Abstract
In the course of developing a low-density lipoprotein receptor (LDLR) gene therapy treatment for homozygous familial hypercholesterolemia (HoFH), we planned to examine the efficacy in a nonhuman primate model, the rhesus macaque heterozygous for an LDL receptor mutation fed a high-fat diet. Unexpectedly, our initial cDNA sequencing studies led to the identification of a heretofore unidentified splicing isoform of the rhesus LDLR gene. Compared with the publicly available GenBank reference sequence of rhesus LDLR, the novel isoform contains a 21 bp in frame insertion. This sequence coincides with part of exon 5 and creates a site for the restriction enzyme MscI. Using this site as a marker for the 21 bp in-frame insertion, we conducted a restriction enzyme screen to examine for the prevalence of the novel isoform in rhesus liver tissue cDNA and its homolog in human liver tissue cDNA. We found that the novel isoform is the predominant LDLR cDNA found in rhesus liver and the sole LDLR cDNA found in human liver. Finally, we compared the in vivo functionality of the novel and previously identified rhesus LDLR splicing isoforms in a mouse model of HoFH.
Keywords: low-density lipoprotein receptor, familial hypercholesterolemia, adeno-associated virus, isoform
the low-density lipoprotein receptor (LDLR) gene family consists of cell surface proteins involved in receptor-mediated endocytosis of cholesterol-carrying lipoprotein particles into cells. The LDLR itself binds low-density lipoprotein (LDL) particles at the plasma membrane, internalizes them, and releases them in the low-pH environment of the endosome where the particles are degraded and the cholesterol is made available for repression of microsomal enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase, the rate-limiting step in cholesterol synthesis (25). Loss-of-function mutations in the LDLR gene cause the autosomal codominant disorder familial hypercholesterolemia (FH). Homozygous FH (HoFH) is associated with severe hypercholesterolemia with LDL-C >500 mg/dl and severe atherosclerotic cardiovascular disease in childhood or adulthood (22). HoFH is a good candidate for liver-directed LDLR-based somatic gene therapy (9).
We have previously demonstrated that adeno-associated virus serotype 8 (AAV8)-mediated delivery of human or mouse LDLR leads to long-term correction and stabilization of cholesterol levels in mouse models of HoFH (9, 13). An important step in the preclinical development of AAV8.LDLR gene therapy for FH is assessment of its efficacy and safety in a nonhuman primate (NHP) model. Previous studies have shown that rhesus macaques simulate humans with respect to the presence of latent AAV genomes widely distributed through multiple tissues and the associated host memory immune responses (6). Within this context, rhesus macaques provide an important NHP animal model enabling for the assessment of host vector interactions on stability of transgene expression and immune based toxicities.
Hummel and colleagues (8) have previously described a family of rhesus macaques that demonstrated persistent elevations in plasma cholesterol. Analysis of skin fibroblasts demonstrated a 50% reduction in the LDLR activity that was subsequently shown to be due to a nonsense mutation in exon 6 of the LDLR gene that led to the expression of a dysfunctional and truncated protein. Attempts to generate a homozygous FH macaque have failed for reasons that are unclear. It has been speculated that there may be a recessive genetic defect linked to this mutation that when bred to homozygosity is associated with embryonic lethality. Feeding a high-fat diet to LDLR+/− rhesus macaques leads to marked downregulation of LDLR expression from the normal allele and induces levels of hypercholesterolemia comparable to those seen in homozygous FH patients (19, 23).
The LDLR gene encoding human LDLR is 45 kb and is localized on chromosome 19p13.1–13.3; it is composed of 18 exons and 17 introns encoding an mRNA of 5.3 kb and a protein of 860 amino acids (25, 29). At the amino acid sequence level, there is 95% homology between human LDLR (GenBank accession no. BT007361) and rhesus macaque LDLR (GenBank accession no. AY466854). By contrast, there is 78% homology between mouse (Z19521) and human LDLR. The LDLR gene encodes a signal peptide (exon 1), a ligand-binding domain (exons 2–6), an epidermal growth factor homology domain (exons 7–14), an O-linked sugar domain (exon 15), a transmembrane domain (exons 16, 17), and a cytoplasmic domain (exons 17, 18). These individual domains exhibit amino acid sequence identities of 85, 95, 97, 85, 90, and 96%, respectively between human and rhesus LDLR. Despite the tremendous homology between human and rhesus LDLR, it is important to use a transgene that is syngeneic to the recipient model; this ensures that host/vector interactions are faithfully modeled with respect to efficacy, dose, and toxicity.
