Abstract
Picture Archiving and Communication Systems (PACS) have been widely deployed in healthcare institutions, and they now constitute a normal commodity for practitioners. However, its installation, maintenance, and utilization are still a burden due to their heavy structures, typically supported by centralized computational solutions. In this paper, we present Dicoogle, a PACS archive supported by a document-based indexing system and by peer-to-peer (P2P) protocols. Replacing the traditional database storage (RDBMS) by a documental organization permits gathering and indexing data from file-based repositories, which allows searching the archive through free text queries. As a direct result of this strategy, more information can be extracted from medical imaging repositories, which clearly increases flexibility when compared with current query and retrieval DICOM services. The inclusion of P2P features allows PACS internetworking without the need for a central management framework. Moreover, Dicoogle is easy to install, manage, and use, and it maintains full interoperability with standard DICOM services.
Keywords: PACS, Digital Imaging and Communications in Medicine (DICOM), Medical imaging, Peer-to-peer, Computer communication networks, Open source, PACS implementation, Information storage and retrieval
Background
Over the last decade, the use of digital medical imaging systems in healthcare institutions has significantly increased, and they constitute valuable tools supporting the medical profession both in decision support and in treatment procedures. Research and industry efforts to develop medical imaging equipment, which evolved gradually to a panoply of imaging resources, have been the major driving forces towards wide acceptance of the Picture Archiving and Communication System (PACS) concept.
PACS gives healthcare practitioners the ability to remotely access multimedia patient information and to set up telemedicine, telework, and collaborative work environments [1]. Currently, there are PACS solutions with different architectures and services, from simple models, typically used in small laboratories, to enterprise-wide platforms, mostly used in large hospital networks. The PACS concept encompasses several technologies that include hardware and software for acquisition, distribution, storage, and analysis of digital images in distributed environments [2].
The Digital Imaging and Communications in Medicine (DICOM) standard architecture was a major contribution to the exchange of structured medical imaging data [3]. Currently, almost all medical imaging equipment manufacturers provide embedded DICOM (version 3) digital output in their products. As a result, large volumes of DICOM data have been produced in the last few years, creating enormous sets of clinical data that in most cases have been stored in local archives without remote indexing and retrieval facilities.
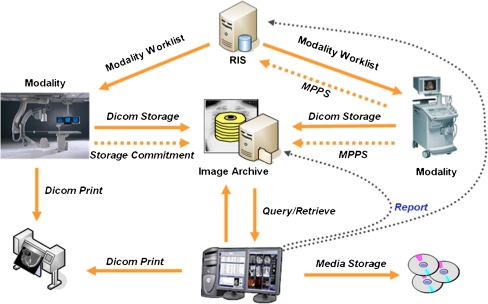
Typically, the core element of PACS is a central server that stores images and the database that contains complementary information about patients and studies (Fig. 1). This system implements the DICOM Storage Service that allows any imaging equipment to directly send the acquired images to the centralized PACS archive. Access to the stored images is then supported by the Query and Retrieve Service [4, 5] or Web Access to DICOM Persistent Objects (WADO) [5, 6]. However, this central server PACS solution has some constraints. The server has to handle the entire DICOM services flow, for instance, receive the uploaded DICOM studies and store them locally, process requested queries, and send the data back to the client (DICOM service class user). To overcome this issue, an average-size medical institution would require a great amount of storage capacity, computational power, and bandwidth, concentrated in a single machine.
Fig. 1.
PACS DICOM services and processes workflow
PACS Data Management
Despite the growing adoption of PACS in healthcare institutions, there are still many limitations hindering widespread and seamless use of these systems. PACS were originally conceived to store the huge amount of images that are generated in a hospital, and the searching mechanisms were rather underestimated. They were specifically chosen to allow a medical specialist to retrieve images based, for instance, on patient name or on patient ID.
A typical DICOM persistent object (i.e., a file) may contain numerous data elements, including the pixel image data, free text, structured reports, and several descriptive attributes such as image modality, equipment reference, acquisition parameters, image resolution, measurements, or clinical trial study data [5]. However, because a typical PACS archive database rarely stores more than the minimal number of fields to support the DICOM Information Model Query/Retrieve (DIM Q/R) model, a great portion of this information is not searchable through this service.
