Abstract
Down syndrome (DS) is caused by trisomy of human chromosome 21 (Hsa21) and results in a large number of phenotypes, including learning difficulties, cardiac defects, distinguishing facial features and leukaemia. These are likely to result from an increased dosage of one or more of the ∼310 genes present on Hsa21. The identification of these dosage-sensitive genes has become a major focus in DS research because it is essential for a full understanding of the molecular mechanisms underlying pathology, and might eventually lead to more effective therapy. The search for these dosage-sensitive genes is being carried out using both human and mouse genetics. Studies of humans with partial trisomy of Hsa21 have identified regions of this chromosome that contribute to different phenotypes. In addition, novel engineered mouse models are being used to map the location of dosage-sensitive genes, which, in a few cases, has led to the identification of individual genes that are causative for certain phenotypes. These studies have revealed a complex genetic interplay, showing that the diverse DS phenotypes are likely to be caused by increased copies of many genes, with individual genes contributing in different proportions to the variance in different aspects of the pathology.
Introduction
Down syndrome (DS) is a complex set of pathologies caused by an extra copy of human chromosome 21 (Hsa21). DS occurs in about 1 in 750 live births and is the most frequent cause of learning difficulties. Although the underlying genetic cause, trisomy Hsa21, is the same in most individuals with DS, the penetrance of the resulting pathologies is varied (Antonarakis et al., 2004). For example, most individuals with DS have memory and learning difficulties, craniofacial alterations and muscle hypotonia, but only some have congenital heart malformations, leukaemia or gut abnormalities. Furthermore, the severity of the defects is variable. For example, the extent of cognitive impairment varies widely between individuals with DS (Pennington et al., 2003).
The prevailing hypothesis for the genetic causes underlying DS pathology is that individual phenotypes are caused by an extra copy of one or more of the ∼310 genes present on Hsa21 (Ensembl release 62, including known and newly identified protein-coding and RNA genes but excluding pseudogenes; http://www.ensembl.org/Homo_sapiens/Location/Chromosome?r=21:1-48129895). Such genes are described as being dosage sensitive, and much effort is being made to identify the dosage-sensitive genes underlying each of the DS phenotypes. The hope is that identification of such genes will lead to a better understanding of the molecular mechanisms underlying the pathologies, and hence to more effective therapy.
The search for these dosage-sensitive genetic culprits has taken advantage of both human and mouse genetics. In humans, rare partial trisomies of Hsa21 have been used to narrow down regions of the chromosome that might contain dosage-sensitive genes. Early studies suggested that a limited region of Hsa21, termed the Down syndrome critical region (DSCR) (Fig. 1), might contain one or more dosage-sensitive genes that contribute to many of the DS phenotypes (McCormick et al., 1989; Rahmani et al., 1989; Korenberg et al., 1990; Sinet et al., 1994). However, further studies that included larger numbers of partial trisomy cases and more-detailed genetic mapping have shown that different regions of Hsa21 contribute to different phenotypes, arguing against a single DSCR (Delabar et al., 1993; Korenberg et al., 1994; Korbel et al., 2009; Lyle et al., 2009). Despite these studies, it is clear that the use of human partial trisomies to identify dosage-sensitive genes is limited by the rarity of partial trisomies, heterogeneity of the specific phenotype and genetic variation between individuals.
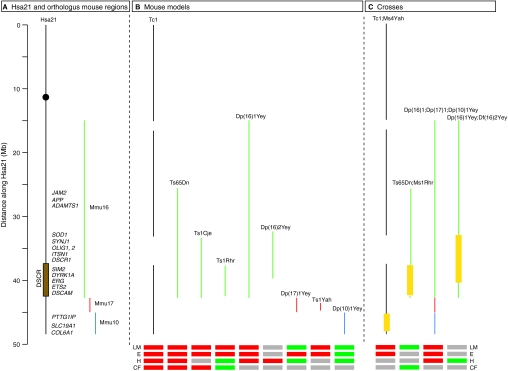
Fig. 1.
Hsa21, orthologous mouse regions and mouse models of DS. (A) A schematic diagram of Hsa21 indicating the approximate positions of candidate dosage-sensitive genes listed in Table 1, and of orthologous regions on mouse chromosomes 10 (blue), 16 (green) and 17 (red). The black circle indicates the centromere and the brown rectangle shows the approximate location of the DSCR. (B) The extent of trisomy in the mouse models of DS discussed in the text is shown. The Tc1 mouse carries a copy of Hsa21 (with some deletions), whereas the other models all contain duplications of mouse regions that are orthologous to Hsa21. (C) Crosses of mouse strains whose analysis has been reported. Where a strain with extra copies of genes has been crossed to a deficiency [Ms1Rhr, Ms4Yah or Df(16)2Yey], the regions where gene dosage has been reduced from three to two copies are indicated by a yellow box. Beneath B and C, red and green boxes indicate the presence or absence, respectively, of phenotypes in the following areas: learning and memory (LM), electrophysiology (E), heart (H) and craniofacial (CF). Grey boxes indicate that no analysis has been reported. Note that, in many cases, different assays have been used in the different models and crosses, making direct comparisons difficult, and in some cases the phenotypes result in an improvement rather than a defect, e.g. Ts1Yah shows improved learning and LTP responses compared with euploid controls. For further details see main text.
Further progress in the search for dosage-sensitive genes underlying DS has been enabled by the generation of several mouse models of DS. Hsa21 shares conserved synteny with orthologous regions on three mouse chromosomes: Mmu10, Mmu16 and Mmu17 (Fig. 1). The first two models that were generated, Ts65Dn and Ts1Cje mice, were animals with duplications of parts of Mmu16 that are orthologous to Hsa21. The Ts65Dn strain carries an extra chromosome that has a region of Mmu16 translocated onto a short segment of Mmu17, and is thus trisomic for ∼104 genes on Mmu16 that are orthologous to Hsa21 genes (Fig. 1) (Davisson et al., 1990; Gardiner et al., 2003; Olson et al., 2004a). However, it should be noted that it also has three copies of 19 genes on Mmu17 that are not orthologues of genes on Hsa21, and thus some phenotypes in this strain might not be related to human DS (Li et al., 2007). By contrast, the Ts1Cje strain has a duplication of a shorter region of Mmu16, encompassing ∼81 genes with orthologues on Hsa21 (Fig. 1) (Sago et al., 1998; Gardiner et al., 2003; Olson et al., 2004a). Both strains show some of the features of DS, including learning difficulties, and have thus been used to narrow down the search for dosage-sensitive genes. More recently, Cre-loxP technology has been used to precisely engineer duplications of the Hsa21-orthologous regions. For example, the Ts1Rhr mouse carries a duplication of the region of Mmu16 that is orthologous to the DSCR, and is thus trisomic for ∼33 genes with orthologues on Hsa21 (Olson et al., 2004a), whereas the Dp(10)1Yey;Dp(16)1Yey;Dp(17)1Yey ‘triple trisomic’ mouse has duplications of Mmu10, Mmu16 and Mmu17, resulting in trisomy for all ∼250 mouse genes that are orthologous to Hsa21 genes (Fig. 1) (Gardiner et al., 2003; Li et al., 2007; Yu et al., 2010b). This latter compound strain is the most complete DS model currently available.
Several years ago, we created a different mouse model of DS, called Tc1, carrying a freely segregating copy of Hsa21 (Fig. 1) (O’Doherty et al., 2005). This chromosome has several deletions, and thus contains ∼83% of the genes on Hsa21 (Susan Gribble, V.L.J.T., E.M.C.F. and Nigel Carter, unpublished data). The Tc1 strain has the advantage of allowing the study of the human genes on Hsa21, including a number that are not found in the orthologous regions of Mmu10, Mmu16 and Mmu17 (Gardiner et al., 2003). Phenotypic characterisation showed that Tc1 mice have a number of DS-like phenotypes, including learning and memory impairment, cardiac defects, craniofacial abnormalities, and decreased tumour angiogenesis (O’Doherty et al., 2005; Morice et al., 2008; Galante et al., 2009; Reynolds et al., 2010).