During our efforts of developing an AAV8-based vector encoding rhesus (rh) LDLR for use in the high-fat diet-fed LDLR−/+ rhesus macaque animal model of HoFH, we sequenced the LDLR cDNA from rhesus liver and unexpectedly identified a novel splice isoform of rhLDLR that is the predominant form in rhesus liver. Using AAV8, we compared the functionality of both the novel and the previously identified rhLDLR isoforms in vivo and found both of them to be equally effective in reducing total cholesterol levels in a mouse model of homozygous FH.
METHODS
RNA isolation and cDNA preparation.
Total RNA from hepatocytes and peripheral blood mononuclear cells (PBMCs) was isolated using QIAamp RNA Blood Mini Kit (Qiagen). Rhesus macaque liver tissues were obtained from an in house colony of animals. Human liver tissues were obtained from National Disease Research Interchange (NDRI). Total RNA (5 μg) was reverse transcribed using Super-Script II reverse transcriptase (Invitrogen) and 100 ng random hexameres (Invitrogen) according to the manufacturer's protocol.
RT-PCR, MscI screen, and sequencing.
Primers were designed to produce PCR products corresponding to exons 4 and 5 of the LDLR open reading frame. PCR was performed using 1 μg total cDNA, 5 pmol primers exons 4 and 5F, 5′-CAAGACGTGCTCCCA-3′ and exons 4 and 5R, 5′-CCAACTTCATCGCTCA-3′ and Qiagen OneStep RT-PCR Kit. The thermal cycling conditions were 38 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 60 s. The amplified products were digested with MscI according to the directions of the manufacturer (New England Biolabs) and electrophoretically separated on 3% NuSieve agarose gels. To characterize the bands, the nondigested amplified products were cloned into the pCR4-TOPO vector using the TOPO TA cloning kit for sequencing (Invitrogen). TOP10 Chemically Competent cells were subsequently transformed. Colonies were picked and cultured overnight in Luria broth containing 50 μg/ml kanamycin. Plasmid DNA was subsequently isolated using a QIAprep Spin Miniprep Kit (Qiagen). The isolated plasmid was sequenced by Genewiz.
Animals.
Male C57BL/6 Ldlr−/−Apobec1−/− mice were bred in house and have been described elsewhere (23). For expression and efficacy studies, mice were given unrestricted access to water and were fed a standard chow diet. The vector was injected via an intravenous tail-vein injection with specified genome copies (GC)/mouse. For all studies, blood was obtained at least one times before and at designated time points after gene transfer. All study protocols were approved by the Institutional Animal Care and Use Committee at The University of Pennsylvania.
Cloning and AAV production.
The rhLDLR and rhLDLR-21 AAV constructs were cloned in an AAV inverted terminal repeat (ITR)-flanked construct named pENN AAV TBG PI construct, which drives expression from a thyroxine-binding globulin (TBG) hepatocyte-specific promoter with a chimeric intron from Promega (Madison, WI) encoding the 5′-donor site from the first intron of the human b-globin and the branch and 3′-acceptor site from the intron located between the leader and body of an immunoglobulin gene heavy chain variable region. The cDNA sequences were amplified from PCR from constructs acquired from openbiosystems.com (GenBank accession no. AY466854) with primers: MLU-mLDLR F (gtaagcACGCGTaggctagcctcgagaattcacgcgt), SALI-mLDLR R (ggaatcGTCGACctcacgccacgtcatcctccagact).
The rhLDLR construct was generated by MluI-SalI insertion of the cDNA insert into pENN AAV TBG PI construct. rhLDLR-21 was synthesized by Genscript based on the available rhLDLR sequence (GenBank accession no. AY466854) and was cloned by MluI-SalI insertion of the cDNA amplicon into the same AAV construct. AAV8 expressing human LDLR was generated as previously described (22). AAV vector was generated by triple transfection of the AAV ITR flanked construct with the packaging plasmid pAAV2/8 and the helper plasmid pdF6 as previously described. AAV vector particles were purified by double CsCl gradient banding as described in Wang et al. (27).
Analytical methods.
