Abstract
Background
Vaccine literature indexing is poorly performed in PubMed due to limited hierarchy of Medical Subject Headings (MeSH) annotation in the vaccine field. Vaccine Ontology (VO) is a community-based biomedical ontology that represents various vaccines and their relations. SciMiner is an in-house literature mining system that supports literature indexing and gene name tagging. We hypothesize that application of VO in SciMiner will aid vaccine literature indexing and mining of vaccine-gene interaction networks. As a test case, we have examined vaccines for Brucella, the causative agent of brucellosis in humans and animals.
Results
The VO-based SciMiner (VO-SciMiner) was developed to incorporate a total of 67 Brucella vaccine terms. A set of rules for term expansion of VO terms were learned from training data, consisting of 90 biomedical articles related to Brucella vaccine terms. VO-SciMiner demonstrated high recall (91%) and precision (99%) from testing a separate set of 100 manually selected biomedical articles. VO-SciMiner indexing exhibited superior performance in retrieving Brucella vaccine-related papers over that obtained with MeSH-based PubMed literature search. For example, a VO-SciMiner search of "live attenuated Brucella vaccine" returned 922 hits as of April 20, 2011, while a PubMed search of the same query resulted in only 74 hits. Using the abstracts of 14,947 Brucella-related papers, VO-SciMiner identified 140 Brucella genes associated with Brucella vaccines. These genes included known protective antigens, virulence factors, and genes closely related to Brucella vaccines. These VO-interacting Brucella genes were significantly over-represented in biological functional categories, including metabolite transport and metabolism, replication and repair, cell wall biogenesis, intracellular trafficking and secretion, posttranslational modification, and chaperones. Furthermore, a comprehensive interaction network of Brucella vaccines and genes were identified. The asserted and inferred VO hierarchies provide semantic support for inferring novel knowledge of association of vaccines and genes from the retrieved data. New hypotheses were generated based on this analysis approach.
Conclusion
VO-SciMiner can be used to improve the efficiency for PubMed searching in the vaccine domain.
Background
Since the introduction of Edward Jenner's vaccine against smallpox in 1796, vaccines have proven useful in their ability to stimulate the immune system and confer protection against infections by pathogenic microorganisms. As such, vaccines provide safe, effective, and cost-effective means to reduce the incidence of infectious diseases. Vaccine research and development has undergone a renaissance in recent years. This is in part attributable to the cost-effectiveness of vaccines and advanced post-genomic technologies [1]. Infectious diseases remain a major source of morbidity and mortality worldwide, causing 14.7 million deaths (26% of total global mortality) in 2001 [2]. Although considerable progress has been made, vaccination against many medically important pathogens, such as Human immunodeficiency virus (HIV) and Brucella, has been unsuccessful due to their unique mechanisms of pathogenesis [3,4]. For rational vaccine design, extensive research is required to better understand the pathogenesis and protective immune responses against such diseases [5].
As the field of vaccine research continues to grow, the amount of vaccine literature is rapidly increasing. Search of the biomedical literature database PubMed (http://www.ncbi.nlm.nih.gov/pubmed) revealed the number of vaccine-related citations has almost doubled over the last decade (from 99,316 papers before 2000 to over 196,055 papers as of April 20, 2011). Due to this rapid growth of published information, it is no longer feasible to keep up to date with all the new literature manually, even within one's own research area. The field of literature mining, a means of computer-assisted information extraction from literature data, is becoming increasingly important to cope with the expanding volume of available biomedical literature. Articles in PubMed are indexed with NLM-developed Medical Subject Headings (MeSH), which is a controlled vocabulary of over 25,000 terms organized in a hierarchical fashion with 15 top-level categories [6]. New articles deposited into PubMed will be read by human experts and indexed with relevant MeSH terms to represent the content. The use of MeSH indexing in PubMed provides a consistent way to retrieve information that may use different terminologies for the same concepts. However, MeSH does not thoroughly cover many biomedical domains, including the vaccine domain. For example, "Brucella vaccine" is currently the lowest level term under 'vaccines' in the MeSH hierarchy. No individual Brucella vaccines are currently indexed by MeSH. Furthermore, as a controlled vocabulary, instead of a biomedical ontology, MeSH does not contain logic definitions for the relations between different terms.
A biomedical ontology represents the consensus-based controlled vocabularies of terms and relations, which are logically formulated to promote intelligent information retrieval and modeling. The Vaccine Ontology (VO; http://www.violinet.org/vaccineontology) [7] is a community-based ontology in the domain of vaccine and vaccination. VO classifies existing vaccines in licensed use, on trial, or in research. The relations between different VO terms have been logically defined and support advanced semantic reasoning. SciMiner is a web-based literature mining tool developed for target (gene and protein) identification and as well for functional enrichment analysis [8,9]. SciMiner uses dictionary and rule-based approaches for literature mining. We hypothesized that a VO-based SciMiner literature mining program would dramatically improve vaccine literature indexing and support literature-based discovery of vaccine-gene interaction networks.
As a case study, we have focused on Brucella, an intracellular bacterium that causes brucellosis, the most common zoonotic disease worldwide [10]. Using the VIOLIN vaccine database and analysis system [11], we previously studied Brucella vaccines using multiple bioinformatics approaches [12]. This study introduced the manual curation of Brucella vaccines in the VIOLIN database and in the VO and the online query system of annotated Brucella vaccine information. However, there has not been a systematic study on how to use the curated Brucella vaccine terms and hierarchy in VO for natural language processing (NLP)-based indexing of Brucella vaccine literature, analysis of vaccine-associated Brucella genes, and the prediction of the interaction network between vaccines and Brucella genes. Such study would significantly improve our understanding of Brucella vaccinology and provide a use case for ontology-based text mining in the field of vaccinology and immunology.
In this study, a VO-based SciMiner approach (VO-SciMiner) was developed, 1) to retrieve and index vaccine names and Brucella genes from PubMed articles, and 2) further identify and analyze interactions between Brucella vaccines and Brucella genes. These studies revealed that VO-SciMiner efficiently indexed Brucella vaccine publications and generated a comprehensive vaccine-gene interaction network.
Methods
Brucella vaccine terms and their semantic relations
All Brucella vaccine terms and their semantic relations available in VO (http://www.violinet.org/vaccineontology) were used in this study. The current version consisting of 67 Brucella VO terms (as of April 2011) and their synonyms were added to the dictionary of VO-SciMiner.
Term expansion rules of Brucella VO terms in SciMiner
A training set consisting of 90 papers with manually confirmed Brucella vaccine VO terms was used to optimize SciMiner for VO term identification. Patterns of variations of VO terms in the actual text were learned and implemented in VO-SciMiner as a series of term expansion rules to maximize the power of term identification. Examples of rules are below:
(1) Variations of functional words are used. For example, "encoding" in the vaccine name can be replaced with "coding for".
(2) Unabbreviated gene names can be used instead of abbreviated gene symbols (e.g., trigger factor for TF).
(3) VO terms with a full pathogen name (e.g., Brucella abortus strain 19) can be abbreviated (e.g., B. abortus strain 19) or abbreviated names can be fully spelled out.
(4) The word "strain(s)" can be added. For example, "Brucella melitensis strain Rev. 1" can be an alternative form for "Brucella melitensis Rev. 1".
(5) Pathogen names can be removed. For example, "Rev. 1" is frequently used without "Brucella melitensis" in the text for "Brucella melitensis Rev. 1".
(6) Spaces and special characters can be altered. For example, "Rev.1", "Rev1", "Rev-1" and "Rev-I" can be used as additional terms for "Rev. 1".
(7) Any SciMiner-identified VO term is ignored if a corresponding genus or disease name does not appear in the text. SciMiner reports identified Brucella VO terms only if there is at least one occurrence of "Brucella" or "brucellosis" in any part of the text. This restriction helps to improve the precision of the analysis by removing potentially non-specific identification from other species.
Performance evaluation of VO-SciMiner
To evaluate the performance of VO-SciMiner in literature indexing, 50 negative and 50 positive papers were manually compiled. Each paper in the positive set had at least one confirmed Brucella VO term and contained a total of 89 confirmed paper-VO associations, defined as the "gold standard". The paper set lacking Brucella vaccine terms, but still in the domain of Brucella or brucellosis, was used as a negative control. Performance was measured by recall, precision, and F-measure (defined below), where the paper-VO associations that matched the gold standard constituted true positives (TP), associations that did not match were false positives (FP), and the gold standard associations that were not matched were false negatives (FN) [13].
