Abstract
Lesion segmentation, which is a critical step in computer-aided diagnosis system, is a challenging task as lesion boundaries are usually obscured, irregular, and low contrast. In this paper, an accurate and robust algorithm for the automatic segmentation of breast lesions in mammograms is proposed. The traditional watershed transformation is applied to the smoothed (by the morphological reconstruction) morphological gradient image to obtain the lesion boundary in the belt between the internal and external markers. To automatically determine the internal and external markers, the rough region of the lesion is identified by a template matching and a thresholding method. Then, the internal marker is determined by performing a distance transform and the external marker by morphological dilation. The proposed algorithm is quantitatively compared to the dynamic programming boundary tracing method and the plane fitting and dynamic programming method on a set of 363 lesions (size range, 5–42 mm in diameter; mean, 15 mm), using the area overlap metric (AOM), Hausdorff distance (HD), and average minimum Euclidean distance (AMED). The mean ± SD of the values of AOM, HD, and AMED for our method were respectively 0.72 ± 0.13, 5.69 ± 2.85 mm, and 1.76 ± 1.04 mm, which is a better performance than two other proposed segmentation methods. The results also confirm the potential of the proposed algorithm to allow reliable segmentation and quantification of breast lesion in mammograms.
Keywords: Watershed, Marker, Lesion, Mammogram, Morphological gradient
Introduction
Breast cancer is the leading cause of death for women all over the world [1]. At present, the major risk factors for breast cancer cannot be avoided and the survival is closely related to early detection [2]. Mammography is the most common modality for detecting the early-stage breast cancer and the only modality recommended for population screening programs [3]. Many computer-aided diagnosis (CAD) systems based on mammograms have been developed to improve the detection rate of breast cancer [4–6]. Lesion segmentation is one of the most important steps in CAD systems. It can produce approximate contours of suspicious regions to provide features which enable discrimination between true lesions and regions corresponding to normal tissue. Since lesion boundaries are usually embedded and hidden in varying densities of parenchyma structures and may become obscured, irregular, and low contrast, lesion segmentation is a challenging task.
In recent years, a number of methods have been developed for lesion segmentation. Yuan et al. presented a dual-stage method for lesion segmentation in a region of interest (ROI) [7]. Their method consisted of a radial gradient index-based segmentation method to yield an initial contour and a geometric active contour model for shape refinement. The boundaries obtained by the segmentation method were compared to hand-drawn results in terms of area overlap metric (AOM). Timp and Karssemeijer presented a robust and automated segmentation technique based on dynamic programming boundary tracing (DPBT) to segment lesions [8]. At the heart of the dynamic programming method is the so-called local cost function, which is used to find the path that most efficiently represents the contour of lesions. Rojas Domínguez and Nandi modified the local cost function and designed two improved DPBT methods [9]. Song et al. segmented breast lesions using plane fitting and dynamic programming (PFDP) [10]. First, the edge candidate points were obtained using a plane fitting method. Then, dynamic programming technique was used to find the optimal contour of a lesion from the edge candidate points. All these dynamic programming-based methods were compared to radiologist’s manual segmentation using the AOM metric. Rojas Domínguez and Nandi’s method is difficult to reproduce; so we compared our proposed algorithm to Timp and Karssemeijer’s method and Song et al.’s method, using the AOM, Hausdorff distance (HD), and average minimum Euclidean distance (AMED) metrics.
The watershed transformation is a powerful tool for image segmentation based on mathematical morphology [11–14]. We can consider the image as a landscape or topographic relief where the gray level of each pixel corresponds to a physical elevation. Immersing the landscape in a lake with holes pierced in local minima, catchment basins will fill up with water starting at these local minima. At points where water coming from different basins would meet, dams are built. This process ends when the water level has reached the highest peak in the landscape. As a result, the landscape is partitioned into regions or basins separated by dams, called watershed lines or simply watersheds [15].
The advantages of the watershed transformation are that it is simple, intuitive, and can be parallelized. The main drawback of this method is the over-segmentation due to the presence of many local minima. To decrease the effect of severe over-segmentation, marker-controlled watershed transformations have been proposed [15–17]. These are robust and flexible methods for segmenting objects with closed contours, such as breast lesions. The internal marker (a region completely inside the lesion) and external marker (a region completely free of pixels containing a lesion) are initially defined. The boundaries, even if not clearly defined, are expressed as ridges between two markers and located. Initial definition of the markers is critical in these methods. Yan and Zhao proposed a marker-controlled watershed method to segment lymphoma in sequential computerized tomography images [16]. In their method, the external marker is obtained manually by drawing a circle enclosing the lymphoma. The internal marker is determined automatically by combining techniques including Canny edge detection, thresholding and morphological operation. Cui et al. also proposed a semi-automated method based on marker-controlled watershed transformation to segment breast lesion volumes on magnetic resonance imaging [17]. They manually selected the ROI in a single image, followed by a Gaussian mixture model applied to a histogram of the pixels inside the ROI to distinguish the lesion class from other tissues. The internal and external markers are determined on the basis of the ROI and the intensity distribution of the lesion, and the lesion contour is delineated using a marker-controlled watershed transform. These methods can hardly be applied to mammogram CAD because of their semi-automated nature.