The plasma total cholesterol levels were measured in individual mice at each time point with an enzymatic assay on a Cobas Fara II (Roche Diagnostic Systems) with the use of Sigma reagents (Sigma Chemical).
Immunoblotting and histochemical analysis.
Levels of hepatic LDLR protein were determined by immunoblotting analysis as previously described (18a) using a polyclonal antisera to rat LDLR (a gift from Dr. Gene C. Ness). Briefly, liver lysates were subjected to SDS-polyacrylamide gel electrophoresis. The proteins were electrophoretically transferred to a polyvinylidene difluoride membrane. The membranes were blocked with 3% gelatin and then incubated with a 1:375 dilution of the rabbit anti-rat LDLR sera. LDLR immunoreactive protein was detected using an alkaline phosphatase-conjugated second antibody.
Statistical analyses.
Atherosclerotic lesion area data were subjected to a one-way ANOVA. Experimental groups were compared with the baseline group by using the Dunnett test. Repeated-measures ANOVA was used to compare cholesterol levels among different groups of mice over time after gene transfer. Statistical significance for all comparisons was assigned at P < 0.05. Graphs represent mean ± SD values.
RESULTS
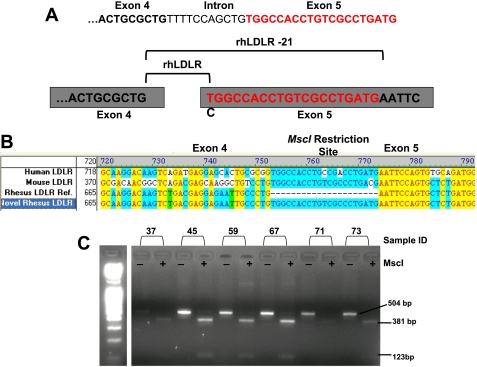
Rhesus macaque LDLR cDNA was cloned by RT-PCR using total cellular RNA prepared from liver. Sequencing of the cloned rhLDLR cDNA revealed a novel splice variant not previously reported. Compared with the previously identified rhLDLR sequence (GenBank accession no. AY466854) (20), the variant of rhLDLR we identified contained a 21 bp sequence at the junction of exons 4 and 5. Upon analysis of the known rhesus genomic DNA sequence, this 21 bp sequence constitutes part of exon 5 and is also found in both human and mouse LDLR genomic DNA sequences (Fig. 1A).
Fig. 1.
Frequency of the novel low-density lipoprotein receptor (LDLR) cDNA in rhesus macaque liver tissues. A: partial genomic sequence of exon 4, the boundary intron sequence, and exon 5. Cartoon diagram depicting site of alternative splice event. B: AlignW analysis comparing reference cDNA sequences of human (GenBank accession no. BT007361), mouse (GenBank accession no. Z19521), and rhesus (rh) LDLR (GenBank accession no. AY466854) to the novel rhesus LDLR described in this paper. C: exon 4 and 5 was amplified from cDNA of normal wild-type rhesus macaques. The PCR products were size-fractionated, gel-purified, and digested with MscI. PCR products from the RhLDLR-21 gene are not digested by MscI. The 21 bp insertion in exon 5 creates a new MscI site. MscI digestion of the PCR products from the novel rhLDLR gene generates fragments of 381 and 123 bp. Numbers above the lanes identify the individual ID of the rhesus macaque. cDNA from a total of 14 rhesus macaques and peripheral blood mononuclear cells (PBMCs) from 5 familial hypercholesterolemia (FH) rhesus macaques were analyzed using this approach.
Analysis of the sequences of the cloned and reference rhLDLR cDNA revealed that the 21 bp sequence encodes an MscI restriction site not present in the reference sequence. We took advantage of this observation to screen cDNA samples from the liver tissues of 14 rhesus macaques and PBMCs from five FH rhesus macaques. Primers were designed to amplify exons 4 and 5 from the liver cDNA. The resultant 504 bp PCR product was digested with MscI and electrophoretically separated on Nu-Sieve agarose gels. If the product contains the 21 bp insert, MscI digestion should produce two bands of ∼381 and 123 bp representing exons 4 and 5, respectively. A representative gel is shown in Fig. 2A. This analysis shows that the predominant isoform contains the 21 bp insert (Fig. 1B).