Indexing of PubMed abstracts with Brucella VO terms using VO-SciMiner
In order to assess the improved information retrieval using ontology-based indexing, VO-SciMiner was applied to a subset of PubMed abstracts. For the present analyses, a total of 14,947 Brucella related papers (defined by "Brucella OR brucellosis" in PubMed as of April 20, 2011) were processed and indexed by SciMiner. The results were compared with PubMed search results using the corresponding VO terms as queries.
Brucella gene name identification from literature
The NCBI RefSeq database contains 10 fully sequenced and annotated Brucella genomes, which share more than 95% sequence homology [14]. However, since these genomes have been independently annotated by multiple research groups, orthologous genes from various Brucella genomes frequently have different gene names (including synonyms) and annotations. To retrieve all relevant literature associated with individual Brucella genes, we used OrthoMCL-DB [15] to identify the orthologs among different Brucella genomes. After removing redundant terms, gene names and annotations were merged for individual orthologs. For genes with variable names across different Brucella genomes, the most frequently used gene name was selected as the representative name; other names were included as synonyms.
Identification of protein localization and over-represented biological functions
The protein subcellular localization of these genes was predicted by our reverse vaccinology program Vaxign [16], which employs PSORTb 3.0, an open-source software for subcellular localization prediction with high accuracy (precision values > 97% and recall values > 94% based on benchmark evaluations) [17]. To determine the over-represented roles of identified Brucella genes associated with Brucella vaccines, an enrichment analysis using the Cluster of Orthologous Group (COG, http://www.ncbi.nlm.nih.gov/COG) information was employed [10,18]. Fisher's exact [19] test was used to calculate the statistical significance of these COG functional groups (p-value < 0.05).
Identification of Brucella vaccines and genes interaction Network
VO-SciMiner was applied to 14,947 Brucella papers to identify Brucella genes and vaccines (VO). If a Brucella gene name and a VO vaccine term were identified in the same document (at the abstract level), a co-citation connection was established between the gene and vaccine. The complete Brucella gene-vaccine network was visualized in Cytoscape [20], an open source bioinformatics software for visualizing molecular interaction networks. Each gene name and VO vaccine term is represented as a node in the network. The number of papers for each co-citation is represented as thickness of each edge.
Web-based implementation of VO-SciMiner
A VO-SciMiner website (http://www.violinet.org/vo-sciminer) has been developed to allow users to search Brucella vaccine papers indexed with VO terms. This website uses PHP on a Linux web server. A MySQL database server is used to store network information. A virtuoso Resource Description Framework (RDF) store is used to store the contents of VO, including asserted and inferred VO hierarchies. A cron (a time-based job scheduler) job has been set up on the web server to retrieve new Brucella-related publications from PubMed on a weekly basis. Once retrieved, the publications will be processed through the VO-SciMiner analysis workflow, and the results will be placed in the MySQL database.
Results
Overall workflow of VO-based literature mining approach
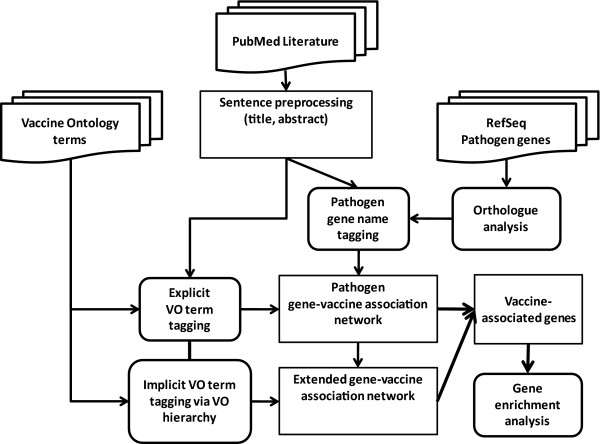
Figure 1 illustrates the proposed workflow of the VO-SciMiner system. Pathogen-related literature is collected through PubMed. The titles and abstracts of the retrieved documents are pre-processed by a sentence splitter, and analyzed by the VO-SciMiner to identify vaccine VO terms and pathogen gene names. The associations of these vaccines and pathogen genes are further analyzed via a vaccine-gene network visualized with Cytoscape. Over-represented biological functions of pathogen genes are examined using the COG information.
Figure 1.
Overall VO-SciMiner workflow.
Asserted and inferred VO hierarchies
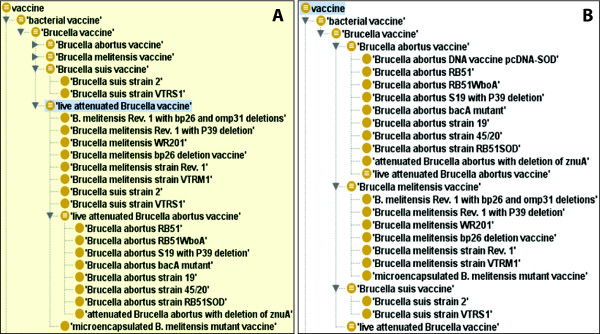
The design of VO is based on OBO (Open Biological and Biomedical Ontologies) Foundry Principles [21]. VO utilizes the Basic Formal Ontology (BFO), a domain-independent ontology, as an upper level ontology [7]. The relation terms defined in the Relation Ontology (RO) are used in VO for representing commonly used relationships. Each subclass found in VO has an "is_a" relationship with its parent class. This characteristic ensures that all vaccine subclasses (e.g., Brucella RB51) are included when a parent class (e.g., "Brucella vaccine") is searched.
Different types of vaccines are classified based on the asserted VO vaccine hierarchy (Figure 2A) and inferred VO vaccine hierarchy (Figure 2B). The asserted ontology hierarchy is an ontology hierarchy specified by ontology developers. The inferred ontology hierarchy, based on the Web Ontology Language (OWL) [20], is generated by a specific ontology reasoner, such as HermiT (http://hermit-reasoner.com). An ontology reasoner infers logical consequences from a set of asserted facts based on necessary and sufficient conditions. If an ontology term is a member of an ontology class, then the term must fulfill the class's necessary conditions. Some classes may also have sufficient conditions, and in those cases the combination of necessary and sufficient conditions makes an ontology class a fully defined class. For example, the class term live attenuated vaccine is fully defined as ['live vaccine' and (has_quality some attenuated)]. Many vaccines have the characteristics (necessary condition) of being attenuated. For example, RB51 has the following necessary condition [has_quality some attenuated]. RB51 and many other vaccines are also live vaccines (another necessary condition). These two necessary conditions are sufficient to define live attenuated vaccine. Therefore, based on the logical definition, the VO term RB51 can be inferred as a live attenuated vaccine. Since RB51 is also a Brucella vaccine, it can be further inferred as a live attenuated Brucella vaccine. Such classification can only be obtained by an ontology inferring since RB51 is not originally asserted as such (Figure 2A).
Figure 2.
Brucella vaccines in VO. (A) Asserted hierarchy; (B) Inferred hierarchy. These are Protégé screenshots of VO without (A) or with reasoning using HermiT 1.3.2 (B).
Performance evaluation of VO term identification using VO-SciMiner
To test the performance of SciMiner, 50 negative and 50 positive papers were manually compiled. The positive set had 89 confirmed paper-VO associations, whereas the negative set contained no VO terms. In this testing set of 100 papers, VO-SciMiner identified 82 paper-VO associations, of which 81 belonged to the gold standard set, thereby achieving 91% recall, 99% precision, and 95% F-measure (Table 1).
Table 1.
Performance of VO-SciMiner literature mining
| Testing set | Gold-Standard VO-Paper association | Identified by VO-SciMiner | True Identification | Recall | Precision | F-measure |
|---|---|---|---|---|---|---|
| Positive 50 | 89 | 81 | 81 | 91% | 100% | 95% |
| Negative 50 | 0 | 1 | 0 | |||
| Total | 89 | 82 | 81 | 91% | 99% | 95% |
VO-based indexing of Brucella vaccine papers using VO-SciMiner
Literature indexing provides researchers with a means to navigate efficiently through a network of scholarly scientific articles in a subject domain. VO-SciMiner was employed to index Brucella papers with Brucella vaccine ontologies. For comparison purposes, typical PubMed searches were conducted using corresponding terms (Table 2). When no hierarchical VO structure information was used, SciMiner performed in a manner similar to that of a typical PubMed search. For example, the query of "Brucella vaccine" yielded 1,379 hits in PubMed Entrez and 1,359 hits in SciMiner. However, when the hierarchical structure of VO was used, VO-SciMiner returned significantly more papers than the typical PubMed searches. A PubMed search of "live attenuated Brucella vaccine" returned 74 hits, while the VO-based SciMiner search returned 922 hits. The primary reason for this increased number of hits is due to the addition of papers indexed with the child (subclass) vaccine terms, belonging to "live attenuated Brucella vaccines" in the VO hierarchy (Figure 2). For some vaccine terms, the VO-based SciMiner search increased the number of positive PubMed paper hits by up to 15-fold (Table 2).
