Abstract
An increasing number of cyclopeptides have been discovered as products of ribosomal synthetic pathway. The biosynthetic study of these cyclopeptides has revealed interesting new mechanisms for cyclization. This review highlighted the recent discoveries in cyclization mechanisms for cyclopeptides synthesized independently of non-ribosomal peptide synthetases, including endopeptidase-catalyzed cyclization, intein-mediated cyclization, and peptide synthetase-catalyzed cyclization. This information may help to design hybrid ribosomal and non-ribosomal biosynthetic systems to produce novel cyclopeptides with various bioactivities.
Keywords: ribosomal peptide, cyclopeptides, non-ribosomal peptide synthetase, cyclization mechanism
Introduction
Cyclopeptides or cyclic peptides represent a family of fantastic compounds with complex structures and various bioactivities, such as antibiotic gramicidin S [1], antitumor RA-VII [2], uterotonic polypeptide kalata B1 [3], antibacterial albonoursin [4,5], and cytotoxic patellamide D [6]. The head-to-tail cyclized backbone protects cyclopeptides against proteolytic cleavage [7]. Most cyclopeptides are products of non-ribosomal peptide synthetases (NRPSs) [8]. These peptides are cyclized by thioesterase (TE) domains located in C-terminal of NRPSs [9]. The cyclization occurs by nucleophilic attacking of nucleophile (e.g. amine group, hydroxyl group, or thiol group) against carbonyl group of ester bond linking to the TE domain. More and more cyclopeptides have been confirmed to be ribosomal products or at least independent of NRPSs, including some cyclopeptides that were previously considered as non-ribosomal peptides (NRPs) [10–12], such as microviridin [12].
Till date, pathways both of NRPS and ribosomal have been discussed [11,13] including the cyclization mechanisms in the NRPS pathway [14]. Some genetic information of these ribosomally synthesized cyclopeptides has been discussed, such as gene clusters of cyanobactins [15] and cDNA sequences of cyclotides [16]. However, no review has specifically discussed the cyclization mechanisms of peptides synthesized independently of NRPSs, although several intriguing mechanisms have been reported for the cyclization of these cyclopeptides, e.g. endopeptidase-catalyzed cyclization of cyclotides [17] and cyanobactins [18], artificial cyclization with a permuted intein in cyanobacterium [19] and peptide synthetase-catalyzed cyclization of albonoursin [20]. These mechanisms of enzymatic cyclization in the ribosomal biosynthetic pathway have extended the knowledge of enzymatic reactions and brought the ribosomal and non-ribosomal pathways together. To modify the ribosomal system is easier than modifying the non-ribosomal system, because the NRPSs are too large to be processed by gene operation. This knowledge will help to develop new technologies in combinatorial biosynthesis and bioengineering to produce novel bioactive compounds.
Endopeptidase-catalyzed Cyclization
Endopeptidases present a family of enzymes, which catalyze the hydrolysis of the peptide bond (or breaking the peptide bond in other words). Now some evidences show that the enzymes of this family also catalyze the transpeptidation by forming a peptide bond, which includes cyclization. Cyclotides and cyanobactins are good examples of endopeptidase-catalyzed cyclization.
Cyclotides belong to plant cyclopeptides type VIII [21], which is a family of mini disulfide-rich peptides derived by plants with ∼30 amino acids and contains a unique protein motif cyclic-cystine-knot. This motif, which includes three disulfide bonds, together with cyclic backbone makes cyclotides exceptionally stable. The first cyclotide kalata B1 was discovered in the African traditional herb Oldenlandia affinis that had been used as uterotonic medicine [22], which showed that cyclotides could be used in clinic safely. Till 2009, various cyclotides with activities of hemolysis, anti-HIV, antimicrobe, cytotoxin, and insecticide have been reported [23]. A study confirmed that the linear analogues lacked bioactivity even if the N-terminals were blocked by the acetyl group [24]. So the cyclic backbone is very important to cyclotides’ bioactivities.