To map dosage-sensitive genes in these trisomic models, the mice are being crossed with strains bearing small deletions, which reduces the number of copies of some of the trisomic genes from three to two. If such a cross rescues a phenotype, this indicates the presence of one or more dosage-sensitive genes in the deleted interval. For example, the Ms4Yah strain, which bears a deletion of the region of Mmu10 that is orthologous to Hsa21 (Fig. 1), was crossed with the Tc1 strain and showed that trisomic genes in this interval do not contribute to learning defects (Duchon et al., 2010). Such mapping crosses can be continued with ever-smaller deletions, until eventually the individual dosage-sensitive genes can be identified by crossing the mouse model with a ‘knockout’ for the relevant gene.
In this review, we present recent progress in the identification of dosage-sensitive genes underlying DS phenotypes that has resulted from the use of both human and mouse genetics.
Learning, memory, and brain development and function
All individuals with DS exhibit some form of learning and memory impairment, which varies in severity (Lott and Dierssen, 2010). Studies of mouse models have indicated that defects in neurogenesis, synaptogenesis, synaptic transmission and cell signalling pathways could all contribute to this phenotype, potentially by causing excessive inhibitory neurotransmission (Belichenko et al., 2004; Kleschevnikov et al., 2004; Harashima et al., 2006; Siarey et al., 2006; Belichenko et al., 2007; Chakrabarti et al., 2007; Morice et al., 2008). Indeed, chronic application of γ-aminobutyric acid (GABA) antagonists to dampen the action of inhibitory neurons can reverse the cognitive deficit in DS mouse models (Fernandez et al., 2007). The analysis and treatment of cognitive defects in mouse models of DS is discussed in detail in the accompanying Perspective featured in this issue of Disease Models & Mechanisms (Das and Reeves, 2011).
Studies of individuals with partial trisomy of Hsa21 demonstrated that multiple regions in Hsa21 contribute to cognitive deficits, indicating that several genes and pathways are involved in this phenotype (Korbel et al., 2009; Lyle et al., 2009). Both studies identified the importance of the proximal portion of 21q. In addition, Lyle et al. identified a contributing region from 37.94 Mb to 38.64 Mb (Lyle et al., 2009), whereas Korbel et al. noted that trisomy of the most telomeric 4.6 Mb of the chromosome resulted in the lowest IQ levels (Korbel et al., 2009). By contrast, analysis of mouse crosses showed that genes on the regions of Mmu10 and Mmu17 that are orthologous to the most telomeric 4.6 Mb of Hsa21 do not contribute to defects in learning and memory (Duchon et al., 2010; Yu et al., 2010a). Indeed, in the Ts1Yah strain, trisomy of 12 mouse genes located on a region of Mmu17 that is orthologous to Hsa21 resulted in improved outcomes in the Morris water maze (a test used to assess spatial learning), and, compared with euploid control mice, the Ts1Yah and Dp(17)1Yey strains show increased hippocampal long-term potentiation (LTP; a form of synaptic plasticity that is thought to be the physiological basis of learning and memory) (Pereira et al., 2009; Yu et al., 2010a). For a more detailed discussion of synaptic plasticity, see Box 1 in the accompanying Perspective (Das and Reeves, 2011).
The evidence connecting trisomy of the DSCR with neurological defects is mixed. Studies of humans with partial trisomy of Hsa21 showed that the DSCR region was not required in three copies to cause intellectual disability, although the data do not exclude the possibility that it contributes to the phenotype (Korbel et al., 2009; Lyle et al., 2009). Analysis of mouse models with or without three copies of the DSCR showed that trisomy of this region was necessary but not sufficient to cause a defect in the Morris water maze test (Olson et al., 2007). However, a separate study using the novel-object-recognition test, a different assay of learning and memory, concluded that trisomy of the DSCR was sufficient to result in cognitive defects (Belichenko et al., 2009). More work is needed to resolve the basis for these distinct conclusions. Nonetheless, a number of genes located in the DSCR have been proposed as candidate dosage-sensitive genes that might contribute to DS-associated brain phenotypes, including DYRK1A, SIM2, DSCAM and KCNJ6.
DYRK1A is expressed in the developing and adult nervous system (Hammerle et al., 2008) and can inhibit cell proliferation and promote premature neuronal differentiation, potentially by regulating NOTCH signalling (Yabut et al., 2010; Hammerle et al., 2011). Together with DSCR1 (also known as RCAN1), a protein also encoded by a gene on Hsa21, DYRK1A inhibits the nuclear translocation of the NFAT family of transcription factors. Thus, it has been proposed that, in DS, excess inhibition of the NFAT pathway might contribute to both neuronal and cardiac defects (Arron et al., 2006). Transgenic mice containing a 180 kb yeast artificial chromosome (YAC) that includes DYRK1A, or mice overexpressing Dyrk1a alone, showed severe learning difficulties and spatial memory deficits (Smith et al., 1997; Altafaj et al., 2001; Ahn et al., 2006). Extracellular hippocampal recordings in DYRK1A transgenic mice demonstrated altered induction of LTP and long term depression (LTD; the selective weakening of synapses during the learning and memory process), although the changes in both processes were in the opposite direction to that seen in other more complete trisomic mouse models, namely Ts65Dn and Ts1Cje (Kleschevnikov et al., 2004; Ahn et al., 2006). Nevertheless, these studies demonstrate that DYRK1A might affect the balance between excitatory and inhibitory transmission. Surprisingly, small human segmental duplications indicate that trisomy of DYRK1A does not lead to severe reductions in IQ levels (Korbel et al., 2009).
The Drosophila single minded transcription factor is a master developmental regulator (Nambu et al., 1991). SIM2, the human orthologue of single minded, is expressed in the developing human brain (Rachidi et al., 2005). Transgenic mice overexpressing Sim2 demonstrate mild learning and memory impairments (Ema et al., 1999); however, no defect was seen in a bacterial artificial chromosome (BAC) transgenic mouse in which Sim2 is expressed from its endogenous promoter, thereby achieving a physiologically more relevant level of overexpression (Chrast et al., 2000). Interestingly, SIM2 has been shown to repress Drebrin expression by directly binding to its promoter (Ooe et al., 2004). Drebrin is known to affect dendritic spine structure and neuritogenesis, and is decreased in the cortex of patients with Alzheimer’s disease (AD) and DS (Hayashi et al., 1996; Hayashi and Shirao, 1999; Shim and Lubec, 2002; Geraldo et al., 2008).
Down syndrome cell adhesion molecule (DSCAM) inhibits dendrite branching when overexpressed in hippocampal neurons in vitro (Alves-Sampaio et al., 2010). NMDA receptor signalling, an important component of learning and memory, leads to local translation of DSCAM protein within neuronal dendrites, a process that is likely to contribute to synaptic plasticity. Interestingly, this local translation of DSCAM is lost in neurons from the Ts1Cje mouse, which contains three copies of Dscam (Alves-Sampaio et al., 2010).
Kcnj6 (GIRK2; G protein-activated inward rectifying potassium channel subunit 2) is overexpressed in the hippocampus of the Ts65Dn mouse model (Harashima et al., 2006), leading to increased GIRK channel density in hippocampal neurons and increased GIRK current in response to inhibitory GABAB signalling (Best et al., 2007). It has been hypothesised that an increased dosage of Kcnj6 together with Dyrk1a might explain the synaptic phenotype of Ts1Rhr mice (Belichenko et al., 2009).