Table 2.
VO-based indexing results using VO-SciMiner and PubMed (as of April 20, 2011)
| VO-SciMiner without child VOs | VO-SciMiner with child VOs | ||||
|---|---|---|---|---|---|
| PubMed Search Keywords | PubMed Entrez |
Total (only by VO-SciMiner) |
Common | Total (only by VO-SciMiner) |
Common |
| Brucella vaccine | 1,379 | 1,359 (0) | 1,359 | 2,155 (790) | 1,365 |
| Live attenuated Brucella vaccine | 74 | 85 (12) | 73 | 922 (849) | 73 |
| Live attenuated Brucella abortus vaccine | 52 | 50 (1) | 49 | 736 (647) | 49 |
| Live attenuated Brucella melitensis vaccine | 36 | 37 (1) | 36 | 204 (168) | 36 |
| Live attenuated Brucella suis vaccine | 5 | 5 (0) | 5 | 23 (18) | 5 |
The differences in literature retrieval can be explained by the differences in how query terms are interpreted. The PubMed Entrez system interprets the query "live attenuated Brucella vaccines" using MeSH-based querying system (Figure 3). VO-SciMiner does not perform such a complicated search. It uses the hierarchical structure of VO to include any subclass terms of a given query. MeSH in the PubMed also has such a feature, but does not include detailed vaccine terminologies in its vocabulary. In addition, as a formal ontology, VO also has logical semantic relations that allow formation of inferred classes for more advanced reasoning.
Figure 3.
Comparison between the Entrez system and VO-based literature search approaches. An example of searching "live attenuated Brucella abortus vaccine" is illustrated.
PubMed searches generally yielded high precision for most of the specific Brucella vaccines (data not shown). In some cases, however, our VO-based approach was able to significantly improve the precision. For example, a typical PubMed search using "Brucella suis (strain 2 OR S2)" had 63 hits as of April 20, 2011, while a corresponding VO (VO_0000722; B. suis strain 2) was identified in only 18 out of 14,947 Brucella related papers. Manual review of the results revealed that the PubMed search had only 12 true positives out of 63 hits, while VO-SciMiner indexing had 17 true positives out of 18 hits. This result demonstrates a significantly improved precision can be obtained using VO-based SciMiner indexing.
Identification of Brucella vaccine-associated genes
From 14,947 Brucella-related papers, 1,009 distinct interactions between 51 Brucella vaccine VO terms and 140 Brucella genes were identified. Grouped based on their functional annotations, the details of these 140 Brucella genes and their various annotations are listed in Table 3. Based on sub-cellular localization annotation, these 140 Brucella proteins are located in different sub-cellular areas, including outer membrane (10 proteins), cytoplasm (75 proteins), periplasm (10 proteins), cytoplasmic membrane (25 proteins), extracellular matrix (2 proteins) and unknown location (18 proteins).
Table 3.
Brucella genes in the gene-VO network
| Locus Tag | Gene symbol | Gene name | # of associated VO terms | # of papers - All VO terms (Live Attenuated vaccines) | Virulence factor or Protective antigen | Sub-cellular localization (PSORTb 3.0 score) |
|---|---|---|---|---|---|---|
| Nucleotide transport and metabolism | ||||||
| BMEI0296 | purE | phosphoribosylaminoimidazole carboxylase, catalytic subunit | 11 | 4 (4) | V | U (2.00) |
| BMEI0295 | purK | phosphoribosylaminoimidazole carboxylase ATPase subunit | 4 | 1 (1) | CM (7.88) | |
| Signal transduction mechanisms | ||||||
| BMEI2035 | bvrS | sensor histidine kinase BvrS, putative | 10 | 3 (1) | V | CM (10.00) |
| BMEI0190 | ptsP | phosphoenolpyruvate-protein phosphotransferase | 5 | 1 (1) | C (9.97) | |
| BMEI2036 | bvrR | DNA-binding response regulator BvrR, putative | 10 | 4 (2) | V | C (9.97) |
| Cell motility | ||||||
| BMEII0150 | fliC | flagellin family protein | 5 | 3 (0) | V | E (9.71) |
| Transcription | ||||||
| BMEII0665 | rnr | exoribonuclease, VacB/RNase II family | 6 | 1 (1) | C (9.97) | |
| BMEII0427 | eryD | erythritol transcriptional regulator | 2 | 1 (0) | C (8.96) | |
| BMEII1116 | vjbR | transcriptional regulator, LuxR family | 10 | 3 (2) | V | U (2.00) |
| BMEI0749 | rpoB | DNA-directed RNA polymerase subunit beta | 6 | 5 (1) | C (9.97) | |
| BMEI1649 | ureG-1 | urease accessory protein UreG | 2 | 1 (0) | C (9.97) | |
| BMEI2036 | bvrR | DNA-binding response regulator BvrR, putative | 10 | 4 (2) | V | C (9.97) |
| Amino acid transport and metabolism | ||||||
| BMEII0404 | leuB | 3-isopropylmalate dehydrogenase | 5 | 1 (1) | C (9.97) | |
| BMEI0101 | cysK | Cysteine synthase A | 6 | 1 (1) | V | C (9.26) |
| BMEI1653 | ureB-1 | urease subunit beta | 3 | 2 (0) | C (9.26) | |
| BMEII0561 | gcvP | glycine dehydrogenase | 5 | 1 (0) | V | C (9.97) |
| BMEI1654 | ureA-1 | urease subunit gamma | 3 | 2 (0) | C (9.26) | |
| BMEII0193 | potA | ABC transporter | 6 | 3 (2) | C (9.12) | |
| BMEII1054 | hisG | ATP phosphoribosyltransferase catalytic subunit | 2 | 1 (0) | C (9.97) | |
| BMEII0205 | dppF | ABC transporter | 6 | 3 (2) | CM (7.88) | |
| BMEI1324 | pepN | aminopeptidase N | 5 | 1 (1) | V | C (9.12) |
| BMEII0407 | asd | aspartate-semialdehyde dehydrogenase | 6 | 1 (0) | C (9.97) | |
| BMEI1652 | ureC-1 | urease subunit alpha | 3 | 3 (0) | C (9.97) | |
| BMEI0933 | cysK | cysteine synthase A | 6 | 1 (1) | V | C (9.97) |
| Secondary metabolites biosynthesis, transport and catabolism | ||||||
| BMEI1111 | acpXL | acyl carrier protein | 5 | 2 (0) | C (9.97) | |
| BMEI1475 | acpP | acyl carrier protein | 5 | 1 (0) | C (9.26) | |
| BMEI0546 | pncA | pyrazinamidase/nicotinamidase | 2 | 1 (0) | V | C (9.97) |
| Cell wall/membrane/envelope biogenesis | ||||||
| BMEI1829 | ropB | outer membrane protein, putative | 23 | 37 (17) | OM (10.00) | |
| BMEI1237 | galE | epimerase/dehydratase family protein, putative | 6 | 3 (1) | V | C (8.96) |
| BMEI1249 | omp25 | outer-membrane protein Omp25 | 11 | 17 (4) | V, P | OM (10.00) |
| BMEI0402 | omp31-1 | outer membrane protein Omp31 | 23 | 22 (9) | P | OM (10.00) |
| BMEII0844 | omp31-2 | outer membrane protein, 31 kDa | 19 | 17 (7) | OM (10.00) | |
| BMEI1413 | gmd | GDP-mannose 4,6-dehydratase | 5 | 2 (0) | V | C (9.97) |
| BMEI0997 | wbdA | glycosyl transferase, group 1 family protein | 16 | 3 (1) | V | U (2.00) |
| BMEI1416 | rfbE | O-antigen export system ATP-binding protein RfbE | 6 | 3 (2) | CM (7.88) | |
| BMEII0847 | wbjE | putative glycosyltransferase | 5 | 1 (0) | U (2.00) | |
| BMEI1393 | wbpZ | glycosyl transferase, group 1 family protein | 5 | 1 (0) | V | C (9.26) |
| BMEI1335 | omp | outer membrane lipoprotein-related protein | 12 | 5 (2) | U (2.00) | |
| BMEI0998 | wboA | glycosyl transferase WboA | 17 | 16 (10) | V | C (8.96) |