Cyclotides were confirmed as gene-coded products [25] and spliced from larger propeptides [26]. The cyclization of backbone occurs after the forming of a cyclic cystine knot in vivo [27], although chemical synthetic research shows that the cyclic backbone is preferred for the generation of cyclic cystine knot in vitro [28,29]. The cyclization of the backbone is catalyzed by asparaginyl endopeptidase (AEP) [30,31]. There are six conserved residues (XXNGLP), which are recognized by AEPs. The reaction is initiated by the electron transferring from histidine to cysteine. Then the thio group will attack the carbonyl group of the asparagine, break the peptide bond, and link the N-terminal of propeptide to the enzyme. The amino group of glycine accepts the proton from histidine and the C-terminal germin-like protein (GLP) tripeptide leaves. Then the N-terminal propeptide folds and the first three residues, which are conserved GLPs, fit into recognizing site S1’, S2’, and S3’. The amino group of glycine residue initiates a nucleophilic attack to form the peptide bond and completes the cyclization. This research gives a novel mechanism of peptidase, in which a pair of reversing activities occurs in a single catalytic site. Peptidases were considered to catalyze the reaction of either breaking or forming a peptide bond [32]. Although the activities both of hydrolysis and transpeptidation share a similar catalytic center (catalytic triad Asp-Ser-His), the nucleophilic attacking groups are different in these two reactions, which are H2O in hydrolysis but NH2 in transpeptidation. If we may define the cyclization of cyclotides as intramolecular transpeptidation, this research is the first example to catalyze both of two reactions in order.
Cyanobactins are small bioactive cyclic peptides produced by cyanobacteria [13]. These cyclic compounds show a wide range of bioactivities such as anti-tumor, anti-HIV, and anti-viral [33]. Both cyclotides and cyanobactins are ribosomal backbone cyclic or N–C cyclic, while the other ribosomal cyclic peptides are side-chain cyclized. Distinguished from cyclotides, cyanobactins are cyclized by subtilisin-like protease which belongs to non-specific serine endopeptidase [18] instead of AEPs [25]. The precursor proteins encoded one or more cyanobactins, which were flanking by the putative recognition sequences [13,20,34]. According to this demonstration, the two proteases encoded by cyanobactin gene cluster were proposed to carry out different functions. One was confirmed to cleave the recognition sequence in the N-terminal, and the other to cleave the C-terminal and catalyze the cyclization [13,20].
In 2009, the cyclization mechanism of cyanobactin with PatG was reported by Lee et al. [20]. An artificial substrate of linear peptide QGGRGDWP-AYDGE could be split and cyclized into cyclo-(QGGRGDWP) by PatG in vitro. PatG is the first cyclase involved in the ribosomal pathway. This enzyme shares the same catalytic triad (Asp-Ser-His) with serine protease. PatG cut AYDGE off from the precursor and loaded QGGRGDWP into the hydroxyl group of Ser in the catalytic triad. The amine group of Gln attacked the carboxyl group as nucleophile to form the cyclic product. No cofactors, extra metals, or energy were required in the reaction. This mechanism is like TE cyclization in NRPS, except that the reaction breaks a peptide bond instead of TE bond.
The substrate selectivity of PatG was fairly flexible. Over 30 natural peptide sequences and over 60 natural products could be cyclized by PatG in vivo or/and in vitro, although the turnover number was as low as 1 per enzyme molecule per day. The authors also excluded the possibility of spontaneous cyclization by feeding the peptide of QGGRGDWP as the substrate [20]. Later in 2010 [18], McIntosh et al. introduced non-proteinogenic amino acids and polyketide-like linkers in the artificial substrates. Results showed that PatG generated products in lengths from 6 to 11 residues, including non-proteinogenic amino acids and polyketide-like linkers. Similar to TE domains, unnatural substrates triggered the hydrolysis of PatG. This result makes it possible to hybrid the ribosomal pathway and non-ribosomal pathway and produces more novel interesting bioactive compounds.
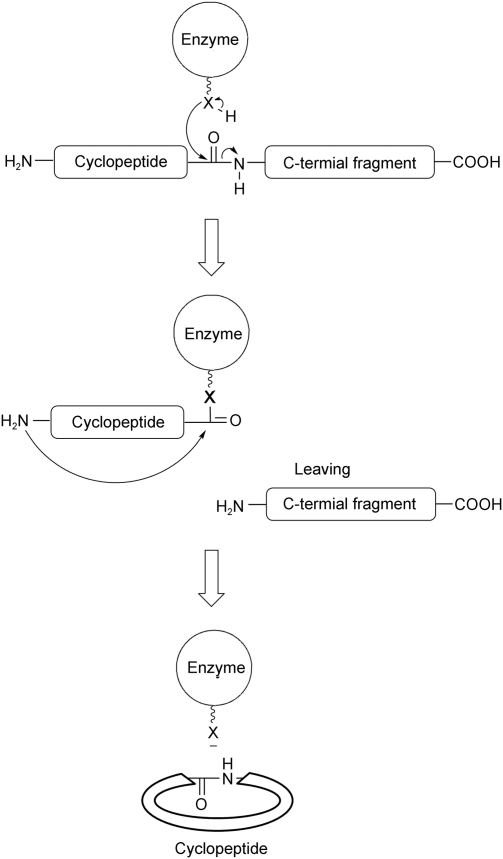
Generally, in the endopeptidase-catalyzed cyclization, the precursors of cyclopeptides are loaded to the catalyst site companying the release of C-terminal fragments. An N-to-X acyl shift transesterification cyclizes the backbones and yields the products (Fig. 1).