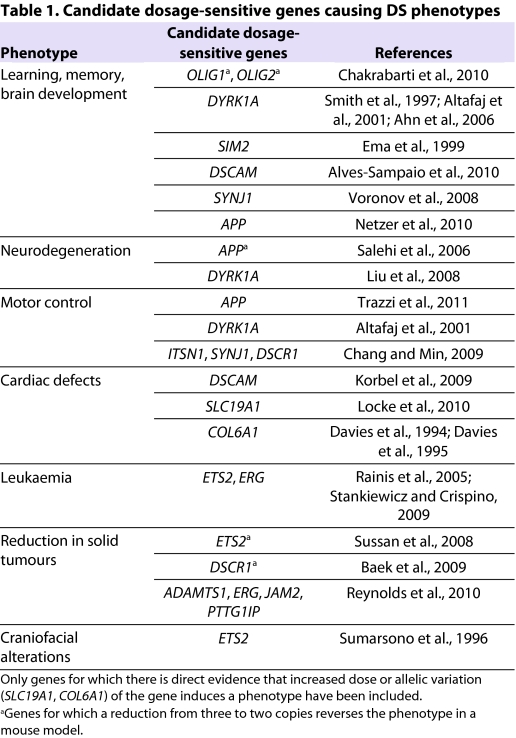
Several genes outside the DSCR have also been implicated in the neurological phenotypes of DS. The Olig1 and Olig2 genes encode transcription factors that have been implicated in neurogenesis and oligodendrogenesis (Takebayashi et al., 2000; Lu et al., 2002; Zhou and Anderson, 2002). Analysis of the Ts65Dn mouse model demonstrated that reduction of both Olig genes from three to two copies corrected the overproduction of inhibitory interneurons and the associated increase in inhibitory neurotransmission in the forebrain, although it is not known whether this resulted in improved behavioural performance (Chakrabarti et al., 2010). This latter study is notable in that it is one of only a few in which specific dosage-sensitive genes contributing to DS phenotypes have been directly identified using the ‘gold standard’ assay of reducing the copy number of the gene from three to two (Table 1).
Table 1.
Candidate dosage-sensitive genes causing DS phenotypes
Synaptojanin 1 (SYNJ1) dephosphorylates phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2], a key lipid involved in normal neurotransmission. PtdIns(4,5)P2 metabolism is altered in the brains of Ts65Dn mice, a defect that is normalised by reducing the Synj1 gene from three to two copies (Voronov et al., 2008). In addition, transgenic mice overexpressing Synj1 show defects in learning and memory (Voronov et al., 2008).
Finally, application of inhibitors to decrease the levels of APP (amyloid precursor protein) metabolites in Ts65Dn mice improves their learning and memory, suggesting that trisomy of APP contributes to neurological phenotypes of DS (Netzer et al., 2010). However, analysis of human segmental trisomies argues against a major role for APP in these defects (Rovelet-Lecrux et al., 2006; Korbel et al., 2009).
Neurodegeneration
DS is characterised by the early onset of the neuropathological features of AD and the eventual onset of dementia. A strong candidate for a dosage-sensitive gene contributing to this phenotype is APP, because proteolysis of APP generates amyloid-β (Aβ), the main constituent of amyloid plaques in AD brains, and mutations or duplications of APP have been associated with early onset AD (Goate et al., 1991; Rovelet-Lecrux et al., 2006; McNaughton et al., 2010).
Degeneration of basal forebrain cholinergic neurons (BFCNs) has been seen both in humans with AD (Lehericy et al., 1993) and in the Ts65Dn mouse (Holtzman et al., 1996). Neurons from Ts65Dn mice have a defect in retrograde transport of the neurotrophic factor nerve growth factor (NGF) to the cell soma, and this might contribute to their degeneration (Cooper et al., 2001). Notably, reduction of the dose of App from three to two copies partially rescues the defective NGF retrograde transport and BFCN degeneration in Ts65Dn mice (Salehi et al., 2006). By contrast, overexpression of APP alone results in defective NGF transport but does not cause BFCN degeneration (Cataldo et al., 2003; Salehi et al., 2006). Together, these studies suggest that increased dosage of APP contributes to BFCN degeneration in DS, but is not sufficient, implying that other Hsa21 genes contribute to the phenotype.
One such gene might be DYRK1A, whose protein product has been shown to phosphorylate APP. Consistent with this, transgenic mice overexpressing Dyrk1a demonstrate higher levels of phospho-APP and Aβ (Ryoo et al., 2008). In addition, DYRK1A might also contribute to the AD-like pathology by phosphorylating Tau, leading to further phosphorylation by glycogen synthase kinase 3β and subsequent aggregation of Tau into neurofibrillary tangles (Liu et al., 2008). Studies of human partial Hsa21 trisomies are consistent with a role of increased dosage of APP but not of DYRK1A in the early onset AD seen in DS (Korbel et al., 2009).
Motor control and hypotonia
Neonates with DS present with muscle hypotonia and many individuals with DS demonstrate some form of motor impairment, often described as clumsiness or deficits in fine motor control (Morris et al., 1982; Shumway-Cook and Woollacott, 1985; Spano et al., 1999; Moldrich et al., 2007). However, in contrast to cognitive and cardiac defects, these motor and muscle phenotypes in DS have not been documented in detail in terms of either penetrance or severity. Studies of human trisomic patients have mapped two regions on Hsa21 that are required in three copies to cause hypotonia: an interval from 37.4 Mb to 38.4 Mb and a second region from 46.5 Mb to 21qter (Lyle et al., 2009).
DS-associated motor deficits have been reproduced in mouse models (O’Doherty et al., 2005; Galante et al., 2009), and both humans with DS and mouse trisomic models exhibit reduced numbers of granule neurons in the cerebellum, a part of the brain that is important for fine motor control (Baxter et al., 2000; Olson et al., 2004b; O’Doherty et al., 2005). Reduced cerebellar neurogenesis might be due to defective sonic hedgehog (SHH) signalling in neuronal precursors (Roper et al., 2006) caused by increased levels of APP, leading to elevated expression of the SHH receptor patched 1, an inhibitor of the SHH signalling pathway (Trazzi et al., 2011).
Transgenic mice overexpressing Dyrk1a (TgDyrk1a) have a delay in neuromotor development, which leads to impaired performance in motor tasks (Altafaj et al., 2001). Interestingly, intrastriatal injections of viral vectors expressing shRNA against Dyrk1a into TgDyrk1a mice rescued this defect (Ortiz-Abalia et al., 2008). By contrast, a BAC transgenic mouse strain overexpressing DYRK1A showed no significant motor deficits, suggesting that the motor phenotypes in TgDyrk1a mice might be a consequence of DYRK1A overexpression at levels well above those seen in DS (Ahn et al., 2006). Alternatively, the differences in phenotype might be due to the use of mouse Dyrk1a versus human DYRK1A, or to effects of the transgene insertion sites.
One possible cause of motor deficits in DS could be defects in synapse morphology and vesicle recycling at the neuromuscular junction (NMJ). In support of this, overexpression of the Drosophila homologues of ITSN1, SYNJ1 and DSCR1 in transgenic flies resulted in locomotor defects and impaired vesicle recycling at the NMJ, suggesting that these three genes, along with APP and DYRK1A, are candidates for the dosage-sensitive genes underlying motor defects in DS (Chang and Min, 2009).
Cardiac defects
A total of 40–61% of individuals with DS present with congenital heart defects (CHDs), a major cause of high morbidity or infant mortality in individuals with DS (Yang et al., 2002; Vis et al., 2009). The most common heart malformation in DS is the atrioventricular septal defect (AVSD), which is considered a hallmark of DS: the incidence of AVSD is 1000-fold increased in individuals with DS compared with the non-DS population (Freeman et al., 1998; Torfs and Christianson, 1998).
