some contexts is substantiated by data in mice indicating that expression of a constitutively active form of RB in the mammary epithelium can promote cancer development (Jiang and Zacksenhaus, 2002). Similarly, loss of function of E2F1, a major mediator of RB action in cells, can also cause tumors (Field et al., 1996; Yamasaki et al., 1996).
Molecular effectors of RB activity
Mitochondrial biogenesis
Recent evidence indicates that RB normally controls mitochondrial biogenesis. In particular, loss of RB function in erythrocytes is associated with impaired erythropoiesis because of mitochondrial defects; although the mechanisms underlying these observations are not completely clear, they might in part involve the transcription factor and RB interactor RB-binding protein 2 (RBP2) (Lopez-Bigas et al., 2008; Sankaran et al., 2008).
Control of cell cycle progression and modulation of chromatin structure
Quiescence is defined as a reversible cell cycle arrest state; by contrast, senescence is defined as a permanent cell cycle arrest associated with changes in chromatin structure (Narita et al., 2003) and is thought to be a potent tumor suppressor mechanism. RB interacts with and controls the activity of several regulators of chromatin structure (Blais and Dynlacht, 2007), including DNMT1 (Robertson et al., 2000). Recent data in human fibroblasts have shown that RB, p130 and p107 can all inhibit the expression of E2F target genes involved in quiescence but that RB specifically controls the transcription of E2F target genes involved in the regulation of cellular senescence; in particular, activation of cyclin E1 upon loss of RB seems to be crucial to bypass senescence (Chicas et al., 2010). p130 (and maybe p107) might also control local chromatin changes during quiescence and senescence via its interactions with members of the DREAM (DP, RB, E2F and MuvB) complex (Litovchick et al., 2011; Tschop et al., 2011). The RB family also regulates overall chromosomal stability by controlling the loading onto chromatin of members of the cohesin and condensin complexes, both of which are important for mitosis (Sage and Straight, 2010).
Lineage-specific transcription factors
Physiological roles of RB during differentiation include interactions with lineage-specific transcription factors, such PU.1 and Id2 (which are required for erythroid development), Runx2 (bone development), MyoD (muscle development) or Pdx1 (pancreas development) (Chinnam and Goodrich, 2011; Kim et al., 2011). Interestingly, these interactions might also play a role during tumorigenesis. For instance, osteosarcoma is characterized by inactivation of both p53 and RB, with loss of p53 function being required for the initiation of the disease in mice. In this context, RB controls the type of tumor formed in p53 mutant mesenchymal progenitors by controlling the fate of these cells: the presence of one copy of the Rb gene in mice leads to osteosarcoma via interactions between RB and Runx2, whereas loss of both Rb alleles leads to the increased expression of PPARγ, which favors the development of hibernomas (fat cell tumors) (Calo et al., 2010; Walkley et al., 2008).
RB and E2F: roles in cancer beyond the control of cell cycle
Recent data have shown that loss of RB in prostate cancer is crucial for the progression of the disease. Whereas RB function seems to be intact in the early stages of prostate cancer, RB inactivation in later stages allows E2F1 to activate the transcription of the gene encoding the androgen receptor (AR), whose functions are essential for tumor progression and metastasis (Sharma et al., 2010).
Cellular mechanisms of tumor suppression by RB
The identification of the cell of origin is an important issue in the cancer field (Visvader, 2011). An attractive view is that tumors are derived from stem cells or progenitor cells. Self-renewing stem cells and progenitor cells have an intrinsic proliferation potential that can readily become unlimited if these cells acquire new tumorigenic mutations. An alternative to this model is that tumors arise from post-mitotic, differentiated cells. In this scenario, the initial cancer-causing alteration might result in dedifferentiation and reacquisition of self-renewal and proliferative capacity. RB plays a crucial role in the control of both cell cycle progression and differentiation, and loss of RB function, in theory, could play an essential role in cancer initiation, both by allowing the expansion of stem cells or progenitor cells and by inducing the reversal of differentiated cells into cancer stem cells. Data from different mouse models suggest that there can be wide variation in the pattern and incidence of cancer initiation in various cell types, tissues and organs upon loss of RB function (see the ‘Mouse models of cancer initiation upon Rb inactivation’ section of the poster, and specific examples and references below).