In this study, a robust and accurate marker-controlled watershed (MCWS) method is proposed to achieve higher segmentation performance and get more accurate lesion contours in mammograms. The focus is mainly on smoothing the gradient image and determining internal and external markers, which is crucial in the marker-controlled watershed method. To assess the performance of this method, we compared it to the DPBT and PFDP methods using the AOM, HD, and AMED metrics.
Materials and Methods
Data Acquisition
Our test set consists of 363 ROIs (173 benign and 190 malignant lesions) of 355 mammograms which were randomly selected from the Digital Database for Screening Mammography [18]. These lesions come from 264 different patients and have different margin types, sizes, and densities, as shown in Table 1. Image analysis of screening mammograms is challenging for the image size. A typical mammographic view ranges from 20 MB to more than 70 MB because of the high spatial requirements. There are two lesion types—masses and microcalcifications in mammograms, we focused on the mass detection in this study. Although the segmentation using the original high-resolution images may help improve accuracy slightly, considering the typical size of masses is greater than 1,000 μm (5,000 μm in this study) in diameter, image reduction is a common step among many different CAD schemes. In our CAD system, taking into account the accuracy and efficiency, the mammograms were reduced by a factor of 8 (from 50 to 400 μm per pixel), and the range of image pixel gray level was reduced from 12 to 8 bits (256 gray levels). The outlines provided by the radiologist were used as ground truth in our experiments to evaluate the performance of the segmentation methods. The geometric center of each lesion based on the radiologist’s marked outline was estimated to be the center of the ROI with a size of 125 × 125 pixels extracted from the reduced image.
Table 1.
Distributions of lesions in different margin types, sizes, and densities in the selected dataset
| Statistics of lesion size in radius (mm) | Number of lesions with various margin type | Number of lesions with various density | |||
|---|---|---|---|---|---|
| Min. | 2.5 | Circumscribed | 90 | Almost entirely fatty | 92 |
| First Qu. | 5.1 | Spiculated | 56 | Scattered fibroglandular | 174 |
| Mean | 7.5 | Ill-defined | 110 | Heterogeneously dense | 64 |
| Median | 6.5 | Microlobulated | 64 | Extremely dense | 33 |
| Third Qu. | 8.8 | Obscured | 43 | ||
| Max | 21.0 | ||||
Overview of Proposed Method
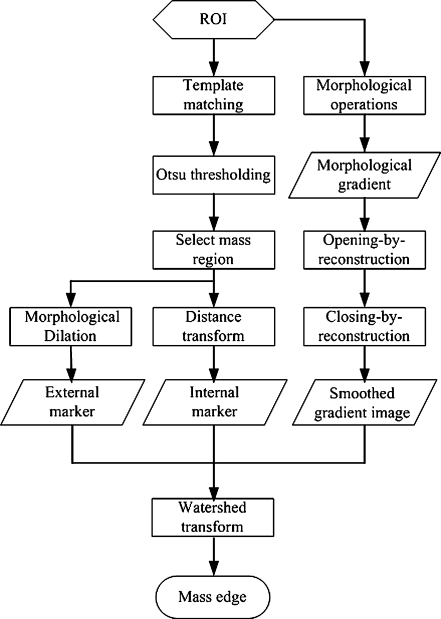
The flow chart of the proposed marker-controlled watershed method is shown as Fig. 1. In this study, we focus mainly on smoothing the gradient image and determining internal and external markers. The morphological gradient image is calculated and smoothed by morphological operations. To automatically determine the internal and external markers, the rough region of the lesion is identified by a template matching and a thresholding method. The internal marker is then determined by performing a distance transform and the external marker is determined by morphological dilation. Details of the algorithm are given in the following sections.
Fig. 1.