| BMEI0340 | pal | lipoprotein, Pal family | 12 | 7 (3) | P | OM (10.00) |
| BMEI0830 | yaeT | bacterial surface antigen | 23 | 37 (17) | OM (10.00) | |
| BMEI1417 | wbkB | wbkB protein | 5 | 2 (0) | V | U (2.00) |
| BMEI0509 | lpcC | lipopolysaccharide core biosynthesis mannosyltransferase LpcC | 9 | 2 (1) | C (9.97) | |
| BMEI1414 | perA | perosamine synthase, putative | 10 | 3 (1) | V | C (9.97) |
| BMEII0837 | hyaD | glycosyl transferase, group 2 family protein | 5 | 1 (0) | CM (9.82) | |
| BMEI1404 | wbkA | mannosyltransferase, putative | 16 | 7 (3) | V | C (9.26) |
| BMEII0253 | mepA | penicillin-insensitive murein endopeptidase | 5 | 1 (1) | P (9.76) | |
| Energy production and conversion | ||||||
| BMEII0404 | leuB | 3-isopropylmalate dehydrogenase | 5 | 1 (1) | C (9.97) | |
| BMEI0474 | petB | ubiquinol-cytochrome c reductase, cytochrome b | 12 | 4 (2) | CM (10.00) | |
| BMEII0429 | eryB | glycerol-3-phosphate dehydrogenase | 2 | 1 (0) | V | C (9.97) |
| BMEI0137 | mdh | malate dehydrogenase | 5 | 1 (1) | U (4.99) | |
| BMEI0140 | kgd | alpha-ketoglutarate decarboxylase | 11 | 8 (4) | C (9.97) | |
| BMEII0076 | tycC | enterobactin synthetase, component F, putative | 2 | 1 (0) | C (9.97) | |
| BMEI1547 | atpI | ATP sythase protein I, putative | 6 | 1 (1) | U (2.00) | |
| Replication, recombination and repair | ||||||
| BMEI0147 | xerC | site-specific tyrosine recombinase XerC | 5 | 1 (0) | C (9.26) | |
| BMEI0215 | ialA | dinucleoside polyphosphate hydrolase | 1 | 1 (0) | C (9.97) | |
| BMEI0884 | gyrA | DNA gyrase subunit A | 2 | 1 (0) | C (9.97) | |
| BMEI0040 | xerD | site-specific tyrosine recombinase XerD | 5 | 2 (0) | C (9.97) | |
| BMEI0880 | ssb | single-stranded DNA-binding protein family | 5 | 2 (1) | C (9.26) | |
| BMEI1823 | gyrB | DNA gyrase subunit B | 2 | 1 (0) | C (9.97) | |
| BMEI1200 | parC | DNA topoisomerase IV subunit A | 2 | 1 (0) | C (9.12) | |
| BMEI0878 | uvrA | excinuclease ABC subunit A | 5 | 2 (1) | V | C (9.97) |
| BMEI1307 | xerC | site-specific recombinase, phage integrase family | 5 | 1 (0) | C (9.97) | |
| BMEII0676 | parE | DNA topoisomerase IV subunit B | 2 | 1 (0) | C (9.97) | |
| BMEI1946 | mutM | formamidopyrimidine-DNA glycosylase | 1 | 1 (0) | V | C (9.97) |
| BMEII0739 | alkB | alkylated DNA repair protein AlkB | 6 | 1 (1) | U (2.00) | |
| BMEII0184 | insN | IS3 family element, transposase orfA | 6 | 1 (1) | U (2.00) | |
| BR1202 | recA | recombinase A | 2 | 1 (0) | V | C (10.00) |
| Posttranslational modification, protein turnover, chaperones | ||||||
| BMEI1650 | ureF | urease accessory protein UreF, putative | 2 | 1 (0) | U (2.00) | |
| BMEII1047 | groES | co-chaperonin GroES | 1 | 1 (0) | C (9.97) | |
| BMEI1041 | sufC | ABC transporter, ATP-binding protein | 6 | 3 (2) | C (9.97) | |
| BMEI2001 | dnaJ | chaperone protein DnaJ | 1 | 1 (0) | C (9.97) | |
| BMEI1330 | htrA | serine protease | 7 | 6 (3) | V | P (9.76) |
| BMEI2002 | dnaK | molecular chaperone DnaK | 12 | 7 (2) | V, P | C (9.97) |
| BMEII1048 | groEL | chaperonin GroEL | 10 | 9 (5) | C (9.97) | |
| BMEI1655 | ureD-1 | urease accessory protein UreD | 3 | 2 (0) | C (9.26) | |
| BMEI1649 | ureG-1 | urease accessory protein UreG | 2 | 1 (0) | C (9.97) | |
| BMEII0401 | trx-2 | thioredoxin | 5 | 2 (1) | C (9.26) | |
| BMEI1069 | tig | trigger factor | 10 | 2 (0) | V, P | C (8.96) |
| BMEI1651 | ureE | urease accessory protein UreE | 2 | 1 (0) | C (9.97) | |
| BMEI1060 | dsbA | outer membrane protein, putative | 23 | 37 (17) | V | U (2.00) |
| BMEI2022 | trx-1 | thioredoxin | 5 | 2 (1) | C (9.26) | |
| BMEI1265 | surA | peptidyl-prolyl cis-trans isomerase, putative | 9 | 1 (1) | P | P (9.76) |
| BMEI1492 | exsA | ABC transporter, ATP-binding/permease protein | 6 | 1 (1) | V | CM (10.00) |
| Translation, ribosomal structure and biogenesis | ||||||
| BMEI2010 | infC | translation initiation factor IF-3 | 2 | 2 (0) | C (9.97) | |
| BMEI0826 | frr | ribosome recycling factor | 5 | 2 (1) | C (9.97) | |
| BMEI0748 | rplL | 50S ribosomal protein L7/L12 | 16 | 17 (6) | P | U (6.49) |
| BMEI0752 | rpsL | 30S ribosomal protein S12 | 5 | 1 (1) | C (9.26) | |
| BMEI1497 | tlyA | hemolysin A | 1 | 2 (0) | C (8.96) | |
| Inorganic ion transport and metabolism | ||||||
| BMEII0003 | modC | molybdenum ABC transporter, ATP-binding protein | 6 | 3 (2) | C (9.12) | |
| BMEII0893 | katA | catalase | 6 | 2 (0) | P (10.00) | |
| BMEII0177 | znuC | zinc ABC transporter, ATP-binding protein | 6 | 3 (2) | V | CM (7.88) |
| BMEI0790 | phoA | bacterial alkaline phosphatase | 9 | 7 (2) | P (10.00) | |
| BMEII0581 | sodC | superoxide dismutase, Cu-Zn | 15 | 14 (8) | V, P | P (10.00) |
| BMEI1292 | fsr | fosmidomycin resistance protein | 2 | 1 (0) | CM (10.00) | |
| BMEII0108 | tauB | taurine ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (9.98) | |
| BMEI0635 | cbiO | cobalt ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (9.82) | |
| BMEII0704 | bfr | bacterioferritin | 12 | 4 (2) | P | C (9.97) |
| BMEII0178 | znuA | zinc ABC transporter, periplasmic zinc-binding protein | 9 | 1 (1) | V | P (9.76) |
| Coenzyme transport and metabolism | ||||||
| BMEII0589 | ribH | riboflavin synthase subunit beta | 12 | 8 (0) | C (9.97) | |
| BMEI2029 | ahcY | S-adenosyl-L-homocysteine hydrolase | 4 | 1 (1) | C (9.97) | |
| BMEI1099 | cobT | nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | 2 | 1 (0) | C (9.97) | |
| BMEI1187 | ribH | riboflavin synthase subunit beta | 12 | 8 (0) | C (9.97) | |
| Cell cycle control, cell division, chromosome partitioning | ||||||
| BMEII0470 | crcB | crcB family protein | 5 | 1 (0) | CM (10.00) | |
| General function prediction only | ||||||
| BMEII0288 | oppF | peptide ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (9.99) | |
| BMEI1081 | surE | stationary phase survival protein SurE | 8 | 5 (2) | C (8.96) | |
| BMEI0215 | ialA | dinucleoside polyphosphate hydrolase | 1 | 1 (0) | C (9.97) | |
| BMEI0920 | mazG | nucleoside triphosphate pyrophosphohydrolase | 5 | 1 (0) | C (8.96) | |
| BMEI1584 | ialB | invasion protein B | 1 | 2 (0) | P | U (2.50) |
| BMEII0355 | gal | D-galactose 1-dehydrogenase, putative | 5 | 1 (0) | C (9.97) | |
| Lipid transport and metabolism | ||||||
| BMEI1111 | acpXL | acyl carrier protein | 5 | 2 (0) | C (9.97) | |
| BMEI1553 | bacA | transport protein | 9 | 5 (4) | V | CM (10.00) |
| BMEI1475 | acpP | acyl carrier protein | 5 | 1 (0) | C (9.26) | |
| Carbohydrate transport and metabolism | ||||||
| BMEII0983 | chvE | sugar ABC transporter, periplasmic sugar-binding protein, putative | 9 | 1 (1) | U (5.02) | |
| BMEI1394 | manA | mannose-6-phosphate isomerase | 5 | 1 (0) | C (8.96) | |