Figure 1.
Mechanism for the cyclization of cyclotides and cyanobactins Propeptide was loaded to the enzyme accompanying the release of C-terminal fragments. The amino group attacked ester or thioester bond leading to the products. X stands for S or O. The illusion was modified from references [17,19,20,30].
Intein-mediated Cyclization
Inteins are internal sequences of proteins that are self-excluded from larger precursor proteins while ligating the remaining parts collectively termed as exteins [35]. There are three types of intein-splicing, which are canonical splicing, intermolecular trans-splicing, and cyclization using permuted inteins. Canonical splicing gives a linear intein with a succinimide residue at the C-terminal. The reaction is initiated by moving the carboxyl group of the last residue of N-terminal extein to the nucleophile group of the first intein residue, which usually are the OH group of serine or threonine and the SH group of cysteine. Then the carboxyl group moves to another nucleophile group of the first residue in C-terminal extein, which are usually also the OH group of serine or threonine and SH group of cysteine, by trans-(thio-)esterification. Succinimide formation of asparagine that is the last residue of the intein wipes the intein off the precursor. Finally, the S-to-N acyl shift generates the peptide and ligates the two exteins together [36]. Intermolecular trans-splicing share a similar mechanism with canonical splicing but takes place between two polypeptides.
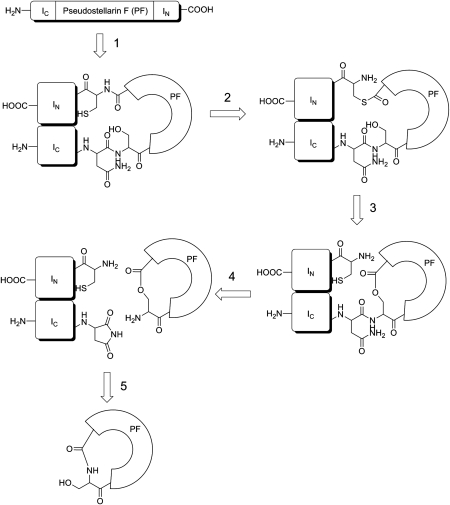
Wu et al. [37] reported that a cyanobacterial intein could splice without an endonuclease part, which was highly conserved in the canonical splicing mechanism. This is the third type. Soon after, Scott et al. [19] reported a novel artificial in vivo system to produce cyclic peptides. With this system, pseudostellarin F, which belonged to Caryophyllaceae type of plant cyclopeptides [21], was expressed at the level of 30 μg/g wet cell mass. It indicated that Caryophyllaceae type of cyclopeptides might be synthesized by a similar biological pathway. A synthesized DNA fragment encoding pseudostellarin F was inserted into Ssp DnaE split intein fragments IC and IN to construct a plasmid, which could be expressed in Escherichia coli. The transformed E. coli produced pseudostellarin F and showed the inhibition to melanin production. The circular ligation mechanism was proposed as follows: the expressed precursor folded to form an active protein ligase (Fig. 2, reaction 1); an N-to-S acyl shift was catalyzed to give a TE intermediate (Fig. 2, reaction 2) that underwent transesterification to form a lariat intermediate (Fig. 2, reaction 3); succinimide formation released the cyclic product as a lactone (Fig. 2, reaction 4) and the final cyclopeptides were generated by an X-to-N acyl shift (Fig. 2, reaction 5).
Figure 2.
Mechanism for intein-mediated cyclization of pseudostellarin F The expressed precursor folded to form an active protein ligase (reaction 1); an N-to-S acyl shift was catalyzed to give a thioester intermediate (reaction 2) that underwent transesterification to form a lariat intermediate (reaction 3); Succinimide formation released the cyclic product as a lactone (reaction 4); the final cyclopeptides was generated by an X-to-N acyl shift (reaction 5). The illusion was modified from references [19,40].