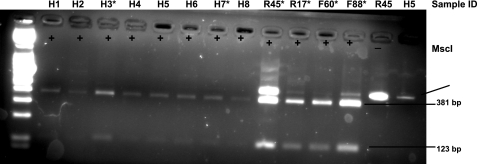
Fig. 2.
Prevalence of rhLDLR and rhLDLR-21 homologs in human liver tissues. Exons 4 and 5 were amplified from cDNA isolated from humans. The PCR products were size-fractionated, gel-purified, and digested with MscI. PCR products lacking the 21 bp sequence coinciding with exon 5 are not digested by MscI. The 21 bp insertion in exon 5 creates a new MscI site. MscI digestion of PCR products containing the 21 bp insertion generates fragments of 381 and 123 bp. Numbers above the lanes identify the individual ID of the human (H), wild-type rhesus (R), and FH rhesus macaques (F). *Nondigested fragment from this sample was subsequently cloned and sequenced. cDNA from a total of 12 human liver samples were analyzed using this approach.
It is estimated that 60% of human genes undergo alternative splicing; this phenomenon plays an important role in expanding the proteome (12). Cross-species conservation has been suggested to be a strong predictor of functional alternative splicing (12). Therefore, we next decided to examine whether human LDLR undergoes alternative splicing similar to that of rhesus LDLR. We obtained 12 human liver samples from the NDRI. cDNA was generated from these samples; exons 4 and 5 were PCR amplified using the same primers as for rhLDLR and subjected to MscI restriction analysis. Using this approach, we found that all human samples examined contained the 21 bp sequence of exon 5 (Fig. 2).
To evaluate the relative abundance of each isoform of rhLDLR, we next decided to clone and sequence the nondigested PCR product. PCR products from four individual rhesus macaques were cloned and sequenced. Of the 27 positively transformed colonies that were picked and sequenced, 23 contained and four of them lacked the 21 bp sequence of exon 5. Nondigested PCR product from two individual human samples were similarly cloned and sequenced. Of the 14 positively transformed colonies that were picked and sequenced, all contained the 21 bp sequence. These data suggest that the predominant human LDLR mRNA isoform contains the 21 bp sequence and that rhLDLR mRNA contains two splice variants: 1) a predominant isoform, newly identified, that is homologous to human LDLR and retains 21 bp of exon 5 (rhLDLR) and 2) a minor isoform, previously identified and listed in the GenBank database, that lacks 21 bp of exon 5 (rhLDLR-21) (20).
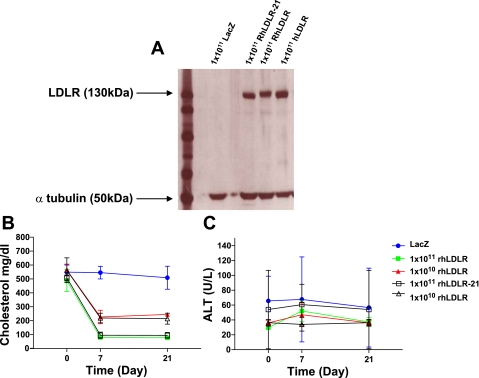
Previous studies by Nomura and colleagues (20) have examined the efficacy of helper-dependent adenovirus-mediated expression of rhLDLR-21 in an Ldlr−/− mouse model of FH. It is indeed surprising that the rhLDLR-21 variant reported by Nomura et al. is functional in correcting the phenotype of FH mice, given that this isoform includes a Cys residue deletion. This deletion results in an unpaired cysteine in the cys-rich repeat encoded by exon 5 and is predicted to disrupt folding and cause mistrafficking and/or endoplasmic reticulum-associated protein degradation (22). Furthermore, computational prediction suggests that alternatively spliced LDLR mRNA could encode receptors functionally different from the normal receptor (26). To evaluate whether rhLDLR-21 might be functionally different from rhLDLR, we conducted a study comparing the efficacy of AAV8-mediated liver-specific expression of rhLDLR-21 or rhLDLR in correcting the hypercholesterolemia of Ldlr−/−Apobec1−/− mice that resemble hoFH humans. Animals were intravenously injected with 1 × 1011 or 1 × 1010 GC of AAV8 encoding rhLDLR-21 or rhLDLR under control of the liver-specific TBG promoter. Protein expression in the liver was examined 21 days after vector injection. As can be seen in Fig. 3A, no differences were observed in terms of total LDLR protein expression between rhLDLR-21 and rhLDLR-injected mice. This suggests that in terms of protein folding and trafficking, the two isoforms are virtually identical. With respect to functionality, both vectors exhibited virtually identical kinetics and have the same minimum dose for cholesterol correction in Ldlr−/− Apobec1−/− mice (Fig. 3B). Importantly, neither vector induced detectable toxicity as measured by serum levels of ALT (Fig. 3B).