Can tumors initiate in differentiated populations following loss of RB function?
In the inner ear, loss of RB results in cell cycle re-entry without affecting the differentiation status of these cells (Sage et al., 2005). Similarly, combined inactivation of RB and the cell cycle regulator ARF in differentiated muscle cells is sufficient to induce cell cycle re-entry and partial dedifferentiation (Pajcini et al., 2010). These observations suggest that RB loss potentially initiates tumor development by triggering cell cycle re-entry of post-mitotic cells, at least in specific contexts. Indeed, loss of RB cooperates with a genotoxic carcinogen to initiate liver cancer development from hepatocytes (Reed et al., 2009). In addition, evidence from RB family mutant mice indicate that retinoblastoma might arise from fully differentiated retinal cells that had previously exited the cell cycle (Ajioka et al., 2007). However, the identity of the cell(s) of origin of retinoblastoma is still an open question and strong evidence also indicates that retinoblastoma might arise from progenitors or precursors in the retina (Macpherson, 2008; Xu et al., 2009). Finally, recent evidence of the emerging role of RB in embryonic cells and during cellular reprogramming supports the possibility that differentiated cells become transformed upon loss of RB function (Conklin and Sage, 2009; Li et al., 2010).
RB suppresses cancer in stem and/or progenitor cells
Given that the chromatin of stem cells is more plastic than that of differentiated cells, stem cells might be more sensitive to RB loss, owing to the capacity of RB to modulate chromatin (Chinnam and Goodrich, 2011). Research performed in Arabidopsis thaliana, in which there is only one member of the RB family, shows that RB inactivation prevents differentiation and leads to an expansion of the stem cell pool, suggesting that RB loss induces uncontrolled proliferation without decreasing self-renewing capacity (Chen, Z. et al., 2009). Recent data obtained in mouse models of osteosarcoma also indicate that different types of progenitors, uncommitted or committed, can serve as tumor-initiating cells (Choi et al., 2010). In a mouse model of breast cancer that is initiated through conditional deletion of Rb in the mammary epithelium, Jiang and colleagues showed that Rb inactivation in bipotential progenitors, but not in downstream committed progenitors, is sufficient to induce tumor growth. Interestingly, in this case, it is p53 status that dictates the type of tumor that develops (Jiang et al., 2010). Similar studies in a mouse model of pituitary adenoma showed that loss of RB in different types of progenitor cells induces tumor formation, although the latency was much shorter when Rb was inactivated in early versus late progenitor cells (Hosoyama et al., 2010). In these examples, inactivation of RB is sufficient to induce uncontrolled proliferation in stem or progenitor cell populations. This is in contrast to what has been observed for stem cells in other compartments, such as in the hematopoietic system, where RB inactivation does not lead to cell cycle entry and does not initiate cancer (Daria et al., 2008; Walkley et al., 2007) unless p107 and p130 are also inactivated (Viatour et al., 2008). Clearly, different mechanisms exist in different populations of stem cells and progenitors to ensure the maintenance of quiescence and the control of the cell cycle.