Scott et al. [38] reported that the IC+1 position that was immediately adjacent to the C-intein could only be occupied by serine, threonine, and cysteine. These residues provided the possibility of nucleophilic transesterification S-to-X shift reaction to generate the branched and lariat intermediates. The IN−1 position that was immediately adjacent to the N-intein showed much less conservation than the IC+1 position; and the cyclic products could be generated while the IN−1 position was occupied by polar (Ser), non-polar (Ala), large (Leu), small (Gly), charged (Lys), aromatic (Tyr), or β-branched (Ile) residues. Other positions showed less effect to the activity of splicing.
With this splicing model, cyclic peptides ranging from four to nine amino acids were generated [38,39]. Interestingly, the precursors were secreted by E. coli prior to cyclization [39]. This subsequent secretion of the cyclopeptides may open a door to screen the libraries of cyclopeptides. The process was reported as a protocol and published in 2007 [40]. Chin et al. [41] reported a method to construct herbicide resistance to Nicotiana tabacum by introducing splicing intein into chloroplast. Although these cyclopeptides were produced in the heterologous hosts ‘artificially’, the process carried out by such organisms as E. coli [39], tobacco [41], and human cells [42] showed that it might occur in microbes, even in planta and mammalian cells.
Peptide Synthetase-catalyzed Cyclization
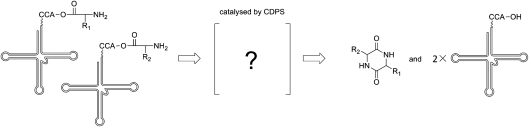
Cyclodipeptides were reported as the products of peptide synthetase by ligating two amino acids together and then cyclizing, which were discovered in diverse bacterial [43], fungi [44], animals [43], and plants [19]. These compounds were believed to be formed from protein hydrolysates as by-products of protein degradation or produced by NRPSs [43]. Some were reported as the products of spontaneous cyclization in the NRPS pathway [43,45]. The isolation of albonoursin biosynthetic gene cluster [43] elucidated a novel non-ribosomal mechanism of cyclodipeptide formation, which was totally independent of NRPSs. A peptide synthetase AlbC was present in this gene cluster, and it was believed to catalyze the formation of cyclodipeptide cyclo-(L-Phe-L-Leu). In further research [46], AlbC was confirmed as a synthetase of cyclodipeptide using tRNA charged amino acids as substrates (Fig. 3). The purified AlbC, which was expressed in E. coli and secreted into a medium, showed the activities of ligation and cyclization to a range of amino acids in vitro. The same activity was also detected in the enzymatic assay with other related proteins. The authors named these proteins as cyclodipeptide synthases (CDPSs). CDPSs were distinguished from other enzymes using amino acyl-tRNA as a substrate to generate peptide bonds by additional cyclization activity. CDPSs use amino acyl-tRNA to generate two peptide bonds and form a cyclodipeptide, while the other enzymes only generate one peptide bond between the amino acyl group of amino acyl-tRNA donor and the N-terminal amine group of peptide or protein acceptor.
Figure 3.
Cyclization of cyclodipeptides via cyclodipeptide synthase (CDPS) Cyclodipeptide is synthesized by CDPS using two amino acyl tRNA as substrates companying with releasing two unloaded tRNAs. The detail of catalytic mechanism is still unknown, which was marked as a question marker in the illustration.
There is a gap between the in vitro activity and in vivo function, which is the specificity of substrate recognition. AlbC showed a tolerant specificity to different amino acyl-tRNA in vitro while only cyclo-(L-Phe-L-Leu) could be generated in vivo. von Dohren [47] proposed that the recognition site might locate on some special tRNA moieties. Although the mechanism of CDPSs remains to be exploited, this new class of enzymes opens a door to the utilization of tRNA charged amino acids to generate a wide range of cyclodipeptides as substrates for more modification which may find novel bioactive compounds.
Final Remark
An increasing number of natural peptides have been confirmed to be synthesized independently of NRPS especially by the ribosomal pathway [11,48]. The understanding of their biosynthetic mechanisms is important for the utility of these compounds, especially the small-molecule peptides that show the outstanding potential as anti-infectious and antitumor agents. The non-ribosomal pathway usually gives complex products while the ribosomal pathway is easier to be modified by genetic operations. The cyclases discovered independent of NRPSs might combine the advantages of both ribosomal and non-ribosomal pathways and provide more novel complex drugs or drug-leads with a simpler molecular biological technique.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (30725048, U1032602, and 91013002), the Chinese Academy of Sciences (KSCX2-YW-R-177), the State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Faculty Seed Grant from Nebraska Research Council, and National Institute of Health of USA (RR015468-01). W.X. was supported in part by China Scholarship Council.