Fig. 3.
In vivo analysis of functionality of rhLDLR-21 and rhLDLR. A: Western blot examining protein expression of rhLDLR and rhLDLR-21 at a dose of 1 × 1011 genome copies (GC)/mouse 21 days after intravenous tail vein injection. Plasma cholesterol and ALT levels (B) in Ldlr−/−Apobec1−/− mice after treatment with 1 × 1011 GC/mouse or 1 × 1010 GC/mouse AAV8.TBG.RhLDLR or AAV8.RhLDLR-21 (n=5 animals per dose group). Each point represents mean ± SD.
DISCUSSION
We have previously demonstrated that AAV8-mediated gene transfer of LDLR leads to significant reductions in plasma cholesterol and non-HDL cholesterol levels in a murine model of HoFH (9). Moreover, this therapeutic approach was found to trigger significant regression and substantial remodeling of atherosclerotic lesions on HoFH mice. Given these positive results, we next decided to examine the safety and efficacy of AAV8 mediated LDLR gene transfer in an NHP.
LDLR+/− rhesus macaques, previously identified by Hummel and colleagues (8), have a 50% reduction in the LDLR activity due to a nonsense mutation in one allele of the LDLR gene. Due to inability to generate homozygotes for this mutation, heterozygotes were fed a high-fat, high-cholesterol diet to downregulate the LDLR expressed by the normal allele and more closely replicate that the phenotype of HoFH patients (4). We decided to begin our macaque gene transfer studies using the reported sequence of LDLR derived from rhesus macaque (20). We undertook studies from various sources of rhesus macaques to delineate the structure of wild-type rhLDLR. We found a predominant form of rhLDLR that was of the same size as human LDLR with 95% homology. It is interesting, however, that the form we cloned from macaque differs from that previously described by Nomura and colleagues (20), who claim that the rhesus receptor is 21 amino acids smaller than human LDLR. We in fact found this smaller isoform by PCR screening of mRNAs as a minor species.
Cross-species conservation has been suggested as a strong indication of functional alternative splicing (12, 24). However, in our analysis we detected no human homolog of rhLDLR-21 among the human tissues that were examined, indicating that the observed splice variant in rhesus is not common under basal conditions in humans. This is not necessarily surprising. Computational searches for alternative splicing events by Pan and colleagues (21) revealed that >11% of human and mouse cassette alternative exons are skipped in one species but used constitutively in the other. Likewise, Calarco et al. (2) studied alternative splicing differences between humans and chimpanzees and found that at least 6% of the exons they tested displayed significant differences in splicing levels between humans and chimpanzees. Blekhman et al. (1) similarly found that large numbers of exons are consistently skipped in livers from humans, chimpanzees, and rhesus macaques, providing evidence for lineage-specific shifts in the composition of alternative transcripts. It must be noted that given the small sample size of our human liver tissue studies, there is a possibility that the human variant of rhLDLR-21 can, under certain circumstances, be expressed. It may not have been detectable using our particular approach or in the specific tissues we examined. Therefore, additional studies will need to be conducted that include a larger sample size of human liver tissues from different patient populations. Moreover, future efforts may benefit from the use of a quantitatively based PCR assay (26) that is more sensitive and can enable for the relative quantification of the two splice variants.
There have been numerous studies demonstrating that alternative splicing can have a significant impact on gene function. For example, alternative splicing of 3-hydroxy-3-methylglutaryl-coenzyme (HMGCR) or LDLR, resulting from single nucleotide polymorphisms (SNPs) in the intron, have been associated with reduced LDL-cholesterol response to statin treatment (4, 10, 15). Likewise, there have been numerous studies examining the role of HMGCR exon 13 alternative splicing as a molecular mechanism underlying the association between HMGCR genetic variation and statin efficacy (16). With respect to LDLR, variation in this locus is known to influence LDL-cholesterol levels (5, 14); common LDLR DNA polymorphisms have been associated with interindividual variation in LDL-cholesterol, as well as other lipid and lipoprotein traits (5, 14, 28), and these variants are thought to mediate changes in LDLR protein expression or regulation. There is recent evidence that SNPs in the 3′-untranslated region of LDLR impact LDLR splicing and impact on LDLR mRNA stability in vitro and LDL-cholesterol levels in vivo (18).