Analysis of humans with partial trisomy of Hsa21 led to the identification of a 2.82 Mb region from DSCAM to CBS that is sufficient to cause CHDs (Korbel et al., 2009). By contrast, other studies found that genetic variation of a more telomeric gene, COL6A1, which is outside this region, was associated with CHDs in trisomy 21 (Davies et al., 1994; Davies et al., 1995). COL6A1 encodes the α1 chain of collagen VI, which is expressed at a higher level in the atrioventricular cushions in the hearts of fetuses with DS compared with those of euploid controls (Gittenberger-de Groot et al., 2003). Genes on Hsa21 as well as on other chromosomes can affect the susceptibility to CHDs in DS. Mutations in the cell adhesion molecule CRELD1 (encoded by the CRELD1 gene on Hsa3) have been postulated as a risk factor for AVSD in the euploid population, and contribute to AVSD in DS (Maslen, 2004). Another study demonstrated that polymorphisms in two folate pathway genes, SLC19A1 and MTHFR (located on Hsa21 and Hsa1, respectively), are associated with AVSD in DS (Locke et al., 2010).
Using high resolution episcopic microscopy (HREM) and three-dimensional (3D) modelling, a range of cardiac defects, including AVSDs similar to those seen in humans with DS, were found in Tc1 mouse embryos (Dunlevy et al., 2010). Cardiac defects have also been reported in the Ts65Dn and Dp(16)1Yey mouse models of DS, but not in Dp(10)1Yey or Dp(17)1Yey mice, indicating that dosage-sensitive genes for CHDs lie on Mmu16 (Li et al., 2007; Williams et al., 2008; Yu et al., 2010b). However, AVSDs were not reported in either the Ts65Dn or the Dp(16)1Yey mouse models. This might reflect trisomy of different sets of genes in these mice compared with Tc1 mice and/or genetic background differences. Alternatively, the difficulty of detecting AVSDs by routine methods might have led to defects being missed in Ts65Dn or Dp(16)1Yey mice; thus, it would be very interesting to apply the HREM technique to embryos from these strains as well.
Korbel et al. combined results from the human segmental trisomy data together with data from mouse models (O’Doherty et al., 2005; Li et al., 2007), and proposed a region of 1.77 Mb from DSCAM to ZNF295 as the ‘heart critical region’ (Korbel et al., 2009). Because DSCAM is the only gene in this region that is known to be expressed in the heart, the authors suggested DSCAM as a candidate dosage-sensitive gene for CHDs in DS. However, no cardiac malformations are seen in the Ts1Rhr strain, which is trisomic for 33 genes in the DSCR, including Dscam (Fig. 1), demonstrating that three copies of Dscam are not sufficient to cause the DS heart phenotype in mice (Dunlevy et al., 2010; Liu et al., 2011). A recent study showed that duplication of a 5.43 Mb region of Mmu16 from Tiam1 to Kcnj6 in the Dp(16)2Yey strain was sufficient to cause cardiac defects (Fig. 1), thereby again excluding Dscam and further narrowing down the region containing causative dosage-sensitive genes (Liu et al., 2011).
Leukaemia
Compared with the non-DS population, individuals with DS have an 18-fold increased risk of developing leukaemia (Hasle et al., 2000). In particular, DS is associated with a 500-fold increased risk of acute megakaryoblastic leukaemia (AMKL). Interestingly, DS-AMKL is always associated with a stereotypical mutation in the X-linked GATA1 gene, which leads to synthesis of a truncated protein termed GATA1S (Wechsler et al., 2002). This has led to the suggestion that the generation of AMKL requires cooperation between trisomy Hsa21 and the GATA1S mutation, together with additional mutations (Hitzler and Zipursky, 2005). Human partial trisomies have mapped an 8.35 Mb region of Hsa21 from RUNX1 to CBS as contributing to AMKL (Korbel et al., 2009). This region includes the RUNX1, ERG and ETS2 genes, all of which have been hypothesised to play a role in DS-AMKL (Hitzler and Zipursky, 2005; Rainis et al., 2005; Stankiewicz and Crispino, 2009).
So far, three mouse models of DS have been tested for leukaemia (Kirsammer et al., 2008; Carmichael et al., 2009; Alford et al., 2010). None showed leukaemia, but all three reported alterations in megakaryopoiesis. Tc1 mice have macrocytic anaemia, and older animals show splenomegaly with increased megakaryopoiesis and erythropoiesis, but they do not develop acute leukaemia, even when crossed with a strain expressing GATA1S (Alford et al., 2010). Both Ts65Dn and Ts1Cje mice also display macrocytic anaemia, whereas only Ts65Dn mice present with a myeloproliferative disorder (MPD) with increased megakaryopoiesis and erythropoiesis (Kirsammer et al., 2008; Carmichael et al., 2009). These results suggest that one or more of the 23 genes that are in three copies in Ts65Dn but not in Ts1Cje are responsible for the MPD, whereas the trisomic region shared between the two mouse models contains a gene(s) that contributes to the macrocytic anaemia.
The Hsa21-encoded RUNX1 transcription factor plays a key role in megakaryopoiesis and haematopoietic stem cell maintenance (Okuda et al., 1996), and hence has been proposed to contribute to the predisposition to AMKL when present in three copies. However, mouse studies showed that reducing Runx1 from three to two copies did not affect MPD in Ts65Dn mice, arguing against a key role for this gene (Kirsammer et al., 2008).
Solid tumours
Individuals with DS are at lower risk of developing almost all types of malignant solid tumours (Yang et al., 2002), indicating that trisomy 21 protects from tumour growth. This observation has led to the suggestion that Hsa21 encodes tumour suppressor genes. A cross of the ApcMin mouse model for colorectal cancer with either the Ts65Dn or Ts1Rhr mouse resulted in fewer tumours in the trisomic mice compared with euploid controls. Conversely, when ApcMin mice were crossed with Ms1Rhr (which have segmental monosomy for the same 33-gene region that is triplicated in the Ts1Rhr strain), a significant increase in the number of tumours was observed compared with controls, demonstrating the influence of gene dosage (Sussan et al., 2008). This effect was mapped to the Ets2 gene, which, when present in three copies, inhibits tumour formation and, conversely, when reduced to one copy, results in increased tumour rates (Sussan et al., 2008). Similar studies using the NPcis mouse model – which develops lymphomas, sarcomas and carcinomas at high frequency – also found that the Ts65Dn genotype delayed tumour development (Yang and Reeves, 2011). However, in this case, no dosage effect of Ets2 was found.
Interestingly, recent data have suggested that reduced solid tumour growth in individuals with DS might also be due to decreased angiogenesis (Baek et al., 2009; Reynolds et al., 2010). Ectopically implanted tumour lines grew more slowly in Ts65Dn mice compared with in euploid controls, indicating that tumour growth was suppressed in the trisomic mice owing to effects in the supporting stroma (Baek et al., 2009). Histological examination of the tumours showed that they had reduced microvessel density, suggesting that Ts65Dn mice have a defect in tumour angiogenesis. The Hsa21-encoded DSCR1 protein had previously been shown to inhibit angiogenesis induced by vascular endothelial growth factor (VEGF) by attenuating NFAT signalling, and was thus a candidate gene for this effect. Indeed, reduction of the dosage of Dscr1 from three to two copies resulted in increased tumour volume and tumour angiogenesis. Another recent study using the Tc1 mouse also proposed that reduced angiogenesis in DS accounts for decreased tumour growth (Reynolds et al., 2010). In this case, the reduced angiogenesis was shown to be due to increased doses of Adamts1, Erg, Jam2 and Pttg1ip. It is interesting to note that the Tc1 model does not have three copies of Dscr1, so this gene cannot account for the reduced angiogenesis in Tc1 mice. In contrast to these two reports, the cross of Ts65Dn to the NPcis mouse model of cancer showed no effect on tumour angiogenesis, showing that this is not a universal mechanism to explain the lower incidence of solid tumours in DS (Yang and Reeves, 2011). Taken together, these studies indicate that the reduced frequency of tumours in DS might be due to increased dosage of several genes, with effects both in the tumour cells and in the supporting stroma.