The flow chart of the marker-controlled watershed method
Morphological Gradient Image Calculation and Smoothing
Generally, the watershed transformation is computed on the gradient image so that the boundaries are located at high gradient points. Several distinct gradients are used in image processing to detect contours. One of them is called the morphological gradient. Its computation is simple: for each point in the image, a structuring element is centered to it and the difference between the maximum and the minimum gray levels inside the structuring element is computed [19]. That is,
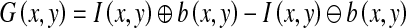 |
1 |
where I, b, and G are the original ROI, the structuring element, and the morphological gradient, respectively. ⊕ and ⊖ are respectively the morphological dilation and erosion operators. In mammograms, lesions are somewhat circular [8]. Therefore, the structuring element b used in this work is a flat, disk-shaped structuring element with a radius of three pixels based on experience. This structuring element is isotropic and can eliminate the dependence on the direction of the edge.
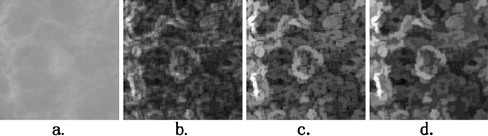
Many regional minima may exist in the morphological gradient image (see Fig. 2b). These regional minima may lead to over-segmentation in the watershed method. To “clean” up the gradient image, “opening by reconstruction” and “closing by reconstruction” operators are employed in this study. Figure 2 shows this process.
Fig. 2.
The process of calculating the morphological gradient image. a An Original ROI image with a lesion at the center. b The morphological gradient image; c sharp peaks in image (b) are reduced using the opening by reconstruction operator; d valleys in image (b) are filled using the closing by reconstruction operator
Morphological reconstruction processes one image, called the marker, based on the characteristics of another image, called the mask. The peaks in the marker image should identify the location of objects to emphasize in the mask image. Denoting Im ⊆ I as the marker, reconstruction operators can be expressed by iterating the dilation of marker Im with a structuring element B, making sure that, at each iteration, the dilation is restricted inside the mask I. The dilations of the marker image are repeated until the contour of the marker image fits under the mask image:
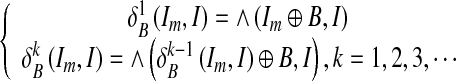 |
2 |
Where 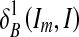 and
and 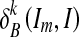 are the dilations of size 1 and k on image Im, respectively, using a structuring element B. The operator “⋀” denotes a point-wise minimum operation. The grayscale reconstruction [20] RB(Im, I) of I from Im is then obtained by iterating grayscale dilations of Im “under” I, until stability is reached:
are the dilations of size 1 and k on image Im, respectively, using a structuring element B. The operator “⋀” denotes a point-wise minimum operation. The grayscale reconstruction [20] RB(Im, I) of I from Im is then obtained by iterating grayscale dilations of Im “under” I, until stability is reached:
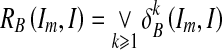 |
3 |
where the operator “⋁” denotes point-wise maximum operation.
In this study, the marker image is obtained by setting Im = G ⊖ b, where G is the image of morphological gradient, and b is the disk-shaped structuring element used previously. The opening by reconstruction operator is used to darken small bright areas and to reduce sharp peaks in the morphological image G (see Fig. 2c):
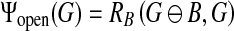 |
4 |
where RB is the morphological reconstruction defined in formula (2). Let Go represent the reconstructed image Ψopen (G). The closing by reconstruction operator is used to brighten small dark areas and to fill valleys in image Go:
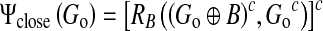 |
5 |
where Go represents the complement of the image Go. Through gradient image reconstruction, important region contours are preserved while most small regular details and noise are removed, as shown in Fig. 2d.
Determination of Internal and External Markers
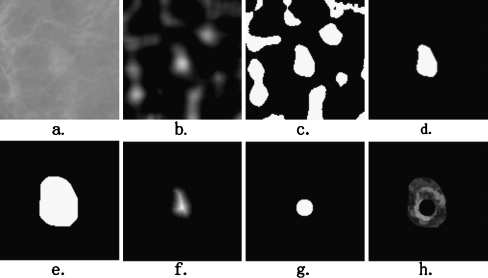
In our segmentation method, we focus mainly on determining the internal marker, which is completely within the lesion, and the external marker, which is completely free of pixels containing a lesion. Figure 3 shows this process. First, a rough region of the lesion is estimated by a template matching [21–23] and a thresholding method. The internal and external markers are then determined by distance transformation and morphological operation, respectively.
Fig. 3.