Functionally, our studies demonstrated that AAV8 mediated liver-specific expression of rhLDLR-21 or rhLDLR led to similar patterns of in vivo correction in a mouse model of FH. We conclude that the previously identified rhLDLR cDNA constitutes a minor splice variant that has relatively little impact on LDLR activity. Given this finding, and the fact that the predominant human LDLR splice variant retains the 21 bp sequence of exon 5, we plan to proceed with rhLDLR in preclinical evaluation of AAV8.LDLR gene therapy in fat-fed LDLR−/+ rhesus macaques.
Genome-wide analysis of alternative splicing indicate that 40–60% of human genes have alternative splice forms; this suggests that alternative splicing is one of the most significant components of the functional complexity of the human genome (11). However, it is not clear how many of the splice variants predicted from expressed sequence tag (EST) cluster analysis are functional and how many represent aberrant splicing noise or EST artifacts such as genomic contamination (17). In this study, we used liver-directed AAV8 gene transfer to examine the functionality of alternative splice variants in vivo. To our knowledge, this is the first report to use somatic gene transfer to study the function of alternative splice variants in vivo. More than 40% of genes in the liver undergo one or more alternative splice events, making it the second highest human tissue, by frequency, to exhibit alternative splicing (30). Within this context, future studies may benefit from the usage of AAV8 to study the function of alternative splice variants in the liver.
GRANTS
This work was supported by National Institutes of Health Grants P01-HL-059407 and P30-DK-047757 to J. M. Wilson. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
J. M. Wilson is a consultant to ReGenX Holdings and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings; in addition, he is an inventor on patents licensed to various biopharmaceutical companies, including affiliates of ReGenX Holdings. This does not alter the authors' adherence to all the policies of Physiological Genomics on sharing data and materials, as detailed online in the guide for authors.
ACKNOWLEDGMENTS
We thank Julie Johnston, Martin Lock, and Arbans Sandhu (Penn Vector Core) for supplying vectors. Also, we are grateful to Debra Cromley, Deirdre McMenamin, and Natalie Ridge for excellent technical support.
Current addresses: S. H. Kassim, Johnson & Johnson, Innate Immunity, 145 King of Prussia Rd., Radnor, PA 19087; L. H. Vandenberghe, F. M. Kirby Center for Molecular Ophthalmology, Univ. of Pennsylvania, Philadelphia, PA 19104.
REFERENCES
- 1. Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res 20: 180–189, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev 21: 2963–2975, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen SJ, Rader DJ, Tazelaar J, Kawashiri MA, Gao GP, Wilson JM. Prolonged correction of hyperlipidemia in mice with familial hypercholesterolemia using an adeno-associated viral vector expressing very-low-density lipoprotein receptor. Mol Ther 2: 256–261, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Donnelly LA, Doney ASF, Dannfald J, Whitley AL, Lang CC, Morris AD, Donnan PT, Palmer CNA. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet Genomics 18: 1021–1026, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Francke U, Brown MS, Goldstein JL. Assignment of the human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand, and a genetic disease. Proc Natl Acad Sci USA 81: 2826–2830, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA 99: 11854–11859, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grossman M, Rader DJ, Muller DWM, Kolansky DM, Kozarsky I, Clark Iii BJ, Stein EA, Lupien PJ, Brewer HB, Jr, Raper SE, Wilson JM. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med 1: 1148–1154, 1995 [DOI] [PubMed] [Google Scholar]
- 8. Hummel M, Li Z, Pfaffinger D, Neven L, Scanu AM. Familial hypercholesterolemia in a rhesus monkey pedigree: Molecular basis of low density lipoprotein receptor deficiency. Proc Natl Acad Sci USA 87: 3122–3126, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kassim SH, Li H, Vandenberghe LH, Hinderer C, Bell P, Marchadier D, Wilson A, Cromley D, Redon V, Yu H, Wilson JM, Rader DJ. Gene therapy in a humanized mouse model of familial hypercholesterolemia leads to marked regression of atherosclerosis. PLoS One 5: e13424, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme A reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation 117: 1537–1544, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, Levine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001 [DOI] [PubMed] [Google Scholar]