Craniofacial alterations
Characteristic facies is one of the few phenotypes seen in all individuals with DS, and is the result of an underlying craniofacial dysmorphology, which includes reduced skull size, flattened occiput (posterior portion of the head), brachycephaly (shortened front-to-back diameter of the skull), small midface and reduced size of the maxilla and the mandible (Richtsmeier et al., 2002). Human partial trisomies have mapped areas of Hsa21 contributing to craniofacial abnormalities, but a discrete region has not been identified (Lyle et al., 2009). Bone morphometric analysis of Ts65Dn mice revealed a pattern of craniofacial anomalies similar to those seen in individuals with DS, including a smaller flattened face, brachycephaly, a flattened occiput and a smaller mandible (Richtsmeier et al., 2000). Subsequent analysis showed that Ts1Cje mice have craniofacial alterations very similar to those seen in Ts65Dn mice (Richtsmeier et al., 2002), indicating that the trisomic region in Ts1Cje mice (Sod1-Znf295) is sufficient to produce the craniofacial phenotype. Overexpression in mice of one of the genes in this interval, Ets2, led to skeletal abnormalities that resembled those in humans with DS (Sumarsono et al., 1996). However, a reduction of the copy number of the Ets2 gene from three to two did not rescue the craniofacial phenotype of Ts65Dn mice, demonstrating that trisomy of this gene is not required for the phenotype (Hill et al., 2009).
Analysis of Ts1Rhr mice, which are trisomic for the DSCR, showed that they had rostrocaudally elongated skulls and larger mandibles than euploid littermates, a phenotype opposite from that seen in individuals with DS and in Ts65Dn mice, demonstrating that trisomy of the DSCR is not sufficient to cause the DS craniofacial phenotype (Olson et al., 2004a). When Ts65Dn mice were crossed to Ms1Rhr to reduce the DSCR from three to two copies, the brachycephaly phenotype was rescued but the overall pattern of dysmorphology was similar to that seen in Ts65Dn mice. Together, these results demonstrate a complex interplay of genetic effects of genes both within and outside the DSCR contributing both positively and negatively to the craniofacial phenotypes.
Congenital gut disorders
Congenital gut disorders have an increased incidence in DS. Patients with DS constitute ∼12% of all cases of Hirschprung’s disease, and duodenal stenosis (DST) and imperforate anus (IA) are 260 and 33 times more likely to occur in DS, respectively (Torfs and Christianson, 1998; Korbel et al., 2009). Hirschprung’s disease arises when a portion of the colon is not innervated correctly by the enteric nervous system; alterations in ∼10 non-Hsa21 genes have thus far been linked to this disease (Amiel et al., 2008). Analysis of human segmental trisomies has defined a 13 Mb critical region for Hirschprung’s disease that contains the DSCAM gene, which is expressed in the neural crest cells that give rise to the enteric nervous system (Korbel et al., 2009). No other Hsa21 genes have been implicated so far. An overlapping critical region was described for DST and IA (Korbel et al., 2009).
Concluding remarks
For most of the last century, the study of the molecular genetics of DS was an undertaking for only the very dedicated, because the resources available for gene mapping were mostly limited to the few known human partial trisomy cases. In the mid-1990s, two new mouse models, Ts65Dn and Ts1Cje, showed that dosage sensitivity could be recapitulated in the mouse, giving rise to phenotypes similar to those seen in human DS. Crucially, these two mouse strains demonstrated that it was possible to use phenotype-genotype comparisons to map causative dosage-sensitive genes, thus establishing the concept of a mouse mapping panel.
These trisomic mouse strains, in combination with gene knockouts and YAC or BAC transgenics, provided new hope for the identification of individual dosage-sensitive genes responsible for different phenotypes. However, the scale of the problem remained immense, given that researchers were attempting to assay an entire chromosome, albeit the smallest autosome, for the effects of individual causative genes. The DNA sequence of Hsa21 generated a list of genes on the chromosome and is an essential tool for the understanding of DS, but it did not give insight into dosage sensitivity (Hattori et al., 2000). Nevertheless, in the last 10 years, the field has again taken a step forward with the advent of chromosome engineering to create mapping panels of trisomic animals that span entire stretches of the human chromosome or the orthologous mouse regions. Thus, it is now realistic to use the resources of mouse genetics to identify the dosage-sensitive genes involved in DS. Results so far suggest that, although some dosage-sensitive genes might have strong effects on their own, it is likely that many of the phenotypes associated with DS are due to complex effects of multiple Hsa21 genes.
An even more challenging aim is to understand the genetic basis of the large variation in DS phenotypes, which is probably caused largely by allelic variation in genes both on Hsa21 and on other chromosomes. In the next 10 years, genome-wide association studies of DNA biobanks from people with DS will be used to identify more candidate genes, and to dissect the effects of individual alleles and combinations of alleles both on Hsa21 and on other chromosomes. These are studies that must be carried out internationally and require high levels of cooperation between different countries to collect samples from individuals with DS and to carry out standardised clinical and psychometric analysis of the participants. There is a huge challenge ahead to create these banks and agree on tests, but already the international community is mobilising and starting to tackle the problem.
This is an exciting time to be involved in dissecting the molecular genetics of DS because we now have the tools to find the individual dosage-sensitive genes and thus to understand the biological effects of trisomy. There is also synergy with human genomic studies that are highlighting the extraordinary variability in copy number that is present in all humans, and hence the importance of gene dosage to everyone, not just to people with DS.
Finally, the molecular genetic study of DS is a challenging endeavour that has given us new insights into euploid and aneuploid biology, but how is the identification of dosage-sensitive genes relevant to people with DS? The disorder is complex, but new therapies that tackle aspects of the syndrome are already being developed by applying knowledge of the dosage-sensitive genes, combined with investigations of their biology (Das and Reeves, 2011). These range from therapies currently in clinical trials to test their ability to enhance learning and memory in people with DS, to novel therapies to combat DS-associated AD. The next 10 years of DS research look very promising indeed.
Acknowledgments
This work was supported by a grant from the Wellcome Trust and by the AnEUploidy integrated project (EU Framework 6). In addition, V.L.J.T. was funded in part by the Medical Research Council (program number U117527252).
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing or financial interests.