Perspectives
For a long time, RB has been viewed as ‘just’ a regulator of cell cycle progression. However, recent observations indicate that RB functions in multiple pathways and biological processes that are deregulated during tumor initiation and progression. RB, similarly to a puppet master, coordinates multiple cellular functions in a large variety of contexts, and its inactivation therefore goes beyond the simple inactivation of a red light on the road to anarchic proliferation. Mouse models in which Rb or RB family proteins are deleted suggest that stem and progenitor cells are particularly sensitive to loss of RB function. However, we still do not understand why loss of RB function leads to cell cycle re-entry in only specific cell populations, highlighting the need for more basic cell cycle research. In particular, future experiments should seek to identify factors that can selectively compensate for loss of RB during cell cycle progression, beyond the well-established compensatory roles of p107 and p130. Furthermore, (re)acquisition or conservation of self-renewal is a key aspect of cancer initiation, but we only have a very limited understanding of how RB can regulate both cell cycle progression and self-renewal activity. Clearly, these last years of research in the RB field have revealed that the picture is more complicated than initially appreciated. It is now evident that RB is central to multiple facets of cancer development, and that a better understanding of RB activity in cells will continue to provide fundamental insights into the biology of cancer.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
REFERENCES
- Ahlander J., Chen X. B., Bosco G. (2008). The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner. PLoS One 3, e2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajioka I., Martins R. A., Bayazitov I. T., Donovan S., Johnson D. A., Frase S., Cicero S. A., Boyd K., Zakharenko S. S., Dyer M. A. (2007). Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 131, 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanian A., Iaquinta P. J., Verona R., Lees J. A. (2004). Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 18, 1413–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A., Dynlacht B. D. (2007). E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 19, 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. R., Deshong A. J., Pelton J. G., Rubin S. M. (2010). Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J. Biol. Chem. 285, 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D. L., Sage J. (2008). Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J. A., Danielian P. S., Nedelcu S., Berman S. D., Lees J. A. (2010). Rb regulates fate choice and lineage commitment in vivo. Nature 466, 1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Pacal M., Wenzel P., Knoepfler P. S., Leone G., Bremner R. (2009). Division and apoptosis of E2f-deficient retinal progenitors. Nature 462, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hafidh S., Poh S. H., Twell D., Berger F. (2009). Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the Retinoblastoma protein. Proc. Natl. Acad. Sci. USA 106, 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R. A., Narita M., Zhang M., Lowe S. W. (2010). Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17, 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnam M., Goodrich D. W. (2011). RB1, development, and cancer. Curr. Top. Dev. Biol. 94, 129–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Curtis S. J., Roy D. M., Flesken-Nikitin A., Nikitin A. Y. (2010). Local mesenchymal stem/progenitor cells are a preferential target for initiation of adult soft tissue sarcomas associated with p53 and Rb deficiency. Am. J. Pathol. 177, 2645–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavarra G., Zacksenhaus E. (2010). Rescue of myogenic defects in Rb-deficient cells by inhibition of autophagy or by hypoxia-induced glycolytic shift. J. Cell Biol. 191, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavarra G., Zacksenhaus E. (2011). Direct and indirect effects of the pRb tumor suppressor on autophagy. Autophagy 7, 544–546 [DOI] [PubMed] [Google Scholar]
- Conklin J. F., Sage J. (2009). Keeping an eye on retinoblastoma control of human embryonic stem cells. J. Cell. Biochem. 108, 1023–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daria D., Filippi M. D., Knudsen E. S., Faccio R., Li Z., Kalfa T., Geiger H. (2008). The retinoblastoma tumor suppressor is a critical intrinsic regulator for hematopoietic stem and progenitor cells under stress. Blood 111, 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field S. J., Tsai F. Y., Kuo F., Zubiaga A. M., Kaelin W. G., Jr, Livingston D. M., Orkin S. H., Greenberg M. E. (1996). E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85, 549–561 [DOI] [PubMed] [Google Scholar]