However, since PACS searching is accomplished mostly through the Query/Retrieve services, information fields other than those defining the DIM (DICOM Information Model) are not easily searchable. For instance, with the more recent digital radiology or computed tomography equipment, radiation dose information is already included in persistent DICOM objects [7, 8]. However, conducting a radiation dose study in traditional PACS solutions is far from being a simple task.
Nevertheless, DICOM metadata already includes exposure index information and, as such, it should be possible, from its analysis, to detect dosage anomalies and to simplify the follow-up of dose adjustments [7–9].
Moreover, current solutions are typically based on three-tier architectures that were mainly conceived to meet the requirements of a single medical center. They are based on centralized repositories supported by RDBMS (Relational Database Management System) technology, and access to data is normally performed through the presentation layer that hides logic and data technology. The trend of the last decade points to somehow associate document-based repositories to traditional image-based information systems. These repositories offer free indexing and searching which are features far from being attained in the PACS environments. Considering the success of indexing and search engines such as Google, an attempt to bring these powerful functions to PACS environments is clearly an opportunity.
Inter-networking
Efforts to deploy PACS-oriented solutions through the Internet have created unforeseen scenarios for the use of this technology. Medical imaging modalities and archive solutions are successively becoming more powerful and less expensive. The result is the proliferation of equipment acquisition by imaging centers, even small ones. As a major consequence, any healthcare institution, even one with limited human or financial resources, is currently able to generate and collect its own medical imaging data [10]. However, the asymmetric distribution of PACS equipment and radiology specialists across geographical areas leads typically to the need to hire third-party service professionals outside the institutions where exams are made [11]. Moreover, there is certainly a vast repository of expert-reviewed medical images that remain locally in hospitals and clinics without being shared. This problem is even more acute in academia where medical researchers gather, over a long time, valuable collections of clinical events. The study of specific diseases such as cancer or cardiovascular diseases would benefit if the knowledge kept enclosed to some extent in the same center could be shared.
Methods
Dicoogle - Indexing Medical Data
We developed a new PACS archive (Dicoogle) that complements and may replace the traditional centralized database with a more agile indexing and retrieving mechanism [12]. With this solution, any document type can be indexed, besides storage and searching of traditional DICOM information model fields. In this way, it is possible to add all other DICOM textual data elements without the need to create new fields, new tables, and new relations that would be necessary in the database approach. Dicoogle can automatically extract, index, and store all metadata detected in modality DICOM header, including private DICOM attribute tags, without re-engineering or re-configuration needs.
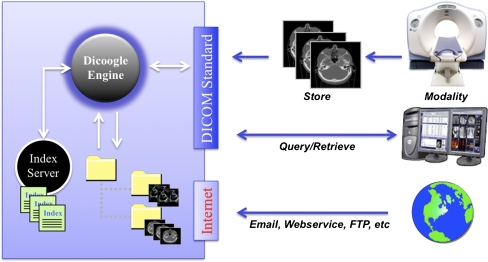
Besides the information exchanged through supported DICOM services (Fig. 2), Dicoogle can also index medical data hosted on the local file system and accessed through common transport protocols such as ftp or http (Fig. 2). With this feature, it is possible to index information that is outside the typical DICOM workflow. The solution includes a file system monitor where all events (file creation, change, deletion, etc.) are intercepted, and according to the kind of data, specific indexing actions will be triggered.
Fig. 2.
Dicoogle components and interfaces
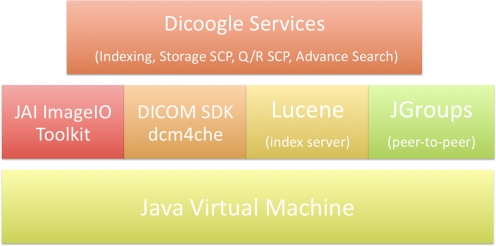
The Dicoogle platform is an open source project developed in Java and planned to run in any common operating system. Figure 3 presents an overview of all Dicoogle components. The indexing system is based on Apache Lucene [13, 14], a public domain document-indexing engine that requires reduced installation and maintenance effort.
Fig. 3.
Dicoogle software components
The implementation of DICOM standard functionalities is supported by the DCM4CHE library [15, 16], a SDK that is used to extract DICOM data elements from persistent objects and to implement the Storage SCP and Query/Retrieve SCP services [4]. The decoded DICOM information is parsed and indexed by a Lucene server according to programmed rules. The Dicoogle user has several possibilities available to configure the indexed elements: (1) DIM fields, (2) DICOM object attributes, and (3) mandatory fields from specific IOD (Information Object Definition) image modality [17]. Finally, Dicoogle uses the JAI ImageIO toolkit [18] to display the medical image thumbnails, improving the usability of the system.