The process of determining of the external and internal markers. a The original ROI of Fig. 2a; b the correlation image; c the binary mask of the correlation image calculated by the Otsu thresholding method; d the nearest isolated region from the center of ROI is selected as the rough region of the lesion; e the rough region dilated with a morphological operator forms the external marker; f the distance image of the rough region; g the internal marker; h the modified morphological gradient image: internal marker (the black region inside the lesion) and external marker (the black region outside the lesion) imposed on the morphological gradient image
In mammograms, lesions tend to have a greater intensity than their neighboring pixels, and are somewhat circular in shape, although they display weak or fading boundaries with neighboring tissue [24]. Based on this prior information of lesions, a hyperbolic secant (Sech) template that exhibits visual and statistical properties of lesions is proposed to locate and obtain the rough region of lesions in this study. Assume that the coordinates of the center of the template image are (0, 0). Then,
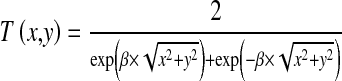 |
6 |
where T(x, y) is the gray level of the template image at the position (x, y). Parameter β controls the rate of change on gray level, and it has been set to 0.08 by making the best match between the template and targets. Suppose the size of the template image is L × L (35 × 35 in this study), then the template image obtains the maximum gray level at the center and the minimum value at a distance of L/2.
To detect the lesion, we use the template image to match with the corresponding region of the ROI image. Let I denote the ROI image, μT and μI (x, y) denote the average gray levels of the template image and the subregion of the ROI image (with the same size as the template image) around the center point (x, y), respectively. The overlapping region of the template and the ROI image is represented by 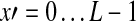 ,
,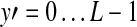 . The similarity measure between the template and the ROI image at point (i, j) is defined as
. The similarity measure between the template and the ROI image at point (i, j) is defined as
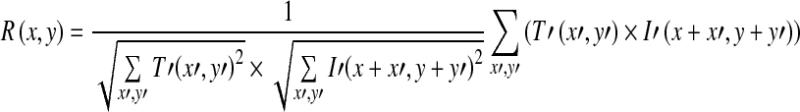 |
7 |
Where
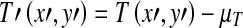 |
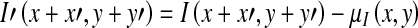 |
The value of the similarity measure of every pixel in the ROI image form a correlation image, as shown in Fig. 3b. To obtain the rough region of the lesion, the correlation image is segmented by the Otsu thresholding method. The pixels with correlation value above the Otsu threshold are set to be the candidate pixels of the lesion, as shown in Fig. 3c.
In mammography CAD systems, the detection method generates some markers/prompts at suspicious regions in mammograms. Then, the segmentation method, which is able to detect the precise outline of the potential lesion, is implemented on the ROI. In our CAD system, the ROI is defined as a rectangular region (125 × 125 pixels) extracted from the mammograms with its center at the center of the marker/prompt. Therefore, by grouping geometrically connected nonzero pixels and assigning an identical number to the pixels in each individual group, the component that is nearest from the center of the ROI is selected as the rough region of the lesion. All other nonzero areas (isolated regions) are set to zero to avoid any further processing, as shown in Fig. 3d. Based on the rough region, the approaches to the obtaining of the external and internal markers are described as follows.
In order to make the external marker that contains the lesion region as complete as possible, the rough region is selected to be the external marker after a dilation with a disk-structuring element with a radius of 10 pixels, as shown in Fig. 3e.
On the other hand, utilizing the rough region of the lesion, a circular internal marker inside the lesion is automatically determined. First, a distance transformation is applied to the binary image that contains the rough region of the lesion. The Euclidean distances from every pixel in the rough region of the lesion to its nearest zero pixel are calculated to form a distance image, as shown in Fig. 3f. Second, on the distance image, the point with the largest distance value is selected to be the center of the circular internal marker. As mentioned previously, the center of the lesion is close to the center point of the ROI. In order to ensure that the center of the internal marker is close to the lesion center, the selection of the marker center is limited in the range of pixels with the distance less than 14 from the ROI center. Finally, to avoid an over-presented internal marker (i.e., the internal marker is too large), which will decrease the performance of the marker-controlled watershed segmentation, the radius of the internal marker is set to be 0.8 (which is obtained by trial and error method) times the value of its center point, as shown in Fig. 3g.
Marker-Controlled Watershed Segmentation
The internal and external markers, and the image of smoothed morphological gradient are obtained by the methods described above. Each marker indicates a specific place within the morphological gradient image (also called “segmentation function”) to force that region to be a global minimum of the image by using the minima imposition technique [15], as shown in Fig. 3h. That is to say, the modified morphological gradient image only has regional minima in the locations of the markers. Finally, the traditional watershed transformation is applied to the modified gradient image to obtain the lesion boundary in the belt between the internal and external markers.