- Firestein R., Bass A. J., Kim S. Y., Dunn I. F., Silver S. J., Guney I., Freed E., Ligon A. H., Vena N., Ogino S., et al. (2008). CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 455, 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoyama T., Nishijo K., Garcia M. M., Schaffer B. S., Ohshima-Hosoyama S., Prajapati S. I., Davis M. D., Grant W. F., Scheithauer B. W., Marks D. L., et al. (2010). A Postnatal Pax7 Progenitor Gives Rise to Pituitary Adenomas. Genes Cancer 1, 388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zacksenhaus E. (2002). Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J. Cell Biol. 156, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Deng T., Jones R., Li H., Herschkowitz J. I., Liu J. C., Weigman V. J., Tsao M. S., Lane T. F., Perou C. M., et al. (2010). Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J. Clin. Invest. 120, 3296–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Kim S. Y., Mellado-Gil J. M., Yadav H., Neidermyer W., Kamaraju A. K., Rane S. G. (2011). RB regulates pancreas development by stabilizing Pdx1. EMBO J. 30, 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen E. S., Knudsen K. E. (2008). Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer 8, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., He Z., Shen J., Huang Q., Li W., Liu X., He Y., Wolf F., Li C. Y. (2010). Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell 7, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L., Florens L. A., Swanson S. K., Washburn M. P., Decaprio J. A. (2011). DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 25, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N., Kisiel T. A., Dewaal D. C., Holmes K. B., Volkert T. L., Gupta S., Love J., Murray H. L., Young R. A., Benevolenskaya E. V. (2008). Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31, 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson D. (2008). Insights from mouse models into human retinoblastoma. Cell Div. 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E. J., Ji J. Y., Yang F., Di Stefano L., Herr A., Moon N. S., Kwon E. J., Haigis K. M., Naar A. M., Dyson N. J. (2008). E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature 455, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- Pajcini K. V., Corbel S. Y., Sage J., Pomerantz J. H., Blau H. M. (2010). Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 7, 198–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. A., Mayhew C. N., McClendon A. K., Yang X., Witkiewicz A., Knudsen E. S. (2009). RB has a critical role in mediating the in vivo checkpoint response, mitigating secondary DNA damage and suppressing liver tumorigenesis initiated by aflatoxin B1. Oncogene 28, 4434–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D. J., Liu C. Y., Lee W. H. (1997). Mutations of N-terminal regions render the retinoblastoma protein insufficient for functions in development and tumor suppression. Mol. Cell Biol. 17, 7342–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D., Ait-Si-Ali S., Yokochi T., Wade P. A., Jones P. L., Wolffe A. P. (2000). DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25, 338–342 [DOI] [PubMed] [Google Scholar]
- Sage C., Huang M., Karimi K., Gutierrez G., Vollrath M. A., Zhang D. S., Garcia-Anoveros J., Hinds P. W., Corwin J. T., Corey D. P., et al. (2005). Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science 307, 1114–1118 [DOI] [PubMed] [Google Scholar]
- Sage J., Straight A. F. (2010). RB’s original CIN? Genes Dev. 24, 1329–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran V. G., Orkin S. H., Walkley C. R. (2008). Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 22, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Yeow W. S., Ertel A., Coleman I., Clegg N., Thangavel C., Morrissey C., Zhang X., Comstock C. E., Witkiewicz A. K., et al. (2010). The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J. Clin. Invest. 120, 4478–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop K., Conery A. R., Litovchick L., Decaprio J. A., Settleman J., Harlow E., Dyson N. (2011). A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 25, 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P., Somervaille T. C., Venkatasubrahmanyam S., Kogan S., McLaughlin M. E., Weissman I. L., Butte A. J., Passegue E., Sage J. (2008). Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell 3, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J. E. (2011). Cells of origin in cancer. Nature 469, 314–322 [DOI] [PubMed] [Google Scholar]
- Walkley C. R., Shea J. M., Sims N. A., Purton L. E., Orkin S. H. (2007). Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell 129, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley C. R., Qudsi R., Sankaran V. G., Perry J. A., Gostissa M., Roth S. I., Rodda S. J., Snay E., Dunning P., Fahey F. H., et al. (2008). Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 22, 1662–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. L., Fang Y., Lee T. C., Forrest D., Gregory-Evans C., Almeida D., Liu A., Jhanwar S. C., Abramson D. H., Cobrinik D. (2009). Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137, 1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki L., Jacks T., Bronson R., Goillot E., Harlow E., Dyson N. J. (1996). Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85, 537–548 [DOI] [PubMed] [Google Scholar]
- Yang H., Williams B. O., Hinds P. W., Shih T. S., Jacks T., Bronson R. T., Livingston D. M. (2002). Tumor suppression by a severely truncated species of retinoblastoma protein. Mol. Cell Biol. 22, 3103–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]