Dicoogle PACS standalone version has proven to be a solution that deals well with complex searching requirements for single desktop environments [12]. However, in distributed scenarios, several weaknesses were clearly identified and further investigation was needed to develop a peer-to-peer application model that best suits the specificity of medical imaging archives (volume data, latency time, etc.).
Building Indexes
To create indexes, Dicoogle works in two phases. The first time the application is executed, it is necessary to scan all files in the computer. During this process, all detected DICOM files are indexed by Lucene [13, 14], much as happens with the files received by standard Storage SCP service. Once the complete scan is finished, the application turns into a “steady state” mode (file system watch) where it is only necessary to detect incremental file changes (create, update, and delete). Dicoogle only needs to monitor directory and file changes using the operating system callback functions that notify the user of directory changes. Using JNI (Java Native Interface) to wrap these native function calls, every time a file is changed an event is generated, and Dicoogle updates the metadata index.
On the other hand, Dicoogle can also work as a traditional PACS solution. In this case, we just need to configure an external DICOM viewer to visualize the selected images. The simplicity of this solution fits well with the requirements of a small imaging center. In an enterprise PACS, Dicoogle can also be used as an image storage archive without affecting existent third services, for instance, the modality worklist, visualization software, or web portal interface.
Searching over Indexes
Dicoogle allows the indexing of DIM Fields, the set of all DICOM attributes, and additional metadata that can be used in association with each file. These parameters can be customized to answer specific user requirements. In general, users do not need complete knowledge of the structure of the DICOM images to retrieve the wanted information. However, skilled users can fine-tune search queries to improve the quality of retrieved results.
The Dicoogle DICOM Query/Retrieve service uses the Lucene indexer to extract the desired information. It applies a specific boolean expression using the indexed DIM fields. For each C-FIND Request [4] (i.e., the DICOM query command), it builds a query using specific keywords such as PatientName, StudyDate, Modality, or other DIM fields. For instance, “PatientName: FELIX* AND StudyDate:[20090101 TO 20090131] AND Modality:CT”.
The query result will include the location of DICOM persistent objects and all other DICOM tags that were indexed. The set of search results are used to build the answer DIM model. For instance, if C-FIND Request level is Study, it creates a list of studies filtered by Study Instance UID with a structure containing the study and patient mandatory fields for C-FIND Response.
The DICOM retrieve service is based on C-MOVE command [4]. Dicoogle uses the Study Instance UID keyword to interrogate the indexer and to identify the DICOM files location that must be sent to the remote host.
The standard DICOM C-FIND query mechanism only uses DIM fields as search criteria to extract information from Dicoogle repository. However, the extended Dicoogle query mechanism can also use all indexed tags, allowing more detailed and powerful searches. Moreover, it is possible to do a free text search over all indexed document. Those enhanced query facilities are transparently available over distributed Dicoogle repositories with the developed P2P network layer.
Extending Dicoogle to Peer-to-Peer Scenarios
Some authors have already proposed several solutions for searching and sharing medical data in collaborative P2P networks, mostly based on proprietary developed applications ([19–21]) or on grid architectures ([22–24]). Those studies show that research efforts in P2P networks to support medical imaging services are residual and focused on localized issues [20, 21]. Blanquer and colleagues [21], for instance, use a hybrid P2P platform to share digital medical information. However, this system is not able to cooperate with standard DICOM peers, which reduces its range of application and makes it a less flexible system.
The paradigm introduced with the Dicoogle index engine also potentiates queries over a set of distributed DICOM repositories, which are logically indexed as a single federated unit. To implement this and other DICOM services in a peers’ federation, it was necessary to have a robust, high-performance and scalable network protocol. Based upon Dicoogle index engine results, we developed a networking platform that allows the medical and academic community to access, share, and discover clinical medical images, through a Peer-to-Peer (P2P) network. These networks offer transparent peer connectivity. Using P2P protocols, peers can cooperate to form self-organized and self-configured peer groups irrespective of their positions in the network [25], and without the need for a centralized management infrastructure. However, current P2P protocol usage is still very much oriented to supporting file-sharing related services [26, 27]. Taking into account the connectivity potential of these networks, the development of P2P technology to support DICOM standard services is a scientific issue that is likely to have a significant impact.