| BMEI1237 | galE | epimerase/dehydratase family protein, putative | 6 | 3 (1) | V | C (8.96) |
| BMEII0430 | eryA | erythritol kinase | 5 | 2 (1) | C (9.26) | |
| BMEI0310 | gap | glyceraldehyde-3-phosphate dehydrogenase | 5 | 1 (1) | C (9.97) | |
| BMEII0750 | smoK | sugar ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (9.99) | |
| BMEI1416 | rfbE | O-antigen export system ATP-binding protein RfbE | 6 | 3 (2) | CM (7.88) | |
| BMEII0940 | smoK | sugar ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (9.99) | |
| BMEII0625 | ugpB | glycerol-3-phosphate ABC transporter, periplasmic glycerol-3-phosphate-binding protein | 1 | 1 (0) | V | P (9.76) |
| BMEI0309 | pgk | phosphoglycerate kinase | 6 | 1 (1) | C (9.97) | |
| BMEII0899 | manB | phosphoglucomutase, putative | 10 | 3 (1) | V | C (9.26) |
| BMEII0428 | eryC | D-erythrulose-1-phosphate dehydrogenase | 9 | 2 (1) | V | C (8.96) |
| ABM67295 | P39 | immunogenic 39-kDa protein | 15 | 7 (3) | P | P (9.44) |
| BMEII0982 | rbsA | sugar ABC transporter, ATP-binding protein, putative | 5 | 1 (1) | CM (9.82) | |
| BMEII0355 | gal | D-galactose 1-dehydrogenase, putative | 5 | 1 (0) | C (9.97) | |
| BMEI1396 | pmm | phosphomannomutase, putative | 10 | 5 (1) | V | C (9.97) |
| BMEII0145 | xylG | D-xylose ABC transporter, ATP-binding protein | 6 | 3 (2) | CM (7.88) | |
| BMEI1886 | pgm | phosphoglucomutase | 9 | 2 (1) | V | C (8.96) |
| BMEII0251 | glk | glucokinase | 5 | 1 (1) | C (9.97) | |
| Intracellular trafficking, secretion, and vesicular transport | ||||||
| BMEI1077 | yajC | preprotein translocase, YajC subunit | 6 | 1 (1) | CM (9.82) | |
| BMEI1076 | secD | protein-export membrane protein, SecD/SecF family | 6 | 1 (1) | CM (10.00) | |
| BMEII0026 | virB2 | type IV secretion system protein VirB2 | 9 | 3 (1) | V | CM (9.46) |
| BMEII0028 | virB4 | type IV secretion system protein VirB4 | 9 | 4 (1) | V | CM (10.00) |
| BMEII0029 | virB5 | type IV secretion system protein VirB5 | 9 | 1 (1) | V | U (2.00) |
| BMEII0032 | virB8 | type IV secretion system protein VirB8 | 9 | 1 (0) | V | U (2.00) |
| BMEII0025 | virB1 | type IV secretion system protein VirB1 | 5 | 1 (0) | V | E (9.64) |
| BMEII0034 | virB10 | type IV secretion system protein VirB10 | 5 | 1 (1) | V | U (4.90) |
| Unclassified (function unknown) | ||||||
| CAA86936 | BLS | Brucella lumazine synthase | 19 | 12 (2) | P | C (9.97) |
| BMEI1305 | omp2b | porin Omp2b | 16 | 21 (9) | OM (9.93) | |
| BMEI1306 | omp2a | porin Omp2a | 13 | 17 (8) | OM (9.93) | |
| BMEI0135 | omp19 | lipoprotein Omp19 | 12 | 10 (5) | V, P | OM (10.00) |
| BMEII0017 | omp10 | lipoprotein Omp10 | 9 | 6 (2) | V | OM (10.00) |
| BMEI0536 | omp28 | immunoreactive 28 kDa outer membrane protein | 19 | 15 (11) | P | P (10.00) |
| BMEI0330 | opgC | opgC protein, putative | 5 | 1 (1) | CM (10.00) | |
| BMEI0634 | crcB | crcB family protein | 5 | 1 (0) | CM (10.00) | |
| BMEI0545 | pncA | hypothetical protein | 5 | 1 (0) | V | CM (10.00) |
Note: Sub-cellular localization was predicted using PSORTb 3.0 implemented in Vaxign. PSORTb score ranges from 0 (0% probability) to 10 (100% probability). Proteins with a score below 7.44 are classed as unknown. The abbreviations in this table are C (Cytoplasmic), CM (CytoplasmicMembrane), E (Extracellular), OM (OuterMembrane), P (Periplasmic) and U (Unknown).
Based on their roles in host-pathogen interactions, the 140 identified Brucella genes associated with Brucella vaccines can be classified into three groups: (1) protective antigens, (2) virulence factors, and (3) unknown. Protective antigens elicit a protective immune response, and are frequently used for vaccine development. VO-SciMiner identified all 14 known protective Brucella antigens [12]. Virulence factors are expressed and secreted by pathogens and often responsible for causing diseases in the host. Out of the 140 Brucella genes associated with Brucella vaccines, 46 are known Brucella virulence factors based on the updated curation information from the Brucella Bioinformatics Portal (BBP; http://www.phidias.us/bbp) [10,22] (Table 3). In total, 81 Brucella genes are neither known protective antigens nor virulence factors. Since these genes have been studied in the context of Brucella vaccines, it seems possible that some of them are potential virulence factors and/or protective antigens. For example, RopB has not been implicated in inducing immune protection. However, our recent reverse vaccinology analysis using Vaxign [16] found that Brucella abortus RopB is a potential Brucella adhesin with a probability of 0.815 [12]. This could make RopB a promising target for future vaccine development as a potential protective antigen.
A functional analysis of the 140 genes and their subgroups is critical to illustrate their association with Brucella vaccine research. Brucella genes have been well categorized with the COG system [18]. We performed a functional enrichment analysis identifying significantly over-represented COG categories within all 140 genes or 46 virulence factors. These include: "carbohydrate transport and metabolism", "cell wall, membrane, and envelope biogenesis", "intracellular trafficking, secretion, and vesicular transport", and "posttranslational modification, protein turnover, chaperones" (Table 4).
Table 4.
COG functional analysis of Brucella genes associated with vaccine research
| VO-associated Genes (n = 140) |
Virulent Genes (n = 46) |
|
|---|---|---|
| COG Description | # of genes (p-value) | # of genes (p-value) |
| Amino acid transport and metabolism | 12 (0.771) | 4 (1.000) |
| Carbohydrate transport and metabolism | 19 (0.000*) | 6 (0.043*) |
| Cell cycle control, cell division, chromosome partitioning | 1 (1.000) | |
| Cell motility | 1 (1.000) | 1 (0.341) |
| Cell wall/membrane/envelope biogenesis | 22 (0.000*) | 9 (0.001*) |
| Coenzyme transport and metabolism | 4 (0.820) | |
| Energy production and conversion | 5 (0.843) | 1 (0.728) |
| Inorganic ion transport and metabolism | 10 (0.149) | 3 (0.475) |
| Intracellular trafficking, secretion, and vesicular transport | 7 (0.003*) | 5 (0.000*) |
| Lipid transport and metabolism | 3 (0.627) | 1 (1.000) |
| Nucleotide transport and metabolism | 2 (0.769) | 1 (1.000) |
| Posttranslational modification, protein turnover, chaperones | 16 (0.000*) | 5 (0.025*) |
| Replication, recombination and repair | 14 (0.001*) | 3 (0.428) |
| Secondary metabolites biosynthesis, transport and catabolism | 3 (0.750) | 1 (0.601) |
| Signal transduction mechanisms | 3 (1.000) | 2 (0.357) |
| Transcription | 6 (0.704) | 2 (1.000) |
| Translation, ribosomal structure and biogenesis | 5 (0.684) | |
Note: * indicates p < 0.05.
