Abstract
We describe a novel computerized system for comparison of RNA secondary structures and demonstrate its use for experimental studies. The system is able to screen a very large number of structures, to cluster similar structures and to detect specific structural motifs. In particular, the system is useful for detecting mutations with specific structural effects among all possible point mutations, and for predicting compensatory mutations that will restore the wild type structure. The algorithms are independent of the folding rules that are used to generate the secondary structures.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baughman G., Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: analysis of the mRNA target site using oligonucleotide-directed mutagenesis. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5389–5393. doi: 10.1073/pnas.81.17.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell G., Schulz M. F., Selzer G., Chollet A., Movva N. R., Semon D., Escanez S., Kawashima E. Optimizing the expression in E. coli of a synthetic gene encoding somatomedin-C (IGF-I). Nucleic Acids Res. 1985 Mar 25;13(6):1923–1938. doi: 10.1093/nar/13.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S., Springer M., Dondon J., Grunberg-Manago M. Posttranscriptional autoregulation of Escherichia coli threonyl tRNA synthetase expression in vivo. J Bacteriol. 1986 Jan;165(1):198–203. doi: 10.1128/jb.165.1.198-203.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza M. C., Kofoid E. C., Marlière P., Louis B. G. Potential secondary structure at translation-initiation sites. Nucleic Acids Res. 1987 Jan 12;15(1):345–360. doi: 10.1093/nar/15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
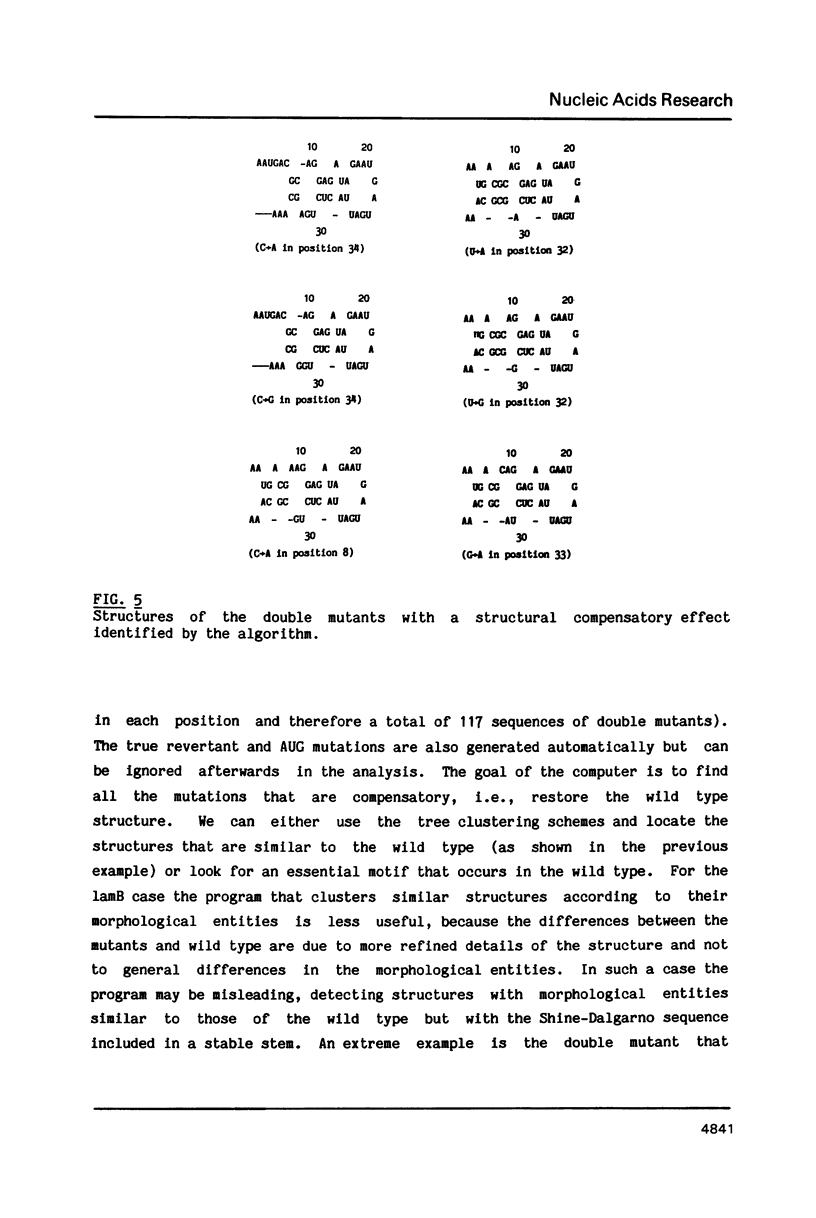
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney K. R., Nomura M. Secondary structure of the autoregulatory mRNA binding site of ribosomal protein L1. Mol Gen Genet. 1987 Nov;210(1):60–68. doi: 10.1007/BF00337759. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Nussinov R., Maizel J. V., Jr An improved secondary structure computation method and its application to intervening sequence in the human alpha-like globin mRNA precursors. Comput Appl Biosci. 1988 Aug;4(3):337–344. doi: 10.1093/bioinformatics/4.3.337. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Currey K. M., Nussinov R., Maizel J. V., Jr Studies of frequently recurring substructures in human alpha-like globin mRNA precursors. Comput Biomed Res. 1987 Dec;20(6):563–582. doi: 10.1016/0010-4809(87)90026-7. [DOI] [PubMed] [Google Scholar]
- Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J. P., Ehresmann C., Ehresmann B. Messenger RNA structure and gene regulation at the translational level in Escherichia coli: the case of threonine:tRNAThr ligase. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7892–7896. doi: 10.1073/pnas.85.21.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Nomura M., Yates J. L., Dean D., Post L. E. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
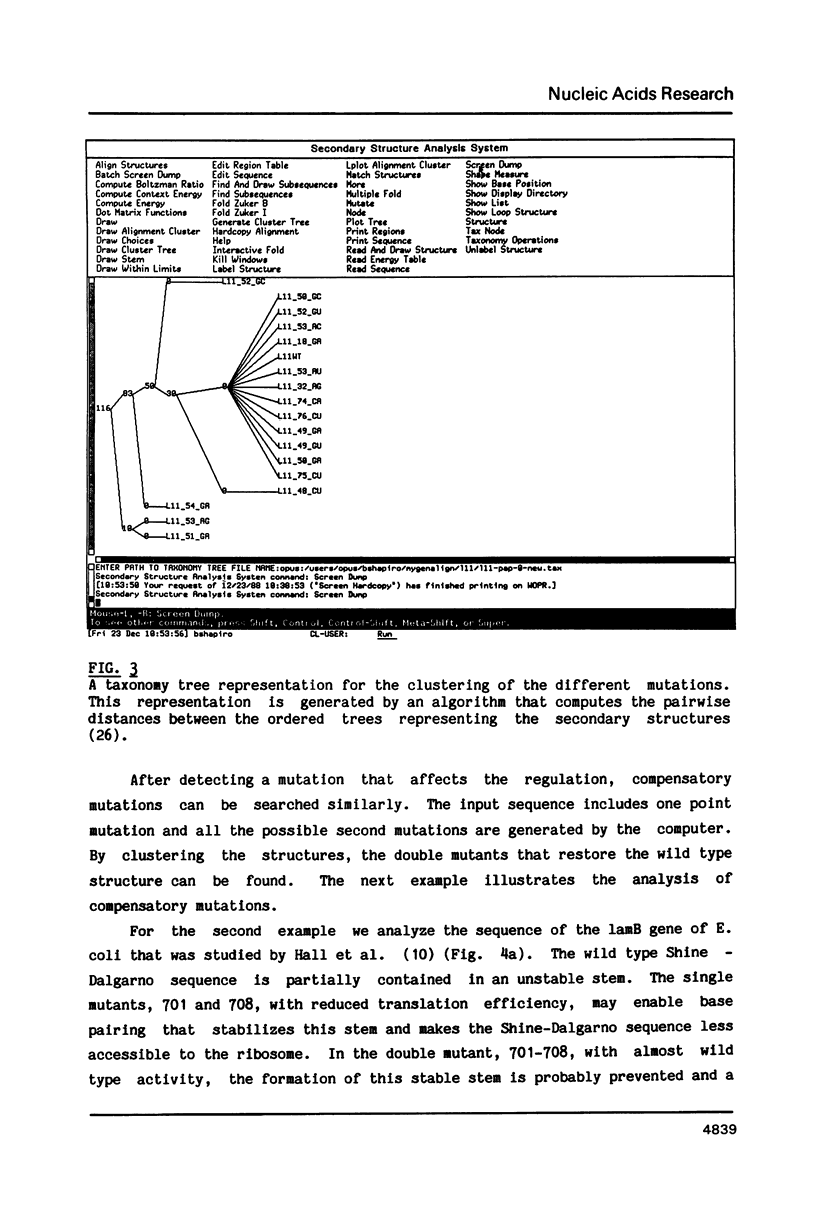
- Shapiro B. A. An algorithm for comparing multiple RNA secondary structures. Comput Appl Biosci. 1988 Aug;4(3):387–393. doi: 10.1093/bioinformatics/4.3.387. [DOI] [PubMed] [Google Scholar]
- Sobel E., Martinez H. M. A multiple sequence alignment program. Nucleic Acids Res. 1986 Jan 10;14(1):363–374. doi: 10.1093/nar/14.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Graffe M., Butler J. S., Grunberg-Manago M. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4384–4388. doi: 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Plumbridge J. A., Butler J. S., Graffe M., Dondon J., Mayaux J. F., Fayat G., Lestienne P., Blanquet S., Grunberg-Manago M. Autogenous control of Escherichia coli threonyl-tRNA synthetase expression in vivo. J Mol Biol. 1985 Sep 5;185(1):93–104. doi: 10.1016/0022-2836(85)90185-8. [DOI] [PubMed] [Google Scholar]