Dicoogle P2P Networking
Several P2P platforms were evaluated, namely JGroups [28], JXTA [29], and XMPP [30], to select the one that best fulfills the requirements of DICOM services, in particular:
Discovering peers: Peers find each other through a common registering mechanism, typically managed by a central server that keeps a list of all peers and resources. However, other methods are available, such as network broadcasting or discovery algorithms [25].
Querying peers for resources (i.e., DICOM services): Once these peers have been discovered, the application can ask them for the content desired by the application.
Sharing resources: In the same way that a peer can ask other peers for specific content, it can also publish and share content after it has been discovered.
The first version of Dicoogle P2P platform was based on Sun’s Microsystems JXTA [29] framework version 2.5. However, successive developments and trials showed that this solution was not stable enough for building a robust P2P network. The final decision was to adopt JGroups [28], an open source toolkit for group communication that assures the messages exchanged between peers arrive at their destination and in the right order.
JGroups was the base platform used to build a decentralized topology, i.e., no server is required to establish the network. The first peer joining the group will be automatically nominated as the leader. Besides the common peer functions, the leader is responsible for accepting the joining peers and distributing a group peer list ordered by the joining time. Therefore, the first entry is the group leader and if the leader crashes or leaves the group, the system will automatically elect the second entry as the new group leader. To connect to Dicoogle, each new peer announces its intention by sending a multicast message. After sending the intention to join the group, one of three things may occur:
The group already exists: in this case, the group leader will reply to the newcomer, accepting or refusing entry to the group. After accepting the new peer, the group leader will disseminate the current view of the system, with the new peer at the end of the list.
The group does not exist: The new peer waits a predefined length of time (10 s by default) for the group leader response. If the peer does not receive any response during this period, it assumes there are no other members in the group and takes on the leader role.
Concurrent leadership candidates: Simultaneous attempts to join an inexistent group may occur. Because there is no group leader yet, this could lead to a situation where the two peers will assume the group’s leadership. To prevent this situation, the peers hear other joining messages during the response period. If a concurrent joining message is detected and no feedback is obtained from a group leader, then we have a situation of concurrent leadership candidates. The leader indecision is solved by electing the peer with the lowest UID (unique identity).
JGroups enabled the development of the Dicoogle peer-to-peer layer, including DIM query/retrieve and images transfer. Dicoogle provides also a network discovery mechanism in order to find all the available resources in the entire group.
Search and Retrieve
Dicoogle searching mechanism has two modes: local search and LAN search. Each mode represents a search domain, and according to its needs the peer user may define which search mode he prefers. Also, the peer administrator may specify which search mode will be executed when the peer is contacted by a third non-Dicoogle station-using the DICOM query service. Each search mode works in the following way:
Local search: This is a straightforward situation where the query is executed upon the local Lucene index.
LAN search: The query is sent to the peers in the same private network. When the other Dicoogle peers receive the search query, they execute it locally over their indexes and then send the result to the calling peer. Finally, the collected results are merged into a single one that is presented to the user or application.
In order to provide the search and the file transfer features, four messages were defined: Query Request, Query Response, File Request, and File Response. The Query Request and the Query Response (Fig. 4) are used for the lookup of resources and the File Request and the File Response are used to retrieve DICOM files.
Fig. 4.
P2P Query request and response message
Query Response is a message that contains the hits of a search. It can contain several fields including the search criteria (<Query>) and a variable number of extra fields <Extrafield> (Fig. 4). When a peer receives a Query Request message, it interrogates its local index engine and sends the results in a Query Response message. The File Request is a message that shall be sent when a peer wants a file of another peer, making use of file hash <FileHash> of query response (Fig. 4). The previous three messages are text-based with a XML structure, while the forth, the File Response message, sends content as binary data. There are several XML limitations to transfer binary files, such as memory size and computation costs, that are associated with the conversion of binary data into base64 format (which is treated as a string). This encoding schema (base64) increases the original object size in 33% and also increases the processing time relative to binary–text–binary conversion.
Results
Dicoogle is a new approach regarding medical image file sharing when compared to the previous Dicoogle desktop version [12]. This new version supports both P2P and client/server usage, and provides a lightweight communication platform based on P2P technology. This PACS is appropriate for small or medium-sized institutions and it can be used in regular clinical workflow, research, or teaching. On other hand, Dicoogle has a search mechanism that can be used in existent repositories to assist in knowledge extraction, a fundamental tool for clinical studies. It can also be installed in parallel with actual institutions PACS, indexing all existent imagiologic information.