References
- 1.Gause GF, Brazhnikova MG. Gramicidin S and its use in the treatment of infect wounds. Nature. 1944;154:703. [Google Scholar]
- 2.Itokawa H, Takeya K, Mori N, Kidokoro S, Yamamoto H. Studies on antitumor cyclic hexapeptides RA obtained from tubiae radix, Rubiaceae (IV): quantitative determination of RA-VII and RA-V in commercial rubiae radix and collected plants. Planta Med. 1984;50:313–316. doi: 10.1055/s-2007-969718. [DOI] [PubMed] [Google Scholar]
- 3.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 4.Khokhoov AS, Lokshin GB, Vul'fson NS, Zaretskii VI. Structure of albonoursin and its natural analog. Russ Chem Bull. 1966;15:1146–1152. [Google Scholar]
- 5.Fukushima K, Yazawa K, Arai T. Biological activities of albonoursin. J Antibiot. 1973;26:175–176. doi: 10.7164/antibiotics.26.175. [DOI] [PubMed] [Google Scholar]
- 6.Williams AB, Jacobs RS. Marine natural product, patellamide D, reverses multidrug resistance in a human leukemic cell line. Cancer Lett. 1993;71:97–102. doi: 10.1016/0304-3835(93)90103-g. [DOI] [PubMed] [Google Scholar]
- 7.Trabi M, Craik DJ. Circular proteins: no end in sight. Trends Biochem Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 8.Mootz H, Marahiel M. Biosynthetic systems for non-ribosomal peptide antibiotic assembly. Curr Opin Chem Biol. 1997;1:543–551. doi: 10.1016/s1367-5931(97)80051-8. [DOI] [PubMed] [Google Scholar]
- 9.Kohli RM, Trauger JW, Schwarzer D, Marahiel MA, Walsh CT. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry-US. 2001;40:7099–7108. doi: 10.1021/bi010036j. [DOI] [PubMed] [Google Scholar]
- 10.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Post-translational modification in microviridin biosynthesis. ChemBioChem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 11.McIntosh JA, Mohamed S, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishitsuka MO, Kusumi T, Kakisawa H, Kaya K, Watanabe MM. Microviridin, a novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis. J Am Chem Soc. 1990;112:8180–8182. [Google Scholar]
- 13.Nolan EM, Walsh CT. How nature morphs peptide scaffolds into antibiotics. ChemBioChem. 2009;10:34–53. doi: 10.1002/cbic.200800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 15.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang CKL, Kaas Q, Chiche L, Craik DJ. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucl Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saska I, Craik DJ. Protease-catalysed protein splicing: a new post-translational modification? Trends Biochem Sci. 2008;33:363–368. doi: 10.1016/j.tibs.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 18.McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular logic: nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J Am Chem Soc. 2010;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Nat Acad Sci USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131:2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan NH, Zhou J. Plant cyclopeptides. Chem Rev. 2006;106:840–895. doi: 10.1021/cr040699h. [DOI] [PubMed] [Google Scholar]
- 22.Gran L. An oxytocic principle found in Oldenlandia affinis DC. An indigenous, Congolese drug “Kalata-Kalata” used to accelerate delivery . Meddelelser Fra Norsk Farmaceutisk Selskap. 1970;32:173–180. [Google Scholar]
- 23.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Adv Drug Deliv Rev. 2009;61:918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Simonsen SM, Daly NL, Craik DJ. Capped acyclic permutants of the circular protein kalata B1. FEBS Lett. 2004;577:399–402. doi: 10.1016/j.febslet.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Jennings C, West J, Waine C, Craik DJ, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc Nat Acad Sci USA. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulvenna JP, Sando L, Craik DJ. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure. 2005;13:691–701. doi: 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Gruber CW, Cemazar M, Clark RJ, Horibe T, Renda RF, Anderson MA, Craik DJ. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J Biol Chem. 2007;282:20435–20446. doi: 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- 28.Daly NL, Craik DJ. Acyclic permutants of naturally occurring cyclic proteins. J Biol Chem. 2000;275:19068–19075. doi: 10.1074/jbc.M000450200. [DOI] [PubMed] [Google Scholar]