- Tessier L. H., Sondermeyer P., Faure T., Dreyer D., Benavente A., Villeval D., Courtney M., Lecocq J. P. The influence of mRNA primary and secondary structure on human IFN-gamma gene expression in E. coli. Nucleic Acids Res. 1984 Oct 25;12(20):7663–7675. doi: 10.1093/nar/12.20.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
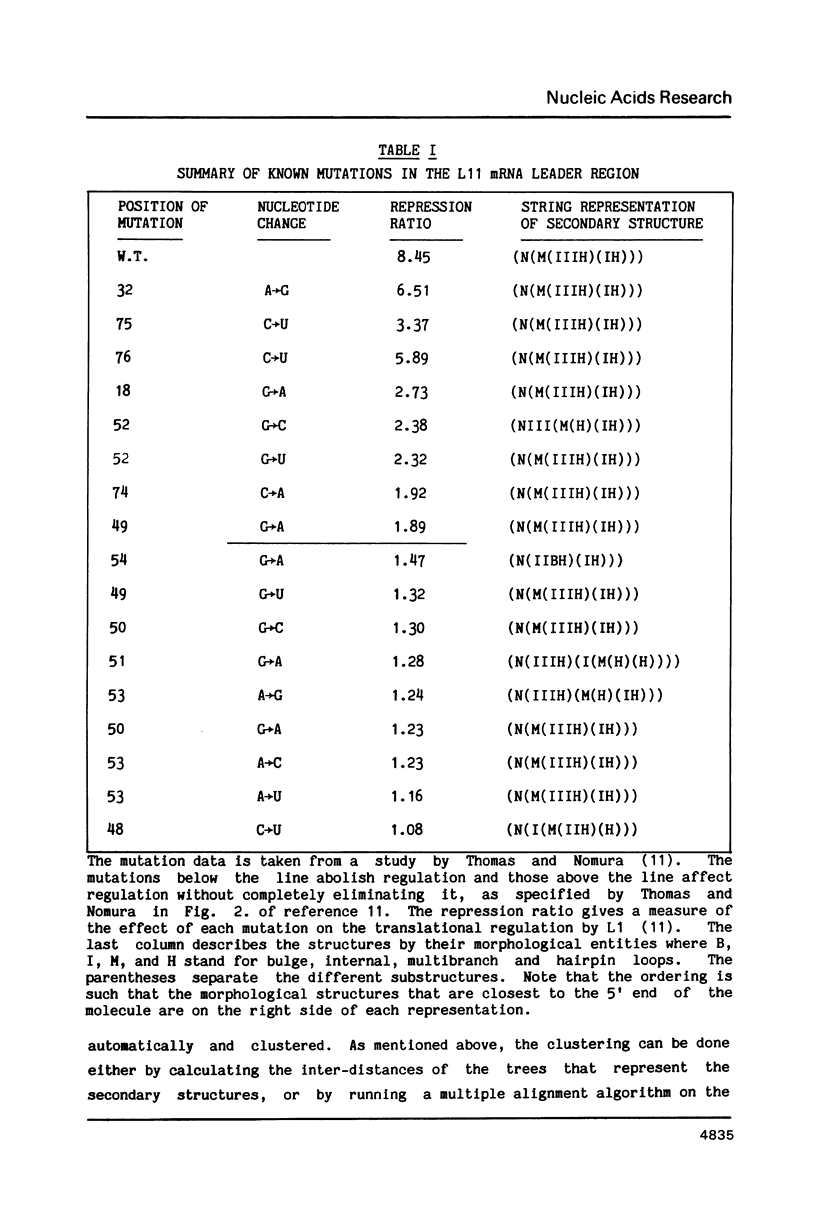
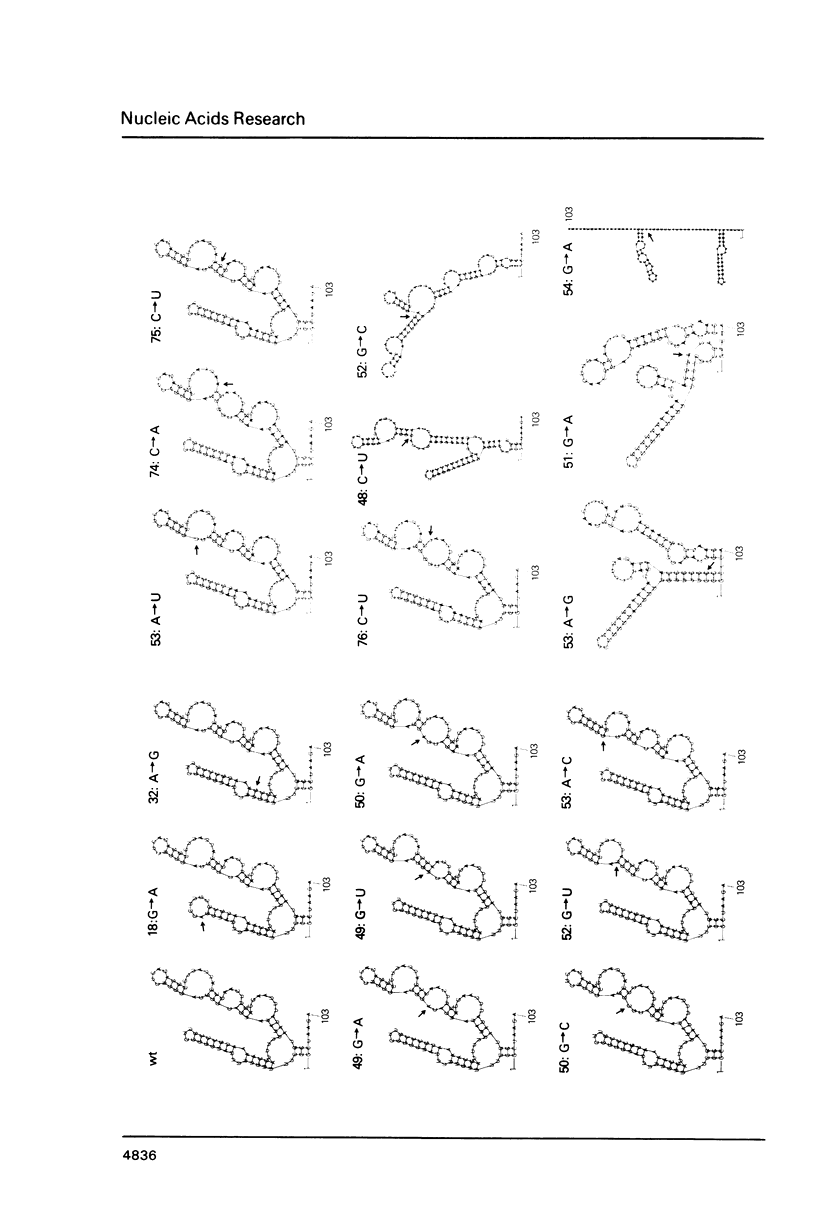
- Thomas M. S., Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987 Apr 10;15(7):3085–3096. doi: 10.1093/nar/15.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Wood C. R., Boss M. A., Patel T. P., Emtage J. S. The influence of messenger RNA secondary structure on expression of an immunoglobulin heavy chain in Escherichia coli. Nucleic Acids Res. 1984 May 11;12(9):3937–3950. doi: 10.1093/nar/12.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Uhlenbeck O. C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987 Dec 15;26(25):8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Zagórska L., Chroboczek J., Klita S., Szafrański P. Effect of secondary structure of messenger ribonucleic acid on the formation of initiation complexes with prokaryotic and eukaryotic ribosomes. Eur J Biochem. 1982 Feb;122(2):265–269. doi: 10.1111/j.1432-1033.1982.tb05876.x. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