With this approach, distinct medical imaging repositories can be viewed as federated PACS. Interoperability with other DICOM-based systems is provided by standard Storage and Query/Retrieve SCP services. Studies can be searched and retrieved from any of the network peers, according to pre-defined access control policies.
Dicoogle conveys several key features regarding new ways of looking into meta-imaging information for retrospective assessments. These may be of relevant usefulness in statistical-oriented management and reporting tasks or wide-scope clinical studies requiring, for example, dose metrics that are now increasingly available in DICOM persistent objects created by the recent models of digital image equipment.
As long as the storage of DICOM files prevails in file-system-oriented archives, our system is able to provide a way to ensure a vendor-independent DIM structure of the whole archive regardless of the particular implementation of the PACS database. This adds up to any particular redundancy policy eventually deployed by the PACS vendor and easily copes with technological migrations whenever and for whatever the motives that may drive the need for deep changes.
Modules and Interface
Dicoogle was developed in Java as an open source platform that can run in distinct operation systems. The engine can be installed as an operating system service since the presentation layer (i.e., the graphical interface) is decoupled from business logic layer (engine).
All Dicoogle functionalities, operational configurations, and security policies can be configured by the user, including:
DICOM services ports and allowed host connections;
Index server file location and basic index operations;
Configuration of index fields: DIM only, all attributes from specific image modality (XA, US, CT, etc.), and all detected textual attributes;
P2P configurations.
As described, Dicoogle indexing scheme allows searching through traditional DICOM information fields and in any other indexed attribute. For every medical imaging modality, the administrator can define the desired indexed mode: DIM fields, selected fields, and full index. In the user interface, the list of all indexed fields of a specific image is available to help the user formulate each query (Fig. 5).
Fig. 5.
Display list of indexed fields
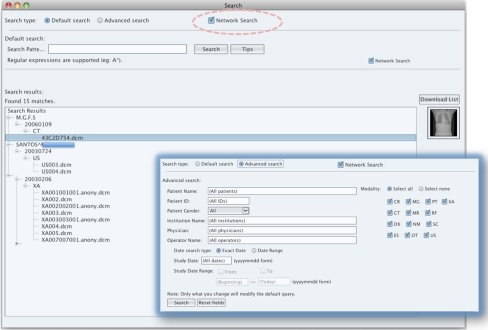
In the search interface (Fig. 6), besides the usual PACS searching features, a free text mode is available allowing users to construct interactively their own query. For instance, it is possible to specify each field, boolean operators to refine the query (Modality:CT and ExposureTime:>700), nearby terms (PatientName:“Robert Laurent” ∼10), and flexible range (StudyDate:[20030801 TO 20040201]). The query results are displayed in a tree-view following the traditional DIM model: patients, studies, series, and image.
Fig. 6.
Search window in P2P mode
When Dicoogle is connected in the P2P network, the search can be executed in all peers. If the user wants to retrieve medical imaging data stored in remote peers, the download manager module will provide feedback about operation progress, thus increasing Dicoogle usability in P2P mode. When a download finishes, the image can be opened and manipulated locally.
Performance Evaluation
In order to evaluate Dicoogle performance in peer-to-peer scenarios, we have used a regular operational 100 Mb LAN and four common desktop computers with distinct computational resources to build a heterogeneous PACS group. The slowest computer was a Pentium4, with a 2.6-GHz processor and 512 MB RAM, while the fastest was an Intel Core Duo E8400, with 3.0-GHz CPUs and 4 GB of memory. In each of these peers, 40,000 DICOM files were indexed for the trial.
To measure Dicoogle response times, we generated successive sets of queries whose responses return an increasing number of results (patients and images), between zero and 41,000. These queries were repeated from one single peer up to the four peers. It must be mentioned that the queries over a single peer were performed locally, using Dicoogle desktop mode. In this way, the results not only allow us to evaluate the scalability of the P2P solution but also the performance of the distributed operation mode against the local one.