Interestingly, the COG category "replication, recombination and repair" is significantly over-represented in the 140 genes (p-value = 0.001); however, this category is not enriched in the virulence factor group (p-value = 0.428). In total, 14 out of 140 Brucella genes belong to this category (Table 3). Among them, three genes (uvrA, mutM, and recA) are known virulence factors. Many of the remaining genes are virulence factors in other pathogens. For example, XerD, an enzyme responsible for resolving chromosomal multimers prior to chromosome segregation, is a virulence factor in E. coli and S. aureus [23,24]. Mutations of gryA and parC are responsible for generation of quinolone-resistance in many bacteria (e.g., Pseudomonas aeruginosa) [25]. These two genes are critical for antibiotics resistance in virulent bacteria. Therefore, it is reasonable to hypothesize that many of the genes in this category may be potential Brucella virulence factors.
Identification of Brucella vaccine-associated gene networks
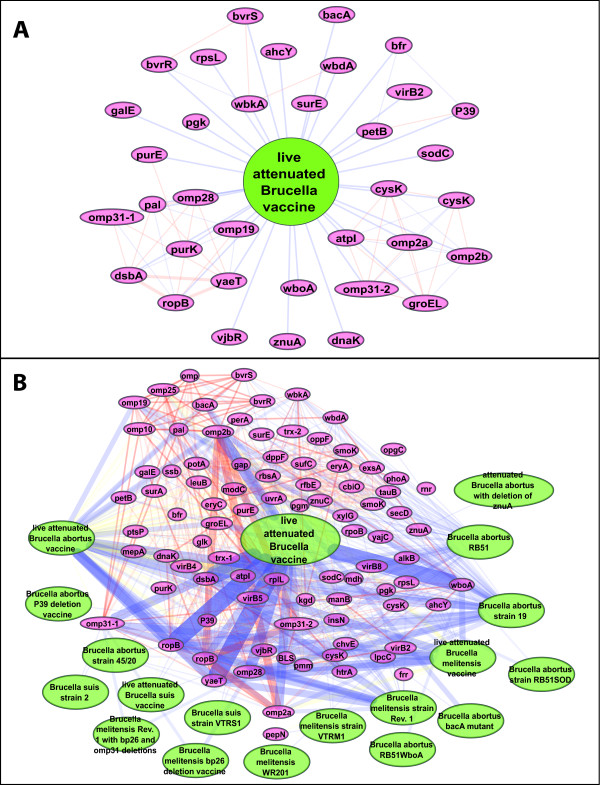
Difference in the volume of the retrieved literature can result in a substantial difference in obtainable knowledge. If one is interested in establishing the interaction network of pathogen genes and vaccines, the networks (co-citations of gene and vaccine), based on the PubMed search and the VO-SciMiner search, can be substantially different (Figure 4). Using 74 papers retrieved by PubMed, the interaction network between genes and live attenuated Brucella vaccines is extremely limited (Figure 4A). Only 35 genes are associated with live attenuated Brucella vaccines. However, when the VO hierarchy of 'live attenuated Brucella vaccine' (Figure 2) is considered, 89 (54 more) genes are identified by VO-SciMiner (Figure 4B).
Figure 4.
Networks between genes and live attenuated Brucella vaccines identified by VO-SciMiner. Searches of "live attenuated Brucella vaccine" using (A) the PubMed Entrez system and (B) the VO-SciMiner system. Edge color represents the types of association; blue for gene-vaccine association and red for gene-gene association.
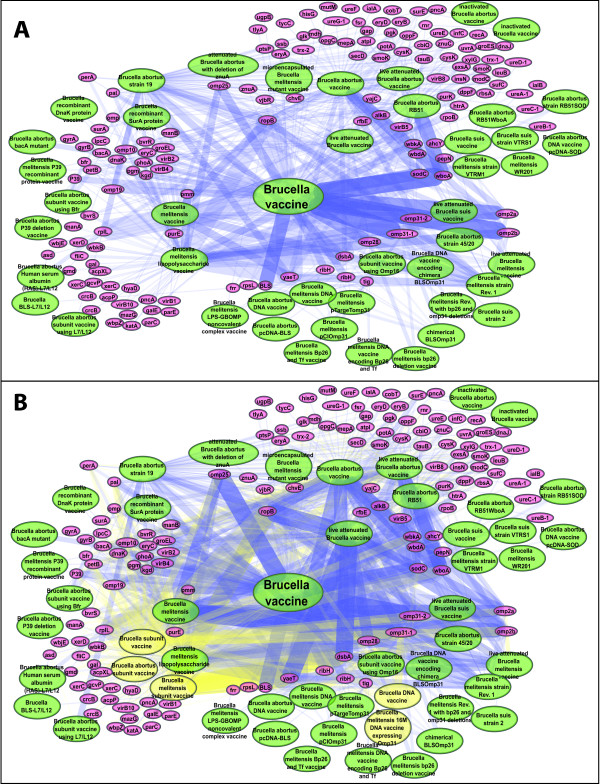
Similarly, without VO hierarchy, the Brucella vaccine-gene network is relatively sparse (Figure 5A), including only 580 vaccine-gene associations. When VO hierarchy is incorporated, the Brucella vaccine-gene network becomes more intense with 1,009 vaccine-gene associations (Figure 5B). This is equivalent to a 74% increase in the number of vaccine-gene associations, resulting in a more comprehensive gene-vaccine association network.
Figure 5.
Brucella vaccine and gene co-citation network. All Brucella vaccine terms in VO were used. This co-citation network was generated without (A) or with (B) VO hierarchy. VO terms are shown in green and Brucella gene in red. The line depth between VO vaccine terms and Brucella gene symbols represents the relative number of documents with each pair of VO vaccine terms and Brucella genes are co-cited. The terms and edges in yellow were inferred from the VO hierarchy.
Three types of interactions are observed in the Brucella vaccine-gene interaction networks (Figures 4 and 5). The first type of interaction is a vaccine-vaccine association, which results from the asserted is_a hierarchy between a vaccine and its parent or child terms as asserted in VO (e.g., Brucella abortus vaccine and Brucella abortus vaccine RB51), or from the inferred VO hierarchy, for example, live attenuated Brucella vaccine and Brucella abortus vaccine RB51 (Figure 2).
Another type of interaction in the Brucella network is vaccine-gene interaction. Many vaccine-gene interactions are based on direct association identified from the literature. Such an example would be an association between Brucella RB51 strain vaccine and the wboA gene [26]. Alternatively, further vaccine-gene interactions are based on asserted or inferred hierarchy by VO reasoning. Since RB51 is inferred to be a live attenuated Brucella vaccine, an inferred association between live attenuated Brucella vaccine and the wboA gene can be made. Indeed, based on the finding of a wboA mutation in RB51, many live attenuated Brucella vaccine candidates, such as VTRS1 and VTRM1 [27], have been developed by mutating the wboA sequence in various wild type Brucella strains.
The present study also reveals that four B. suis vaccine-gene interactions are isolated from the other interactions, suggesting that these vaccine-gene interactions were only studied in the context of B. suis, but not for other Brucella species. The inferred vaccine-gene associations are potentially valuable for systematic analysis of genes associated with less-specific type of vaccines (e.g., live attenuated Brucella vaccines) instead of specific vaccines such as RB51.
The last type of interaction in the Brucella network is gene-gene interaction. The obvious gene-gene interactions in a network are found between those genes co-cited in the same paper(s). In addition, many new gene-gene interactions can be inferred through their association with the same vaccines. In a gene-gene interaction network under the scope of live attenuated Brucella vaccine based on PubMed search (Figure 4A), two genes in a two-component system, bvrR and bvrS are associated, and are linked to one gene, wbkA (Figure 4A). When the vaccine hierarchy was used in VO-SciMiner, eight more genes were detected that are associated with bvrR and bvrS under the same scope of live attenuated Brucella vaccine. VO-SciMiner also detects what specific live attenuated Brucella vaccines (e.g., B. abortus vaccine RB51) interact with a gene (e.g., wboA). New hypotheses can also be generated based on the gene-gene network under a Brucella vaccine domain. For example, Brucella bvrS and bvrR are only associated directly with B. abortus vaccines (Figure 4A and 4B). However, many genes (e.g., wbdA and wbkA) that interact with bvrS and bvrR are also associated with live attenuated B. melitensis and B. suis vaccines. Therefore, it is reasonable to hypothesize that the mutants of bvrR or bvrS gene in B. melitensis and B. suis may also be candidates for live attenuated Brucella vaccines.