Evaluation
The widely used performance evaluation methods in image segmentation can be grouped into two categories: one is based on matching of the regions obtained with different methods; the other is in terms of the distance between boundaries obtained with different methods. We use the area overlap metric [9, 10, 25], the Hausdorff distance metric, and the average minimum Euclidean distance metric [26–28] to quantify the consistency between the segmented results and the ground truth provided by a radiologist’s manually delineated outlines. The AOM metric is defined as the ratio of the intersection to the union of the two areas to be compared:
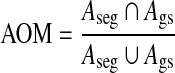 |
8 |
where Aseg denotes the area of the automatically segmented lesion, and Ags is the corresponding ground truth area. The HD metric is defined as the maximum distance between the contours obtained with the computerized segmentation method and the radiologist’s marked outlines which are denoted as 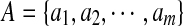 and
and 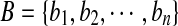 , respectively. And the AMED metric computes the average distance:
, respectively. And the AMED metric computes the average distance:
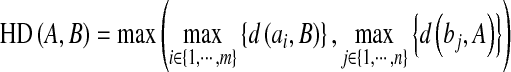 |
9 |
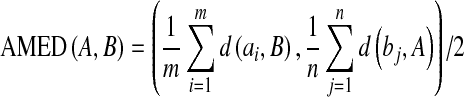 |
10 |
where 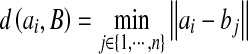 is the distance from ai to the closest point on the contour B.
is the distance from ai to the closest point on the contour B.
Results and Discussion
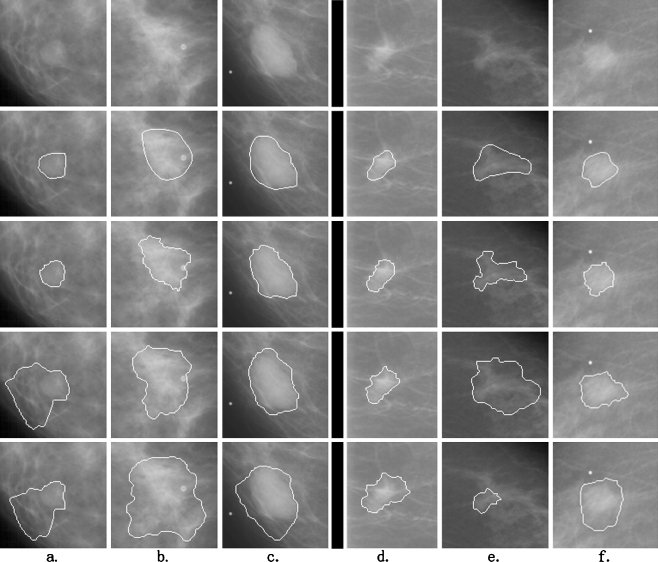
To test the performance of the method, all the 363 ROIs were segmented by our proposed method and the DPBT and PFDP methods. The methods of DPBT and PFDP were implemented on the basis of the descriptions in Timp and Karssemeijer’s study [8] and Song et al.’s study [10], respectively. Figure 4 shows six results of these segmentation methods: the lesions of the left three columns are benign, and the other three are malignant. The results of our method are, visually inspected, closer to the radiologist’s outlines than the other methods.
Fig. 4.
Six examples of segmentation results by the three segmentation methods. a–c Three examples of benign lesions; d–f three examples of malignant lesions. The top line is the original ROIs. The next four lines are the corresponding contours marked by radiologist, our marker-controlled watershed method, the DPBT method and PFDP method, respectively
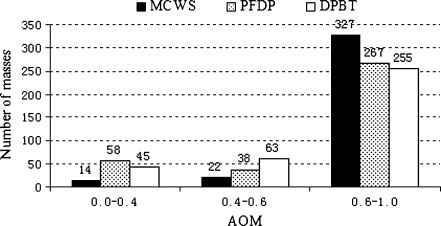
For each lesion in the test dataset, AOM, HD, and AMED metrics are calculated with each of the three segmentation methods. Generally, the segmentation result is considered to be good when the value of AOM is higher than 0.6, and poor when lower than 0.4. Figures 5 shows the numbers of lesions correctly segmented at overlap threshold levels of 0.4 and 0.6 for each of the three segmentation methods. It can be seen that the proposed method can achieve more good results and less poor results than the other two methods. For instance, the number of lesions with good/poor segmentation result for the MCWS method is 327 (90%)/14 (4%), for the PFDP 267 (74%)/58 (16%), and DPBT 255 (70%)/45 (12%), respectively. The distributions, means, and standard deviation of the HD and AMED metrics for all the three methods are given in Table 2. The mean ± SD of the values of AOM, HD, and AMED for our method are 0.72 ± 0.13, 5.69 ± 2.85 mm, and 1.76 ± 1.04 mm, whereas those for PFDP and DPBT methods are 0.67 ± 0.22, 7.28 ± 4.43 mm, and 2.98 ± 3.16 mm and 0.65 ± 0.19, 7.07 ± 4.27 mm, and 2.77 ± 2.61 mm, respectively. The higher value of AOM and lower values of HD and AMED indicate that the results of the proposed method are closer to the radiologist’s outlines than the other two methods. All these indicate that the performance of our proposed segmentation algorithm is better than the other two methods.