- 29.Aboye TL, Clark RJ, Burman R, Roig MB, Craik DJ, Goransson U. Interlocking disulfides in circluar proteins: toward efficient oxidative folding of cyclotides. Antioxid Redox Signal. 2011;14:77–86. doi: 10.1089/ars.2010.3112. [DOI] [PubMed] [Google Scholar]
- 30.Conlan BF, Gillon AD, Craik DJ, Anderson MA. Circular proteins and mechanisms of cyclization. Biopolymers. 2010;94:573–583. doi: 10.1002/bip.21422. [DOI] [PubMed] [Google Scholar]
- 31.Mylne JS, Colgrave ML, Daly NL, Chanson AH, Elliott AG, McCallum EJ, Jones A, et al. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011;7:257–259. doi: 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]
- 32.Jakubke H, Kuhl P, Konnecke A. Basic principles of protease-catalyzed peptide bond formation. Angew Chem-Int Edit. 1985;24:85–93. [Google Scholar]
- 33.Burja AM, Banaigs B, Abou-Masnsour E, Burgess JG, Wright PC. Marine cyanobacteria: a profile source of natural products. Tetrahedron. 2001;57:9347–9377. [Google Scholar]
- 34.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanbacterial symbiont of Lissoclinum patella. Proc Nat Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perler FB, Davis EO, Dean GE, Gimble FS, Jack WE, Neff N, Noren CJ, et al. Protein splicing elements: inteins and exteins—a definition of terms and recommended nomenclature. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Xu M, Liu M. Protein trans-splicing and funtional mini-inteins of a cyanobacterial dnab intein. Biochim Biophys Acta. 1998;1387:422–432. doi: 10.1016/s0167-4838(98)00157-5. [DOI] [PubMed] [Google Scholar]
- 38.Scott CP, Abel-Santos E, Jones AD, Benkovic SJ. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem Biol. 2001;8:801–815. doi: 10.1016/s1074-5521(01)00052-7. [DOI] [PubMed] [Google Scholar]
- 39.Deschuyteneer G, Garcia S, Michiels B, Baudoux B, Degand H, Morsomme P, Soumillion P. Intein-mediated cyclization of randomized peptides in the periplasm of Escherichia coli and their extracellular secretion. ACS Chem Biol. 2010;5:691–700. doi: 10.1021/cb100072u. [DOI] [PubMed] [Google Scholar]
- 40.Tavassoli A, Benkovic SJ. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat Protoc. 2007;2:1126–1133. doi: 10.1038/nprot.2007.152. [DOI] [PubMed] [Google Scholar]
- 41.Chin HG, Kim G, Marin I, Mersha F, Evans TC, Chen L, Xu MQ, et al. Protein trans-splicing in trangenic plant chloroplast: reconstruction of herbicide resistance from split genes. Proc Nat Acad Sci USA. 2003;100:4510–4515. doi: 10.1073/pnas.0736538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinsella TM, Ohashi CT, Harder AG, Yam GC, Li W, Peelle B, Pali ES, et al. Retrovirally delivered random cyclic peptide libraries yield inhibitors of interleukin-4 signaling in human B cells. J Biol Chem. 2002;277:37512–37518. doi: 10.1074/jbc.M206162200. [DOI] [PubMed] [Google Scholar]
- 43.Lautru S, Gondry M, Genet R, Pernodet J-L. The albonoursin gene cluster of S. noursei: biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem Biol. 2002;9:1355–1364. doi: 10.1016/s1074-5521(02)00285-5. [DOI] [PubMed] [Google Scholar]
- 44.Walzel B, Riederer B, Keller U. Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea. Chem Biol. 1997;4:223–230. doi: 10.1016/s1074-5521(97)90292-1. [DOI] [PubMed] [Google Scholar]
- 45.Schwarzer D, Mootz H, Marahiel MA. Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetase. Chem Biol. 2001;8:997–1010. doi: 10.1016/s1074-5521(01)00068-0. [DOI] [PubMed] [Google Scholar]
- 46.Gondry M, Sauguest L, Belin P, Thai R, Amouroux R, Tellier C, Tuphile K, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 47.von Dohren H. Charged tRNAs charge into secondary metabolism. Nat Chem Biol. 2009;5:374–375. doi: 10.1038/nchembio0609-374. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt EW. The hidden diversity of ribosomal peptide natural products. BMC Biol. 2010;8:83–86. doi: 10.1186/1741-7007-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]