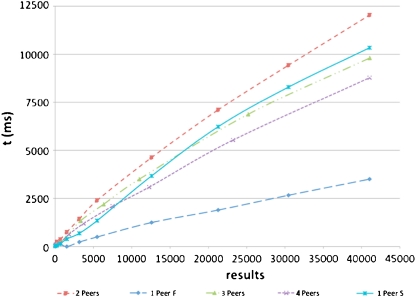
A total of 3,000 queries were generated, and for each sub-set the mean was calculated. The final results are shown in Figure 7. The purpose of each experiment was to compare search times and to establish a relationship between the number of results found and the time needed to get results through a query among peers.
Fig. 7.
Dicoogle query response times
These results show that Dicoogle can be used effectively to search studies in a P2P network. For a query result associated, for instance, with a list of 1,600 images, this represents in our repository a response time that varies between 136 ms (one fast peer) and 750 ms (two peers). In P2P group search, the impact of the querying processing distribution may reduce even more this response time (Fig. 7). In this case, the P2P threshold will be the network bandwidth.
Conclusions
We have presented the peer-to-peer Dicoogle solution, a DICOM conformant PACS archive that incorporates two new concepts.
First, a new storage and retrieval approach based on document indexing that can replace or extend the traditional PACS RDBMS. With Dicoogle, complex search requirements can be satisfied because more and better information can be extracted from DICOM persistent objects. Moreover, to boost the informative potential of distributed medical image repositories, we developed an indexing engine able to support distributed query and retrieval of medical imaging data. Large-scale imaging studies requiring DICOM tagged information are now feasible in a much more straightforward manner.
A second important feature is the support of P2P technology. With this solution, it is possible to establish an on-demand connection, at any time, with any partner, without a complex and unfriendly set-up time. This will turn large volumes of clinical information and analytical tools, currently hidden in clinical units, into shared repositories and high-quality collaborative environments for medical applications, education, and research.
P2P architectures appear as natural off-the-shelf solutions to the increasing dynamics of DICOM nodes. This strategy is also more likely to provide peer distributed redundancy and disaster recovery solutions than traditional and often expensive vendor-specific approaches.
Acknowledgment
The research leading to these results has received funding from Fundação para a Ciência e Tecnologia (FCT) under grant agreement PTDC/EIA-EIA/104428/2008.
Contributor Information
Carlos Costa, Phone: +351-234-370500, FAX: +351-234-370545, Email: carlos.costa@ua.pt.
Carlos Ferreira, Email: c.ferreira@ua.pt.
Luís Bastião, Email: bastiao@ua.pt.
Luís Ribeiro, Email: luisribeiro@ua.pt.
Augusto Silva, Email: augusto.silva@ua.pt.
José Luís Oliveira, Email: jlo@ua.pt.
References
- 1.Costa C, Silva A, Oliveira JL: Current perspectives on PACS and cardiology case study. In: Vaidya S, Jain LC, Yoshida H Eds. Studies in Computational Intelligence: Advanced Computational Intelligence Paradigms in Healthcare. Berlin: Springer, p. capt. 5, 2007, pp 79–108
- 2.Huang HK: PACS and Imaging Informatics: Basic Principles and Applications. New York: Wiley, 2004
- 3.Oosterwijk H. Dicom Basics, third edition. Aubrey: OTech; 2005. [Google Scholar]
- 4.DICOM-P4: Digital Imaging and Communications in Medicine (DICOM), Part 4: Service Class Specifications, National Electrical Manufacturers Association, 2009
- 5.Pianykh OS, H.M.S., Boston MA: USA Digital Imaging and Communications in Medicine (DICOM)—A Practical Introduction and Survival Guide. Berlin: Springer, 2008
- 6.DICOM-P18: Digital Imaging and Communications in Medicine (DICOM), Part 18: Web Access to DICOM Persistent Objects (WADO), National Electrical Manufacturers Association, 2009
- 7.Warren-Forward H, et al. An assessment of exposure indices in computed radiography for the posterior–anterior chest and the lateral lumbar spine. Br J Radiol. 2007;80(949):26–31. doi: 10.1259/bjr/59538862. [DOI] [PubMed] [Google Scholar]