Fig. 5.
Histograms of AOM values for the lesions in the test dataset. AOM area overlap metric, MCWS marker-controlled watershed, PFDP plane fitting and dynamic programming, DPBT dynamic programming boundary tracing
Table 2.
Distributions of the HD and AMED criteria for the three segmentation methods
| Method | Min. | First Qu. | Median | Third Qu. | Max. | Mean | SD |
|---|---|---|---|---|---|---|---|
| HD (mm) | |||||||
| MCWS | 1.60 | 3.62 | 5.24 | 7.21 | 18.03 | 5.69 | 2.85 |
| PFDP | 1.69 | 4.45 | 6.73 | 8.86 | 20.83 | 7.28 | 4.43 |
| DPBT | 1.25 | 4.16 | 5.61 | 8.80 | 20.35 | 7.07 | 4.27 |
| AMED (mm) | |||||||
| MCWS | 0.48 | 1.17 | 1.54 | 2.04 | 9.18 | 1.76 | 1.04 |
| PFDP | 0.50 | 1.21 | 1.67 | 2.90 | 16.49 | 2.98 | 3.16 |
| DPBT | 0.50 | 1.27 | 1.83 | 3.06 | 14.07 | 2.77 | 2.61 |
MCWS marker-controlled watershed, PFDP plane fitting and dynamic programming, DPBT dynamic programming boundary tracing, HD Hausdorff distance, area overlap metric, AMED average minimum Euclidean distance
Table 3 shows the results of the Wilcoxon statistic. The differences between the segmentation-metrics distribution of MCWS and the other two methods are found to be statistically significant (P values are no more than 0.001). It is clear from the test that a significant increase in all metrics has been obtained with the marker-controlled watershed method compared with the other two segmentation methods.
Table 3.
The Wilcoxon statistic for the statistical differences between the segmentation-metrics distribution of MCWS, PFDP, and DPBT methods
| Method | P value for AOM | P value for HD | P value for AMED |
|---|---|---|---|
| MCWS–DPBT | <0.001 | <0.001 | <0.001 |
| MCWS–PFDP | <0.001 | <0.001 | <0.001 |
| PFDP–DPBT | 0.253 | 0.521 | 0.340 |
MCWS marker-controlled watershed, PFDP plane fitting and dynamic programming, DPBT dynamic programming boundary tracing, AOM area overlap metric, HD Hausdorff distance, area overlap metric, AMED average minimum Euclidean distance
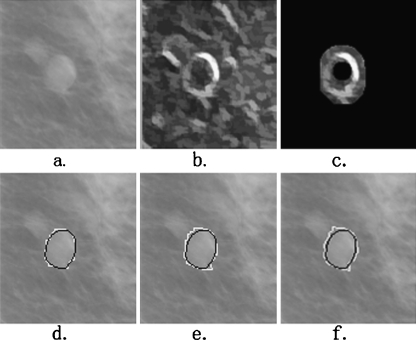
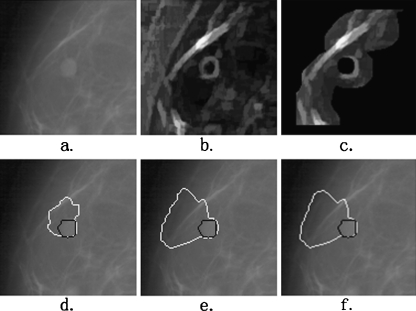
The main reason for this improved performance is the accurate positioning of the internal and external markers, between which there is the contour of the lesion. However, there are rare cases in which the performance of our segmentation algorithm is not ideal. Figures 6 and Fig. 7 illustrate an almost perfect segmentation and a very poor segmentation, respectively. In Fig. 6, the AOM and AMED metrics for our segmentation method are 0.93 and 1.29, respectively (whereas those for PFDP and DPBT methods are 0.85 and 2.17 and 0.83 and 2.18, respectively). The internal and external markers reduce the regional minima and sharp peaks very well and the segmented region is almost identical to the ground truth. In the poor performance example shown in Fig. 7, the values of the AOM and AMED metrics for our proposed method are 0.30 and 11.97, respectively (whereas those for PFDP and DPBT methods are 0.13 and 27.73 and 0.13 and 28.49, respectively). It can be observed that the main reason for the failure of the algorithm in this case is the surrounding dense background tissues inside the external marker. As can be seen, a section of the lesion boundary is correctly located, but the other section is pulled away from the lesion by the edge of the dense tissues. The marker-controlled watershed method would possibly perform better, if all this dense tissue can be excluded from the external marker. This aspect will be evaluated in further research.