REFERENCES
- Ahn K. J., Jeong H. K., Choi H. S., Ryoo S. R., Kim Y. J., Goo J. S., Choi S. Y., Han J. S., Ha I., Song W. J. (2006). DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 22, 463–472 [DOI] [PubMed] [Google Scholar]
- Alford K. A., Slender A., Vanes L., Li Z., Fisher E. M., Nizetic D., Orkin S. H., Roberts I., Tybulewicz V. L. (2010). Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood 115, 2928–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altafaj X., Dierssen M., Baamonde C., Marti E., Visa J., Guimera J., Oset M., Gonzalez J. R., Florez J., Fillat C., et al. (2001). Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum. Mol. Genet. 10, 1915–1923 [DOI] [PubMed] [Google Scholar]
- Alves-Sampaio A., Troca-Marin J. A., Montesinos M. L. (2010). NMDA-mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down’s syndrome. J. Neurosci. 30, 13537–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J., Sproat-Emison E., Garcia-Barcelo M., Lantieri F., Burzynski G., Borrego S., Pelet A., Arnold S., Miao X., Griseri P., et al. (2008). Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet. 45, 1–14 [DOI] [PubMed] [Google Scholar]
- Antonarakis S. E., Lyle R., Dermitzakis E. T., Reymond A., Deutsch S. (2004). Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 5, 725–738 [DOI] [PubMed] [Google Scholar]
- Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., et al. (2006). NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- Baek K. H., Zaslavsky A., Lynch R. C., Britt C., Okada Y., Siarey R. J., Lensch M. W., Park I. H., Yoon S. S., Minami T., et al. (2009). Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L. L., Moran T. H., Richtsmeier J. T., Troncoso J., Reeves R. H. (2000). Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum. Mol. Genet. 9, 195–202 [DOI] [PubMed] [Google Scholar]
- Belichenko P. V., Masliah E., Kleschevnikov A. M., Villar A. J., Epstein C. J., Salehi A., Mobley W. C. (2004). Synaptic structural abnormalities in the Ts65Dn mouse model of Down Syndrome. J. Comp. Neurol. 480, 281–298 [DOI] [PubMed] [Google Scholar]
- Belichenko P. V., Kleschevnikov A. M., Salehi A., Epstein C. J., Mobley W. C. (2007). Synaptic and cognitive abnormalities in mouse models of Down syndrome: exploring genotype-phenotype relationships. J. Comp. Neurol. 504, 329–345 [DOI] [PubMed] [Google Scholar]
- Belichenko N. P., Belichenko P. V., Kleschevnikov A. M., Salehi A., Reeves R. H., Mobley W. C. (2009). The “Down syndrome critical region” is sufficient in the mouse model to confer behavioral, neurophysiological, and synaptic phenotypes characteristic of Down syndrome. J. Neurosci. 29, 5938–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best T. K., Siarey R. J., Galdzicki Z. (2007). Ts65Dn, a mouse model of Down syndrome, exhibits increased GABAB-induced potassium current. J. Neurophysiol. 97, 892–900 [DOI] [PubMed] [Google Scholar]
- Carmichael C. L., Majewski I. J., Alexander W. S., Metcalf D., Hilton D. J., Hewitt C. A., Scott H. S. (2009). Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood 113, 1929–1937 [DOI] [PubMed] [Google Scholar]
- Cataldo A. M., Petanceska S., Peterhoff C. M., Terio N. B., Epstein C. J., Villar A., Carlson E. J., Staufenbiel M., Nixon R. A. (2003). App gene dosage modulates endosomal abnormalities of Alzheimer’s disease in a segmental trisomy 16 mouse model of down syndrome. J. Neurosci. 23, 6788–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Galdzicki Z., Haydar T. F. (2007). Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J. Neurosci. 27, 11483–11495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Best T. K., Cramer N. P., Carney R. S., Isaac J. T., Galdzicki Z., Haydar T. F. (2010). Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 13, 927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. T., Min K. T. (2009). Upregulation of three Drosophila homologs of human chromosome 21 genes alters synaptic function: implications for Down syndrome. Proc. Natl. Acad. Sci. USA 106, 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R., Scott H. S., Madani R., Huber L., Wolfer D. P., Prinz M., Aguzzi A., Lipp H. P., Antonarakis S. E. (2000). Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 9, 1853–1864 [DOI] [PubMed] [Google Scholar]
- Cooper J. D., Salehi A., Delcroix J. D., Howe C. L., Belichenko P. V., Chua-Couzens J., Kilbridge J. F., Carlson E. J., Epstein C. J., Mobley W. C. (2001). Failed retrograde transport of NGF in a mouse model of Down’s syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc. Natl. Acad. Sci. USA 98, 10439–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I., Reeves R. H. (2011). The use of mouse models to understand and improve cognitive deficits in Down syndrome. Dis. Model. Mech. 4, 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. E., Howard C. M., Farrer M. J., Coleman M. M., Cullen L. M., Williamson R., Wyse R. K., Kessling A. M. (1994). Unusual genotypes in the COL6A1 gene in parents of children with trisomy 21 and major congenital heart defects. Hum. Genet. 93, 443–446 [DOI] [PubMed] [Google Scholar]
- Davies G. E., Howard C. M., Farrer M. J., Coleman M. M., Bennett L. B., Cullen L. M., Wyse R. K., Burn J., Williamson R., Kessling A. M. (1995). Genetic variation in the COL6A1 region is associated with congenital heart defects in trisomy 21 (Down’s syndrome). Ann. Hum. Genet. 59, 253–269 [DOI] [PubMed] [Google Scholar]
- Davisson M. T., Schmidt C., Akeson E. C. (1990). Segmental trisomy of murine chromosome 16, a new model system for studying Down syndrome. Prog. Clin. Biol. Res. 360, 263–280 [PubMed] [Google Scholar]
- Delabar J. M., Theophile D., Rahmani Z., Chettouh Z., Blouin J. L., Prieur M., Noel B., Sinet P. M. (1993). Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1, 114–124 [DOI] [PubMed] [Google Scholar]
- Duchon A., Pothion S., Brault V., Sharp A. J., Tybulewicz V. L., Fisher E. M., Herault Y. (2010). The telomeric part of the human chromosome 21 from Cstb to Prmt2 is not necessary for the locomotor and short-term memory deficits observed in the Tc1 mouse model of Down syndrome. Behav. Brain Res. 217, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlevy L., Bennett M., Slender A., Lana-Elola E., Tybulewicz V. L., Fisher E. M., Mohun T. (2010). Down’s syndrome-like cardiac developmental defects in embryos of the transchromosomic Tc1 mouse. Cardiovasc. Res. 88, 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M., Ikegami S., Hosoya T., Mimura J., Ohtani H., Nakao K., Inokuchi K., Katsuki M., Fujii-Kuriyama Y. (1999). Mild impairment of learning and memory in mice overexpressing the mSim2 gene located on chromosome 16, an animal model of Down’s syndrome. Hum. Mol. Genet. 8, 1409–1415 [DOI] [PubMed] [Google Scholar]
- Fernandez F., Morishita W., Zuniga E., Nguyen J., Blank M., Malenka R. C., Garner C. C. (2007). Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat. Neurosci. 10, 411–413 [DOI] [PubMed] [Google Scholar]
- Freeman S. B., Taft L. F., Dooley K. J., Allran K., Sherman S. L., Hassold T. J., Khoury M. J., Saker D. M. (1998). Population-based study of congenital heart defects in Down syndrome. Am. J. Med. Genet. 80, 213–217 [PubMed] [Google Scholar]
- Galante M., Jani H., Vanes L., Daniel H., Fisher E. M., Tybulewicz V. L., Bliss T. V., Morice E. (2009). Impairments in motor coordination without major changes in cerebellar plasticity in the Tc1 mouse model of Down syndrome. Hum. Mol. Genet. 18, 1449–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K., Fortna A., Bechtel L., Davisson M. T. (2003). Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene 318, 137–147 [DOI] [PubMed] [Google Scholar]
- Geraldo S., Khanzada U. K., Parsons M., Chilton J. K., Gordon-Weeks P. R. (2008). Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell. Biol. 10, 1181–1189 [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot A. C., Bartram U., Oosthoek P. W., Bartelings M. M., Hogers B., Poelmann R. E., Jongewaard I. N., Klewer S. E. (2003). Collagen type VI expression during cardiac development and in human fetuses with trisomy 21. Anat. Rec. A. Discov. Mol. Cell Evol. Biol. 275, 1109–1116 [DOI] [PubMed] [Google Scholar]
- Goate A., Chartier-Harlin M. C., Mullan M., Brown J., Crawford F., Fidani L., Giuffra L., Haynes A., Irving N., James L., et al. (1991). Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349, 704–706 [DOI] [PubMed] [Google Scholar]