- 12. Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr Opin Struct Biol 14: 273–282, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Lebherz C, Gao G, Louboutin JP, Millar J, Rader D, Wilson JM. Gene therapy with novel adeno-associated virus vectors substantially diminishes atherosclerosis in a murine model of familial hypercholesterolemia. J Gene Med 6: 663–672, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Linsel-Nitschke P, Götz A, Erdmann J, Braenne I, Braund P, Hengstenberg C, Stark K, Fischer M, Schreiber S, El Mokhtari NE, Schaefer A, Schrezenmeier J, Rubin D, Hinney A, Reinehr T, Roth C, Ortleep J, Hanrath P, Hall AS, Mangino M, Lieb W, Lamina C, Heid IM, Doering A, Gieger C, Peters A, Meitinger T, Wichmann HE, König IR, Ziegler A, Kronenberg F, Samani NJ, Schunkert H. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease - a Mendelian randomisation study. PLoS ONE 3: e2986, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mangravite LM, Medina MW, Cui J, Pressman S, Smith JD, Rieder MJ, Guo X, Nickerson DA, Rotter JI, Krauss RM. Combined influence of LDLR and HMGCR sequence variation on lipid-lowering response to simvastatin. Arterioscler Thromb Vasc Biol 30:1485–1492, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Medina MW. The relationship between HMGCR genetic variation, alternative splicing, and statin efficacy. Disc Med 9:495–499, 2010 [PubMed] [Google Scholar]
- 17. Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet 30: 13–19, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Muallem H, North KE, Kakoki M, Wojczynski MK, Li X, Grove M, Boerwinkle E, Wilhelmsen KC, Heiss G, Maeda N. Quantitative effects of common genetic variations in the 3′UTR of the human LDL-receptor gene and their associations with plasma lipid levels in the Atherosclerosis Risk in Communities study. Hum Genet 121: 421–431, 2007 [DOI] [PubMed] [Google Scholar]
- 18a. Ness GC, Zhao Z. Thyroid hormone rapidly induces hepatic LDL receptor mRNA levels in hypophysectomized rats. Arch Biochem Biophys 315: 199–202, 1994 [DOI] [PubMed] [Google Scholar]
- 19. Neven L, Khalil A, Pfaffinger D, Fless GM, Jackson E, Scanu AM. Rhesus monkey model of familial hypercholesterolemia: relation between plasma Lp[a] levels, apo[a] isoforms, and LDL-receptor function. J Lipid Res 31: 633–643, 1990 [PubMed] [Google Scholar]
- 20. Nomura S, Merched A, Nour E, Dieker C, Oka K, Chan L. Low-density lipoprotein receptor gene therapy using helper-dependent adenovirus produces long-term protection against atherosclerosis in a mouse model of familial hypercholesterolemia. Gene Ther 11: 1540–1548, 2004 [DOI] [PubMed] [Google Scholar]
- 21. Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21: 73–77, 2005 [DOI] [PubMed] [Google Scholar]
- 22. Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: New insights in pathogenesis and treatment. J Clin Invest 111: 1795–1803, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scanu AM, Khalil A, Neven L, Tidore M, Dawson G, Pfaffinger D, Jackson E, Carey KD, McGill HC, Fless GM. Genetically determined hypercholesterolemia in a rhesus monkey family due to a deficiency of the LDL receptor. J Lipid Res 29: 1671–1681, 1988 [PubMed] [Google Scholar]
- 24. Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet 20: 68–71, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science 228: 815–822, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tveten K, Ranheim T, Berge KE, Leren TP, Kulseth MA. Analysis of alternatively spliced isoforms of human LDL receptor mRNA. Clinica Chimica Acta 373: 151–157, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Wang L, Wang H, Bell P, McCarter RJ, He J, Calcedo R, Vandenberghe LH, Morizono H, Batshaw ML, Wilson JM. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol Ther 18: 118–125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamamoto T, Davis CG, Brown MS. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 39: 27–38, 1984 [DOI] [PubMed] [Google Scholar]
- 30. Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome biology 5: R74, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]