Fig. 6.
An example of good performance of segmentation. a The original ROI; b the smoothed morphological gradient image; c the extracted markers imposed on the gradient image. The segmentation results using the MCWS, PFDP, and DPBT methods are shown in (d–f), respectively, with the white overlay. The black overlays are the ground truth. The AOM values for these three segmentation results are 0.93, 0.85, and 0.83, respectively. The AMED values are 1.29, 2.17, and 2.18, respectively
Fig. 7.
An example of poor performance of segmentation. a The original ROI; b the smoothed morphological gradient image; c the extracted markers imposed on the gradient image. The segmentation results using the MCWS, PFDP, and DPBT methods are shown in (d–f), respectively, with the white overlay. The black overlays are the ground truth. The AOM values for these three segmentation results are 0.30, 0.13, and 0.13, respectively. The AMED values are 11.97, 27.73, and 28.49, respectively
Conclusion
Lesion segmentation can produce approximate contours to provide features, and plays a crucial role in CAD systems for classification of suspicious regions. In this study, we propose an automatic, accurate, and robust marker-controlled watershed algorithm to segment breast lesions in mammograms. The morphological gradient image is smoothed by reconstruction. The internal and external markers are automatically obtained by combining techniques including template matching, thresholding, distance transformation, and morphological operation. The segmentation algorithm has been applied to 363 lesions and resulted in AOM, HD, and AMED of 0.72, 5.69 mm, and 1.76 mm, respectively. The experimental results have confirmed the potential of the proposed method to allow reliable segmentation and quantification of breast lesions in mammograms.
The influence of the segmentation result on the performance of a CAD system has not been studied yet. We will concentrate our future work on studying the influence of the proposed segmentation method on the performance of our CAD system.
Contributor Information
Shengzhou Xu, Phone: + 86-139-71238526, FAX: +86-27-87545004, Email: xushengzhou2008@163.com.
Hong Liu, Phone: +86-27-87792212, FAX: +86-27-87545004, Email: whkaociya@gmail.com.
Enmin Song, Phone: +86-27-87792211, FAX: +86-27-87545004, Email: esong@mail.hust.edu.cn.
References
- 1.Nishikawa RM. Current status and future directions of computer-aided diagnosis in mammography. Comput Med Imaging Graph. 2007;31:224–235. doi: 10.1016/j.compmedimag.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Reddy M, Given-Wilson R. Screening for breast cancer. Women’s Health Medicine. 2006;3:22–27. doi: 10.1383/wohm.2006.3.1.22. [DOI] [Google Scholar]
- 3.Dubey RB, Hanmandlu M, Gupta SK. A comparison of two methods for the segmentation of masses in the digital mammograms. Comput Med Imaging Graph. 2010;34:185–191. doi: 10.1016/j.compmedimag.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Morton MJ, Whaley DH, Brandt KR, Amrami KK. Screening mammograms: Interpretation with computer-aided detection—prospective evaluation. Radiology. 2006;239:375–383. doi: 10.1148/radiol.2392042121. [DOI] [PubMed] [Google Scholar]
- 5.Masotti M, Lanconelli N, Campanini R. Computer-aided mass detection in mammography: False positive reduction via gray-scale invariant ranklet texture features. Medical physics. 2009;36:311–316. doi: 10.1118/1.3049588. [DOI] [PubMed] [Google Scholar]
- 6.Eltonsy NH, Tourassi GD, Elmaghraby AS. A concentric morphology model for the detection of masses in mammography. IEEE Trans Med Imaging. 2007;26:880–889. doi: 10.1109/TMI.2007.895460. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y, Giger ML, Li H, Suzuki K, Sennett C. A dual-stage method for lesion segmentation on digital mammograms. Med Phys. 2007;34:4180–4193. doi: 10.1118/1.2790837. [DOI] [PubMed] [Google Scholar]
- 8.Timp S, Karssemeijer N. A new 2D segmentation method based on dynamic programming applied to computer aided detection in mammography. Med Phys. 2004;31:958–971. doi: 10.1118/1.1688039. [DOI] [PubMed] [Google Scholar]