A web server for browsing and analyzing VO-SciMiner literature mining results
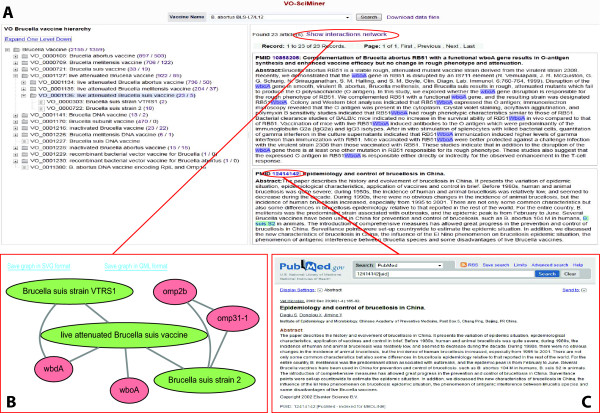
A web-based application of VO-SciMiner-based Brucella paper indexing has been developed (http://www.violinet.org/vo-sciminer). This webpage provides two ways to explore the Brucella vaccine literature (Figure 6A). The first option is to select a vaccine using the dropdown list. A list of publications related to the selected vaccine will be displayed on the lower right panel when the search button is clicked. For each publication, VO-SciMiner lists the PubMed ID, the title, and an abstract if available. In the abstract section, all the related vaccines and genes are highlighted and linked to a page containing detailed information of corresponding vaccines and genes. Another way to select a vaccine is by navigating through the hierarchical structure of different vaccines on the lower left panel. This panel lists all the Brucella vaccines on an inferred tree structure. If available, two numbers will be shown for each node. The first number is the total number of publications related to this node and any of its child nodes. The second number shows the number of publications related only to this node. If any of the numbers are clicked, the lower right panel will display a detailed list of publications. A hyperlink 'Show interactions network' will appear at the top of the screen. This will bring up a new display of the vaccine and gene interaction network (Figure 6B).
Figure 6.
Screen shot of VO-SciMiner webpage. Two panels are illustrated in this web page with "VO_0001136: live attenuated Brucella suis vaccine" selected. The left panel shows the hierarchical structure of the ontology term 'Brucella vaccine' and its child terms in VO. Two numbers are displayed next to each of the VO terms. The first number represents the total number of search hits including the research results from all the child terms (if any) and the second number is the number of hits for the current term. Once either number is clicked, the abstract details are displayed on the right panel. The identified VO terms and Brucella genes are highlighted.
Discussion
The present study demonstrates that the application of Vaccine Ontology in literature mining enhances the retrieval of Brucella vaccine literature. Term variation rules, inclusion vaccine synonyms, and the inference of vaccine terms through the hierarchy increases the amount of literature that can be retrieved. VO-based SciMiner is capable of identifying up to 15 times more Brucella vaccine papers than the typical PubMed search using the Entrez system. The increase in the retrieved literature results in the generation of a comprehensive co-citation network of Brucella genes and Brucella vaccines, revealing key vaccine-gene associations and provides potential valuable opportunities for the development of new vaccines.
To our knowledge, this study is the first manuscript that compares MeSH and specific domain ontology (e.g., Vaccine Ontology in this case) for literature indexing. The results show that VO can be used to improve literature searching for Brucella vaccines. While MeSH provides reasonable searching specificity, its level of sensitivity may be very low for many biomedical research areas (e.g., vaccines). For example, "Brucella vaccine" is currently one of the lowest level (the most specific) terms in MeSH, and no individual Brucella vaccines such as RB51 are indexed by MeSH. In contrast, VO-based SciMiner contains detailed vaccine terminologies in a controlled hierarchical structure, allowing searches across the hierarchy, thereby achieving a very significant recall.
Therefore, it is possible to substantially increase PubMed searching sensitivity by incorporating ontology information. Increased sensitivity in turn will result in increasing the amounts of information obtained on any given search topic. As illustrated in the gene-vaccine networks in Figure 4, the level of obtainable information from papers either retrieved by PubMed (A) and VO-SciMiner (B) is substantial. Based on the hierarchy structure of VO, the VO-based approach can retrieve a more comprehensive network. Therefore, it is possible to use a community-based ontology (e.g., VO) to replace, or enhance, a part of a MeSH-based document retrieval system such as PubMed in some domains (e.g., in vaccine).
Using ontologies to support literature mining has become an active area of research over the last decade. Multiple text-mining tools and web-resources, including GoPubMed [28], SEGOPubMed [29], and GoWeb [30] have been developed to link PubMed papers to ontologies, particularly Gene Ontology, within the biomedical data. Using the domain knowledge and hierarchical organization, these tools allow users to perform ontology-based PubMed document browsing. However, they are limited by lack of coverage in the vaccine domain as in the PubMed Entrez search system. BioPortal (http://bioportal.bioontology.org), a steadily growing online resource developed by the National Center for Biomedical Ontology (NCBO), currently indexes multiple biomedical public resources. These include gene expression and protein-protein interaction data sets, which are comprised of over 250 ontologies and 4 million ontology terms. The BioPortal provides a convenient web interface. Unfortunately, PubMed biomedical literature is not supported in the BioPortal yet. In this study, we demonstrated that the newly developed VO provides a powerful system for ontology-based vaccine literature indexing, through the dictionary- and rule-based VO-SciMiner system. It offers the further advance of a significantly enhanced recall and precision. The dictionary- and rule-based literature mining system for optimization of a vaccine term searching is not presently found in the BioPortal or any other literature mining system.
In an effort to improve vaccine literature mining and to support the vaccine research community, VO-SciMiner has been integrated into the VIOLIN vaccine research database and data analysis system (http://www.violinet.org)[11]. VIOLIN includes all the vaccine literature from PubMed and provides multiple tools for navigating various vaccines and relevant literature. These tools include (1) Litesearch providing basic vaccine literature keyword searching feature; (2) Vaxmesh, a MeSH-based visualization tool for displaying the MeSH hierarchy and the numbers of vaccine papers associated with each MeSH terms; (3) Vaxpresso, a VIOLIN vaccine literature mining program powered by Textpresso natural language processing (NLP) program [31], for identifying genes and proteins in literature data. Initially, we intended to integrate VO into our Vaxpresso. However, Textpresso, the base system of Vaxpresso, had a limitation of slow-execution since it is a plain text file-based system rather than a relational database-based program. Compared to these existing VIOLIN text mining tools, VO-SciMiner is a significant extension because it is the first NLP literature mining program that integrates VO terms and VO hierarchy for efficient vaccine literature indexing. This study also represents the first effort to apply VO to studying the interaction network between vaccines and microbial genes.
In addition to improving the retrieval of Brucella vaccine-related papers by incorporating VO into SciMiner, SciMiner was extended to identify Brucella pathogen genes reported in the literature. Comprehensive co-citation networks of Brucella vaccines and Brucella genes were generated as illustrated in Figures 4 and 5. These comprehensive networks not only illustrate direct gene-vaccine associations co-cited in the literature, but also reveal indirect associations inferred through the VO hierarchy. These literature-based networks can enhance vaccine development, since they allow the generation of additional hypotheses. As demonstrated in this paper, many Brucella genes are potential new protective antigens and/or virulence factors. Protective antigens are defined as those genes that are able to induce protection in vivo. Mutation of a virulence factor gene in Brucella results in an attenuated mutant strain, which can be a potential live attenuated vaccine. Therefore, it is suggestive that genes identified by the presently described methodology can serve possible targets for development of new Brucella vaccines.
It should be noted that the text mining approach implemented in VO-SciMiner has its own limitations. The co-citation-based VO-SciMiner is still in an early stage of being applied to the field of vaccinology research. The identification of potential new gene-vaccine associations based on a text mining method may include false-positives due to excellent but still imperfect performance (an F-measure of 95%). Besides, co-citation of both a gene and a vaccine in the same document does not necessarily mean that they are functionally associated. Therefore, any interesting association will require careful consideration and experimental investigation. In spite of these limitations, text mining studies have proven useful in identifying previously unknown knowledge such as predicted combinatorial binding of transcriptional factors to regulatory elements [32], and reconstructing protein-protein interactions and pathways [33].
Our current study has also generated predictions that form new hypotheses for further experimental studies. To make our current approach more robust, the gene-vaccine text mining method may be improved in the future by retrieving additional data, including the types of interactions, the animal or cell types used, and specific experimental designs and conditions. Furthermore, the text mining prediction approach becomes more powerful and specific when it is integrated with other computational and experimental methods such as microarray and proteomics studies [34-36].
Brucella vaccines were used as examples in this study. In the near future, other bacterial vaccines will be added to the VO-SciMiner system. Incorporating additional rules for species other than Brucella may be challenging if other bacterial vaccines have substantially different structures than Brucella VOs. However, the Brucella case study as presented provides a general strategy and framework to address these issues. The VO-SciMiner indexing strategy can also be used to improve other gene interaction network studies. For example, previous studies presented the retrieval of an IFN-γ and vaccine-mediated immune network based on a genome-wide centrality-based literature discovery method [37], which was improved by application of VO [38]. However, it is suggestive that further improvements may be made in the identification of vaccine and gene entities contained in the literature using the VO-SciMiner indexing approaches. The indexing of human and mouse genes for studying the interactions between Brucella vaccines and host genes is currently under investigation. The approach taken in this study is generic and can be applied to analyze other vaccines listed in VO.