- Hammerle B., Elizalde C., Tejedor F. J. (2008). The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur. J. Neurosci. 27, 1061–1074 [DOI] [PubMed] [Google Scholar]
- Hammerle B., Ulin E., Guimera J., Becker W., Guillemot F., Tejedor F. J. (2011). Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development 138, 2543–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima C., Jacobowitz D. M., Witta J., Borke R. C., Best T. K., Siarey R. J., Galdzicki Z. (2006). Abnormal expression of the G-protein-activated inwardly rectifying potassium channel 2 (GIRK2) in hippocampus, frontal cortex, and substantia nigra of Ts65Dn mouse: a model of Down syndrome. J. Comp. Neurol. 494, 815–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle H., Clemmensen I. H., Mikkelsen M. (2000). Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet 355, 165–169 [DOI] [PubMed] [Google Scholar]
- Hattori M., Fujiyama A., Taylor T. D., Watanabe H., Yada T., Park H. S., Toyoda A., Ishii K., Totoki Y., Choi D. K., et al. (2000). The DNA sequence of human chromosome 21. Nature 405, 311–319 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Shirao T. (1999). Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J. Neurosci. 19, 3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ishikawa R., Ye L. H., He X. L., Takata K., Kohama K., Shirao T. (1996). Modulatory role of drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J. Neurosci. 16, 7161–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. A., Sussan T. E., Reeves R. H., Richtsmeier J. T. (2009). Complex contributions of Ets2 to craniofacial and thymus phenotypes of trisomic “Down syndrome” mice. Am. J. Med. Genet. A 149A, 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzler J. K., Zipursky A. (2005). Origins of leukaemia in children with Down syndrome. Nat. Rev. Cancer 5, 11–20 [DOI] [PubMed] [Google Scholar]
- Holtzman D. M., Santucci D., Kilbridge J., Chua-Couzens J., Fontana D. J., Daniels S. E., Johnson R. M., Chen K., Sun Y., Carlson E., et al. (1996). Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl. Acad. Sci. USA 93, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsammer G., Jilani S., Liu H., Davis E., Gurbuxani S., Le Beau M. M., Crispino J. D. (2008). Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood 111, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleschevnikov A. M., Belichenko P. V., Villar A. J., Epstein C. J., Malenka R. C., Mobley W. C. (2004). Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J. Neurosci. 24, 8153–8160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O., Tirosh-Wagner T., Urban A. E., Chen X. N., Kasowski M., Dai L., Grubert F., Erdman C., Gao M. C., Lange K., et al. (2009). The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA 106, 12031–12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Kawashima H., Pulst S. M., Ikeuchi T., Ogasawara N., Yamamoto K., Schonberg S. A., West R., Allen L., Magenis E., et al. (1990). Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am. J. Hum. Genet. 47, 236–246 [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Chen X. N., Schipper R., Sun Z., Gonsky R., Gerwehr S., Carpenter N., Daumer C., Dignan P., Disteche C., et al. (1994). Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl. Acad. Sci. USA 91, 4997–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehericy S., Hirsch E. C., Cervera-Pierot P., Hersh L. B., Bakchine S., Piette F., Duyckaerts C., Hauw J. J., Javoy-Agid F., Agid Y. (1993). Heterogeneity and selectivity of the degeneration of cholinergic neurons in the basal forebrain of patients with Alzheimer’s disease. J. Comp. Neurol. 330, 15–31 [DOI] [PubMed] [Google Scholar]
- Li Z., Yu T., Morishima M., Pao A., LaDuca J., Conroy J., Nowak N., Matsui S., Shiraishi I., Yu Y. E. (2007). Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum. Mol. Genet. 16, 1359–1366 [DOI] [PubMed] [Google Scholar]
- Liu C., Morishima M., Yu T., Matsui S. I., Zhang L., Fu D., Pao A., Costa A. C., Gardiner K. J., Cowell J. K., et al. (2011). Genetic analysis of Down syndrome-associated heart defects in mice. Hum. Genet. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Liang Z., Wegiel J., Hwang Y. W., Iqbal K., Grundke-Iqbal I., Ramakrishna N., Gong C. X. (2008). Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J. 22, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke A. E., Dooley K. J., Tinker S. W., Cheong S. Y., Feingold E., Allen E. G., Freeman S. B., Torfs C. P., Cua C. L., Epstein M. P., et al. (2010). Variation in folate pathway genes contributes to risk of congenital heart defects among individuals with Down syndrome. Genet. Epidemiol. 34, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott I. T., Dierssen M. (2010). Cognitive deficits and associated neurological complications in individuals with Down’s syndrome. Lancet Neurol. 9, 623–633 [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D., Rowitch D. H. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 [DOI] [PubMed] [Google Scholar]
- Lyle R., Bena F., Gagos S., Gehrig C., Lopez G., Schinzel A., Lespinasse J., Bottani A., Dahoun S., Taine L., et al. (2009). Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 17, 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslen C. L. (2004). Molecular genetics of atrioventricular septal defects. Curr. Opin. Cardiol. 19, 205–210 [DOI] [PubMed] [Google Scholar]
- McCormick M. K., Schinzel A., Petersen M. B., Stetten G., Driscoll D. J., Cantu E. S., Tranebjaerg L., Mikkelsen M., Watkins P. C., Antonarakis S. E. (1989). Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics 5, 325–331 [DOI] [PubMed] [Google Scholar]
- McNaughton D., Knight W., Guerreiro R., Ryan N., Lowe J., Poulter M., Nicholl D. J., Hardy J., Revesz T., Rossor M., et al. (2010). Duplication of amyloid precursor protein (APP), but not prion protein (PRNP) gene is a significant cause of early onset dementia in a large UK series. Neurobiol. Aging, [Epub ahead of print] 10.1016/j.neurobiolaging.2010.1010.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich R. X., Dauphinot L., Laffaire J., Rossier J., Potier M. C. (2007). Down syndrome gene dosage imbalance on cerebellum development. Prog. Neurobiol. 82, 87–94 [DOI] [PubMed] [Google Scholar]
- Morice E., Andreae L. C., Cooke S. F., Vanes L., Fisher E. M., Tybulewicz V. L., Bliss T. V. (2008). Preservation of long-term memory and synaptic plasticity despite short-term impairments in the Tc1 mouse model of Down syndrome. Learn. Mem. 15, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. F., Vaughan S. E., Vaccaro P. (1982). Measurements of neuromuscular tone and strength in Down’s syndrome children. J. Ment. Defic. Res. 26, 41–46 [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A., Jr, Crews S. T. (1991). The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67, 1157–1167 [DOI] [PubMed] [Google Scholar]
- Netzer W. J., Powell C., Nong Y., Blundell J., Wong L., Duff K., Flajolet M., Greengard P. (2010). Lowering beta-amyloid levels rescues learning and memory in a Down syndrome mouse model. PLoS One 5, e10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty A., Ruf S., Mulligan C., Hildreth V., Errington M. L., Cooke S., Sesay A., Modino S., Vanes L., Hernandez D., et al. (2005). An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309, 2033–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. (1996). AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 [DOI] [PubMed] [Google Scholar]
- Olson L. E., Richtsmeier J. T., Leszl J., Reeves R. H. (2004a). A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science 306, 687–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson L. E., Roper R. J., Baxter L. L., Carlson E. J., Epstein C. J., Reeves R. H. (2004b). Down syndrome mouse models Ts65Dn, Ts1Cje, and Ms1Cje/Ts65Dn exhibit variable severity of cerebellar phenotypes. Dev. Dyn. 230, 581–589 [DOI] [PubMed] [Google Scholar]
- Olson L. E., Roper R. J., Sengstaken C. L., Peterson E. A., Aquino V., Galdzicki Z., Siarey R., Pletnikov M., Moran T. H., Reeves R. H. (2007). Trisomy for the Down syndrome ‘critical region’ is necessary but not sufficient for brain phenotypes of trisomic mice. Hum. Mol. Genet. 16, 774–782 [DOI] [PubMed] [Google Scholar]