- 8.Kallman HE, et al. DICOM metadata repository for technical information in digital medical images. Acta Oncol. 2009;48(2):285–288. doi: 10.1080/02841860802258786. [DOI] [PubMed] [Google Scholar]
- 9.Vano E, et al. Paediatric entrance doses from exposure index in computed radiography. Phys Med Biol. 2008;53(12):3365–3380. doi: 10.1088/0031-9155/53/12/020. [DOI] [PubMed] [Google Scholar]
- 10.Reiner BI, et al. Multi-institutional analysis of computed and direct radiography—part II. Economic analysis. Radiology. 2005;236:420–426. doi: 10.1148/radiol.2362040673. [DOI] [PubMed] [Google Scholar]
- 11.Polónia DF, et al: Inequality problems in the distribution of radiologists in Portugal: requirements for the creation of an imaging marketplace. In eChallenges, Istanbul—Turkey, 2009
- 12.Costa C, et al. Indexing and retrieving DICOM data in disperse and unstructured archives. Int J Comput Assist Radiol Surg. 2009;4(1):71–77. doi: 10.1007/s11548-008-0269-7. [DOI] [PubMed] [Google Scholar]
- 13.Apache SF: Lucene Index Server—http://lucene.apache.org, 2010
- 14.McCandless M, Hatcher E, Gospodnetic O: Lucene in Action, second edition, Greenwich: Manning, 2010
- 15.dcm4che. sourceforge project: http://sourceforge.net/projects/dcm4che/, 2007
- 16.Warnock MJ, et al: Benefits of Using the DCM4CHE DICOM Archive. Journal of Digital Imaging—Springer New York, 20—Supplement 1: pp 125–129, 2007 [DOI] [PMC free article] [PubMed]
- 17.DICOM-P3: Digital Imaging and Communications in Medicine (DICOM), Part 3: Information Object Definitions, National Electrical Manufacturers Association, 2009
- 18.SUN JAI ImageIO Project, https://jai-imageio.dev.java.net/
- 19.Fiaidhi J, Orabi M, Mohammed S. Sharing DICOM learning objects within a mobile peer-to-peer podacasting environment. Int J Mob Commun. 2008;6(4):417–435. doi: 10.1504/IJMC.2008.018051. [DOI] [Google Scholar]
- 20.Maglogiannis I, Constantinos D, Kazatzopoulos L. Enabling collaborative medical diagnosis over the Internet via peer-to-peer distribution of electronic health records. J Med Syst. 2006;30(2):107–116. doi: 10.1007/s10916-005-7984-1. [DOI] [PubMed] [Google Scholar]
- 21.Blanquer I, Hernadez V, Mas F: A peer-to-peer environment to share medical images and diagnoses providing context-based searching. In Proceedings of the 13th Euromicro Conference on Parallel, Distributed and Network-Based Processing. IEEE Computer Society, 2005
- 22.Pan TC, et al. Informatics in radiology: GridCAD: grid-based computer-aided detection system. Radiographics. 2007;27(3):889–897. doi: 10.1148/rg.273065153. [DOI] [PubMed] [Google Scholar]
- 23.Blanquer I, Hernandez V, Segrelles D: TRENCADIS—A Grid Architecture for Creating Virtual Repositories of DICOM Objects in an OGSA-Based Ontological Framework in Biological and Medical Data Analysis—Lecture Notes in Computer Science. Berlin: Springer, 2006, pp 183–194
- 24.Michal V, et al: A Reliable DICOM Transfer Grid Service Based on Petri Net Workflows. Proceedings of the 2008 Eighth IEEE International Symposium on Cluster Computing and the Grid (CCGRID)—Volume 00, IEEE Computer Society, 2008
- 25.Stephanos A-T, Diomidis S. A survey of peer-to-peer content distribution technologies. ACM Comput Surv. 2004;36(4):335–371. doi: 10.1145/1041680.1041681. [DOI] [Google Scholar]
- 26.Madhukar A. Williamson C: A Longitudinal Study of P2P Traffic Classification. Modeling, Analysis, and Simulation of Computer and Telecommunication Systems, 2006. MASCOTS 2006. 14th IEEE International Symposium, 2006
- 27.Naimul B, et al: A comparative analysis of web and peer-to-peer traffic. Proceedings of the 17th International Conference on World Wide Web. Beijing: ACM, 2008
- 28.JGroups P, JGroups—A Toolkit for Reliable Multicast Communication. 2010; available from: http://www.jgroups.org/
- 29.Traversat B, et al: Project JXTA 2.0 Super-Peer Virtual Network, Project JXTA—Sun Microsystems, Inc., 2003
- 30.P. Saint-Andre, E: RFC3920—Extensible Messaging and Presence Protocol (XMPP): Core, IETF, 2004