Conclusions
We have developed a methodology that incorporates Vaccine Ontology into literature mining to improve relevant paper retrieval in the domain of vaccine research. To test our methodology, Brucella vaccines were used. Our analyses indicate that the VO-based SciMiner method substantially increases retrieval of associated data, leading to an improved analysis and understanding of the vaccine-gene networks.
List of abbreviations used
MeSH: Medical Subject Headings; VO: Vaccine Ontology; OBO: Open Biological and Biomedical Ontologies; BFO: Basic Formal Ontology; COG: Clusters of Orthologous Groups.
Authors' contributions
JH conceived and coordinated the project, developed the VO-SciMiner, generated all figures and tables, and drafted the manuscript. ZX implemented VO-SciMiner web server, and participated in writing of the manuscript. ELF supervised the project and edited the manuscript. YH conceived and supervised the project, served as the domain expert in interpretation of the results, and drafted and edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Junguk Hur, Email: juhur@umich.edu.
Zuoshuang Xiang, Email: zxiang@med.umich.edu.
Eva L Feldman, Email: efeldman@med.umich.edu.
Yongqun He, Email: yongqunh@umich.edu.
Acknowledgements
This work was supported by grant funding from National Institutes of Health [U54-DA021519 to ELF, R01-AI081062 to YH]; and a Rackham Pilot Research grant at the University of Michigan.
References
- Almond JW. Vaccine renaissance. Nat Rev Microbiol. 2007;5(7):478–481. doi: 10.1038/nrmicro1702. [DOI] [PubMed] [Google Scholar]
- American-Diabetes-Association. Economic costs of diabetes in the U.S. In 2007. Diabetes Care. 2008;31(3):596–615. doi: 10.2337/dc08-9017. [DOI] [PubMed] [Google Scholar]
- Bradac J, Dieffenbach CW. HIV vaccine development: Lessons from the past, informing the future. IDrugs. 2009;12(7):435–439. [PubMed] [Google Scholar]
- Perkins SD, Smither SJ, Atkins HS. Towards a Brucella vaccine for humans. FEMS Microbiol Rev. 2010. [DOI] [PubMed]
- Disease NIoAaI. The Jordan Report, Accelerated Development of Vaccines 2007. 2007.
- Medical Subject Headings - Fact Sheet. http://www.nlm.nih.gov/pubs/factsheets/mesh.html
- He Y, Cowell L, Diehl AD, Mobley H, Peters B, Ruttenberg A, Scheuermann RH, Brinkman RR, Courtot M, Mungall C, VO: Vaccine Ontology. International Conference on Biomedical Ontology: 2009; Buffalo, NY. http://dx.doi.org/10.1038/npre.2009.3552.1
- Hur J, Schuyler AD, States DJ, Feldman EL. SciMiner: web-based literature mining tool for target identification and functional enrichment analysis. Bioinformatics. 2009;25(6):838–840. doi: 10.1093/bioinformatics/btp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J, Sullivan KA, Schuyler AD, Hong Y, Pande M, States DJ, Jagadish HV, Feldman EL. Literature-based discovery of diabetes- and ROS-related targets. BMC Medical Genomics. 2010;3:49. doi: 10.1186/1755-8794-3-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Zheng W, He Y. BBP: Brucella genome annotation with literature mining and curation. BMC Bioinformatics. 2006;7:347. doi: 10.1186/1471-2105-7-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Todd T, Ku KP, Kovacic BL, Larson CB, Chen F, Hodges AP, Tian Y, Olenzek EA, Zhao B, VIOLIN: vaccine investigation and online information network. Nucleic acids research. 2008. pp. D923–928. [DOI] [PMC free article] [PubMed]
- He Y, Xiang Z. Bioinformatics analysis of Brucella vaccines and vaccine targets using VIOLIN. Immunome Res. 2010;6(Suppl 1):S5. doi: 10.1186/1745-7580-6-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A, Lu Z, Wang X, Cohen A, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J. et al. Overview of BioCreative II gene normalization. Genome Biology. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta OR, Folkerts O, Fei Z, Mane SP, Evans C, Martino-Catt S, Bricker B, Yu G, Du L, Sobral BW. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS One. 2008;3(5):e2193. doi: 10.1371/journal.pone.0002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006. pp. D363–368. [DOI] [PMC free article] [PubMed]
- He Y, Xiang Z, Mobley HL. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol. 2010;2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ. et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. Journal of the Royal Statistical Society. 1922;85(1):87–94. doi: 10.2307/2340521. [DOI] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ. et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Tian Y, He Y. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol. 2007;8(7):R150. doi: 10.1186/gb-2007-8-7-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker AF, Lupas A, Ingraham K, So CY, Lunsford RD, Li T, Bryant A, Holmes DJ, Marra A, Pearson SC. et al. Genetic characterization of gram-positive homologs of the XerCD site-specific recombinases. J Mol Microbiol Biotechnol. 2000;2(2):225–233. [PubMed] [Google Scholar]
- Eskra L, Canavessi A, Carey M, Splitter G. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect Immun. 2001;69(12):7736–7742. doi: 10.1128/IAI.69.12.7736-7742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran QT, Nawaz MS, Deck J, Foley S, Nguyen K, Cerniglia CE. Detection of type III secretion system virulence and mutations in gyrA and parC genes among quinolone-resistant strains of Pseudomonas aeruginosa isolated from imported shrimp. Foodborne Pathog Dis. 2011;8(3):451–453. doi: 10.1089/fpd.2010.0687. [DOI] [PubMed] [Google Scholar]
- Olsen SC, Boyle SM, Schurig GG, Sriranganathan NN. Immune responses and protection against experimental challenge after vaccination of bison with Brucella abortus strain RB51 or RB51 overexpressing superoxide dismutase and glycosyltransferase genes. Clin Vaccine Immunol. 2009;16(4):535–540. doi: 10.1128/CVI.00419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AJ, Schurig GG, Boyle SM, Sriranganathan N, Bevins JS, Enright FM, Elzer PH, Kopec JD. Protection of BALB/c mice against homologous and heterologous species of Brucella by rough strain vaccines derived from Brucella melitensis and Brucella suis biovar 4. Am J Vet Res. 1996;57(5):677–683. [PubMed] [Google Scholar]
- Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005. pp. W783–786. [DOI] [PMC free article] [PubMed]
- Vanteru BC, Shaik JS, Yeasin M. Semantically linking and browsing PubMed abstracts with gene ontology. BMC Genomics. 2008;9(Suppl 1):S10. doi: 10.1186/1471-2164-9-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietze H, Schroeder M. GoWeb: a semantic search engine for the life science web. BMC Bioinformatics. 2009;10(Suppl 10):S7. doi: 10.1186/1471-2105-10-S10-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2(11):e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Ni S, Miranker DP, Iyer VR. Predicting combinatorial binding of transcription factors to regulatory elements in the human genome by association rule mining. BMC Bioinformatics. 2007;8:445. doi: 10.1186/1471-2105-8-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiadou S, Pyysalo S, Tsujii J, Kell DB. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28(7):381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Faro A, Giordano D, Spampinato C. Combining literature text mining with microarray data: advances for system biology modeling. Briefings in bioinformatics. 2011. [DOI] [PubMed]
- Hodges AP, Dai D, Xiang Z, Woolf P, Xi C, He Y. Bayesian network expansion identifies new ROS and biofilm regulators. PLoS One. 2010;5(3):e9513. doi: 10.1371/journal.pone.0009513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, Erhardt RA, Valencia A. Text-mining approaches in molecular biology and biomedicine. Drug discovery today. 2005;10(6):439–445. doi: 10.1016/S1359-6446(05)03376-3. [DOI] [PubMed] [Google Scholar]
- Ozgur A, Xiang Z, Radev DR, He Y. Literature-based discovery of IFN-gamma and vaccine-mediated gene interaction networks. J Biomed Biotechnol. 2010;2010:426479. doi: 10.1155/2010/426479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur A, Xiang Z, Radev DR, He Y. Mining of vaccine-associated IFN-gamma gene interaction networks using the Vaccine Ontology. Journal of Biomedical Semantics. 2011;2(Suppl 2):S8. doi: 10.1186/2041-1480-2-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]