- 9.Rojas DA, Nandi AK. Improved dynamic-programming-based algorithms for segmentation of masses in mammograms. Med Phys. 2007;34:4256–4269. doi: 10.1118/1.2791034. [DOI] [PubMed] [Google Scholar]
- 10.Song E, Jiang L, Jin R, Zhang L, Yuan Y, Li Q. Breast mass segmentation in mammography using plane fitting and dynamic programming. Acad Radiol. 2009;16:826–835. doi: 10.1016/j.acra.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Beucher S, Meyer F: The morphological approach to segmentation: the watershed transformation. Mathematical Morphology in Image Processing, Marcel Dekker Inc 433–481, 1992
- 12.Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based onimmersion simulations. IEEE transactions on pattern analysis and machine intelligence. 1991;13:583–598. doi: 10.1109/34.87344. [DOI] [Google Scholar]
- 13.Meyer F, Beucher S. Morphological segmentation. Journal of visual communication and image representation. 1990;1:21–46. doi: 10.1016/1047-3203(90)90014-M. [DOI] [Google Scholar]
- 14.Johnson P, Hahn H, Heath D, Fishman E. Automated multidetector row CT dataset segmentation with an interactive watershed transform (IWT) algorithm: Part 2—body CT angiographic and orthopedic applications. J Digit Imaging. 2008;21:413–421. doi: 10.1007/s10278-007-9087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez W, Leija L, Alvarenga AV, Infantosi AFC, Pereira WCA. Computerized lesion segmentation of breast ultrasound based on marker-controlled watershed transformation. Med Phys. 2010;37:82–95. doi: 10.1118/1.3265959. [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Zhao B, Wang L, Zelenetz A, Schwartz LH. Marker-controlled watershed for lymphoma segmentation in sequential CT images. Med Phys. 2006;33:2452–2460. doi: 10.1118/1.2207133. [DOI] [PubMed] [Google Scholar]
- 17.Cui YF, et al. Malignant lesion segmentation in contrast-enhanced breast MR images based on the marker-controlled watershed. Med Phys. 2009;36:4359–4369. doi: 10.1118/1.3213514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heath M, et al. Current Status of the Digital Database for Screening Mammography. Dordrecht: Kluwer Academic; 1998. [Google Scholar]
- 19.Rittner L, de Alencar Lotufo R: Segmentation of DTI based on tensorial morphological gradient. Medical Imaging 2009: Image Processing, Lake Buena Vista, USA, 2009
- 20.Vincent L. Morphological grayscale reconstruction in image analysis: Applications and efficient algorithms. IEEE transactions on image processing. 1993;2:176–201. doi: 10.1109/83.217222. [DOI] [PubMed] [Google Scholar]
- 21.Lin Z, Davis LS, Doermann D, DeMenthon D: Hierarchical part-template matching for human detection and segmentation. Proc. 11th IEEE International Conference on Computer Vision, Rio de Janeiro, 2007
- 22.Brake GM, Karssemeijer N. Single and multiscale detection of masses in digital mammograms. IEEE Transaction on Medical Imaging. 1999;18:628–639. doi: 10.1109/42.790462. [DOI] [PubMed] [Google Scholar]
- 23.Morrison S, Linnett LM. A Model Based Approach to Object Detection in Digital Mammography. Japan: Kobe; 1999. [Google Scholar]
- 24.Sample JT: Computer Assisted Screening of Digital Mammogram Images. Louisiana State University, Department of Computer Science, Doctor of Philosophy2003
- 25.Wang Q, et al. Segmentation of lung nodules in computed tomography images using dynamic programming and multidirection fusion techniques1. Academic Radiology. 2009;16:678–688. doi: 10.1016/j.acra.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Chao WH, Chen YY, Lin SH, Shih YYI, Tsang S. Automatic segmentation of magnetic resonance images using a decision tree with spatial information. Comput Med Imaging Graph. 2009;33:111–121. doi: 10.1016/j.compmedimag.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Wang Y, Shi X. An improved graph cut segmentation method for cervical lymph nodes on sonograms and its relationship with node’s shape assessment. Comput Med Imaging Graph. 2009;33:602–607. doi: 10.1016/j.compmedimag.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, et al. Characterization of mammographic masses based on level set segmentation with new image features and patient information. Med Phys. 2008;35:280–290. doi: 10.1118/1.2820630. [DOI] [PMC free article] [PubMed] [Google Scholar]