- Ooe N., Saito K., Mikami N., Nakatuka I., Kaneko H. (2004). Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 24, 608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Abalia J., Sahun I., Altafaj X., Andreu N., Estivill X., Dierssen M., Fillat C. (2008). Targeting Dyrk1A with AAVshRNA attenuates motor alterations in TgDyrk1A, a mouse model of Down syndrome. Am. J. Hum. Genet. 83, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington B. F., Moon J., Edgin J., Stedron J., Nadel L. (2003). The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child Dev. 74, 75–93 [DOI] [PubMed] [Google Scholar]
- Pereira P. L., Magnol L., Sahún I., Brault V., Duchon A., Prandini P., Gruart A., Bizot J.-C., Chadefaux-Vekemans B., Deutsch S., et al. (2009). A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum. Mol. Genet. 18, 4756–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi M., Lopes C., Charron G., Delezoide A. L., Paly E., Bloch B., Delabar J. M. (2005). Spatial and temporal localization during embryonic and fetal human development of the transcription factor SIM2 in brain regions altered in Down syndrome. Int. J. Dev. Neurosci. 23, 475–484 [DOI] [PubMed] [Google Scholar]
- Rahmani Z., Blouin J. L., Creau-Goldberg N., Watkins P. C., Mattei J. F., Poissonnier M., Prieur M., Chettouh Z., Nicole A., Aurias A., et al. (1989). Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc. Natl. Acad. Sci. USA 86, 5958–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainis L., Toki T., Pimanda J. E., Rosenthal E., Machol K., Strehl S., Gottgens B., Ito E., Izraeli S. (2005). The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 65, 7596–7602 [DOI] [PubMed] [Google Scholar]
- Reynolds L. E., Watson A. R., Baker M., Jones T. A., D’Amico G., Robinson S. D., Joffre C., Garrido-Urbani S., Rodriguez-Manzaneque J. C., Martino-Echarri E., et al. (2010). Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature 465, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtsmeier J. T., Baxter L. L., Reeves R. H. (2000). Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev. Dyn. 217, 137–145 [DOI] [PubMed] [Google Scholar]
- Richtsmeier J. T., Zumwalt A., Carlson E. J., Epstein C. J., Reeves R. H. (2002). Craniofacial phenotypes in segmentally trisomic mouse models for Down syndrome. Am. J. Med. Genet. 107, 317–324 [DOI] [PubMed] [Google Scholar]
- Roper R. J., Baxter L. L., Saran N. G., Klinedinst D. K., Beachy P. A., Reeves R. H. (2006). Defective cerebellar response to mitogenic Hedgehog signaling in Down [corrected] syndrome mice. Proc. Natl. Acad. Sci. USA 103, 1452–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. (2006). APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 38, 24–26 [DOI] [PubMed] [Google Scholar]
- Ryoo S. R., Cho H. J., Lee H. W., Jeong H. K., Radnaabazar C., Kim Y. S., Kim M. J., Son M. Y., Seo H., Chung S. H., et al. (2008). Dual-specificity tyrosine(Y)-phosphorylation regulated kinase 1A-mediated phosphorylation of amyloid precursor protein: evidence for a functional link between Down syndrome and Alzheimer’s disease. J. Neurochem. 104, 1333–1344 [DOI] [PubMed] [Google Scholar]
- Sago H., Carlson E. J., Smith D. J., Kilbridge J., Rubin E. M., Mobley W. C., Epstein C. J., Huang T. T. (1998). Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl. Acad. Sci. USA 95, 6256–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi A., Delcroix J. D., Belichenko P. V., Zhan K., Wu C., Valletta J. S., Takimoto-Kimura R., Kleschevnikov A. M., Sambamurti K., Chung P. P., et al. (2006). Increased App expression in a mouse model of Down’s syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron 51, 29–42 [DOI] [PubMed] [Google Scholar]
- Shim K. S., Lubec G. (2002). Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer’s disease and Down syndrome. Neurosci. Lett. 324, 209–212 [DOI] [PubMed] [Google Scholar]
- Shumway-Cook A., Woollacott M. H. (1985). Dynamics of postural control in the child with Down syndrome. Phys. Ther. 65, 1315–1322 [DOI] [PubMed] [Google Scholar]
- Siarey R. J., Kline-Burgess A., Cho M., Balbo A., Best T. K., Harashima C., Klann E., Galdzicki Z. (2006). Altered signaling pathways underlying abnormal hippocampal synaptic plasticity in the Ts65Dn mouse model of Down syndrome. J. Neurochem. 98, 1266–1277 [DOI] [PubMed] [Google Scholar]
- Sinet P. M., Theophile D., Rahmani Z., Chettouh Z., Blouin J. L., Prieur M., Noel B., Delabar J. M. (1994). Mapping of the Down syndrome phenotype on chromosome 21 at the molecular level. Biomed. Pharmacother. 48, 247–252 [DOI] [PubMed] [Google Scholar]
- Smith D. J., Stevens M. E., Sudanagunta S. P., Bronson R. T., Makhinson M., Watabe A. M., O’Dell T. J., Fung J., Weier H. U., Cheng J. F., et al. (1997). Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 16, 28–36 [DOI] [PubMed] [Google Scholar]
- Spano M., Mercuri E., Rando T., Panto T., Gagliano A., Henderson S., Guzzetta F. (1999). Motor and perceptual-motor competence in children with Down syndrome: variation in performance with age. Eur. J. Paediatr. Neurol. 3, 7–13 [DOI] [PubMed] [Google Scholar]
- Stankiewicz M. J., Crispino J. D. (2009). ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood 113, 3337–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumarsono S. H., Wilson T. J., Tymms M. J., Venter D. J., Corrick C. M., Kola R., Lahoud M. H., Papas T. S., Seth A., Kola I. (1996). Down’s syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature 379, 534–537 [DOI] [PubMed] [Google Scholar]
- Sussan T. E., Yang A., Li F., Ostrowski M. C., Reeves R. H. (2008). Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature 451, 73–75 [DOI] [PubMed] [Google Scholar]
- Takebayashi H., Yoshida S., Sugimori M., Kosako H., Kominami R., Nakafuku M., Nabeshima Y. (2000). Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech. Dev. 99, 143–148 [DOI] [PubMed] [Google Scholar]
- Torfs C. P., Christianson R. E. (1998). Anomalies in Down syndrome individuals in a large population-based registry. Am. J. Med. Genet. 77, 431–438 [PubMed] [Google Scholar]
- Trazzi S., Mitrugno V. M., Valli E., Fuchs C., Rizzi S., Guidi S., Perini G., Bartesaghi R., Ciani E. (2011). APP-dependent up regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum. Mol. Genet. 20, 1560–1573 [DOI] [PubMed] [Google Scholar]
- Vis J. C., Duffels M. G., Winter M. M., Weijerman M. E., Cobben J. M., Huisman S. A., Mulder B. J. (2009). Down syndrome: a cardiovascular perspective. J. Intellect. Disabil. Res. 53, 419–425 [DOI] [PubMed] [Google Scholar]
- Voronov S. V., Frere S. G., Giovedi S., Pollina E. A., Borel C., Zhang H., Schmidt C., Akeson E. C., Wenk M. R., Cimasoni L., et al. (2008). Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc. Natl. Acad. Sci. USA 105, 9415–9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J., Greene M., McDevitt M. A., Anastasi J., Karp J. E., Le Beau M. M., Crispino J. D. (2002). Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32, 148–152 [DOI] [PubMed] [Google Scholar]
- Williams A. D., Mjaatvedt C. H., Moore C. S. (2008). Characterization of the cardiac phenotype in neonatal Ts65Dn mice. Dev. Dyn. 237, 426–435 [DOI] [PubMed] [Google Scholar]
- Yabut O., Domogauer J., D’Arcangelo G. (2010). Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J. Neurosci. 30, 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Reeves R. H. (2011). Increased survival following tumorigenesis in Ts65Dn mice that model Down Syndrome. Cancer Res. 71, 3573–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rasmussen S. A., Friedman J. M. (2002). Mortality associated with Down’s syndrome in the USA from 1983 to 1997, a population-based study. Lancet 359, 1019–1025 [DOI] [PubMed] [Google Scholar]
- Yu T., Clapcote S. J., Liu C., Li S., Pao A., Bechard A. R., Belichenko P., Kleschevnikov A., Asrar S., Chen R., et al. (2010a). Effects of individual segmental trisomies of human chromosome 21 syntenic regions on hippocampal long-term potentiation and cognitive behaviors in mice. Brain Res. 1366, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Li Z., Jia Z., Clapcote S. J., Liu C., Li S., Asrar S., Pao A., Chen R., Fan N., et al. (2010b). A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet. 19, 2780–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Anderson D. J. (2002). The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109, 61–73 [DOI] [PubMed] [Google Scholar]