Abstract
Muscle strength is an important determinant in elite sports performance as well as in the activities of daily living. Muscle metabolism also plays a role in the genesis, and therefore prevention, of common pathological conditions and chronic diseases. Even though heritability estimates between 31 and 78% suggest a significant genetic component in muscle strength, only a limited number of genes influencing muscle strength have been identified. This study aimed to identify and prioritize positional candidate genes within a skeletal muscle strength quantitative trait locus on chromosome 12q22-23 for follow-up. A two-staged gene-centered fine-mapping approach using 122 single nucleotide polymorphisms (SNPs) in stage 1 identified a familybased association (n = 500) between several tagSNPs located in the ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 (ATP2A2; rs3026468), the NUAK family, SNF1-like kinase, 1 (NUAK1; rs10861553 and rs3741886), and the protein phosphatase 1, catalytic subunit, gamma isoform (PPP1CC; rs1050587 and rs7901769) genes and knee torque production (P values up to 0.00092). In stage 2, family-based association tests on additional putatively functional SNPs (e.g., exonic SNPs, SNPs in transcription factor binding sites or in conserved regions) in an enlarged sample (n = 536; 464 individuals overlap with stage 1) did not identify additional associations with muscle strength characteristics. Further in-depth analyses will be necessary to elucidate the exact role of ATP2A2, PPP1CC, and NUAK1 in muscle strength and to find out which functional polymorphisms are at the base of the interindividual strength differences.
Keywords: complex trait, family-based association, genotype/phenotype association
muscular fitness is an important factor in the elite sports performance as well as in the activities of daily living (31). Muscle metabolism also plays an important role in the genesis, and therefore prevention, of many common pathological conditions and chronic diseases like cardiovascular disease (41, 42), osteoporosis (27), metabolic syndrome (58), obesity (16), and diabetes (42, 62). Given the fact that the Western society is aging, the progressive loss of muscle mass and function that occurs with aging (sarcopenia) will become a major health issue as it has devastating effects on quality of life (e.g., institutionalization, dependence of the elderly) and eventually on survival (35).
Several studies suggest that apart from environmental influences such as training, social status, or nutrition, a large genetic component is involved in the interindividual variation in muscle strength. Heritability estimates range between 31 and 78% and differ between muscle groups, contraction velocities, and contraction angles (25, 33, 44, 49–51, 53). Despite this significant genetic determination of muscle strength, only a limited number of studies showed significant association of genetic variants in quantitative trait loci (QTL) with muscle strength phenotypes, and even fewer associations have consistently been replicated (reviewed in Ref. 11).
Studies investigating genetic linkage with human muscle strength measures are even scarcer. To date only Tiainen et al. (52) and our research group (14, 15, 23, 24, 26, 48) have reported genetic linkage results on skeletal muscle strength characteristics. Our research group was the first to perform linkage analyses on a unique collection of young male Caucasian siblings drawn from the Leuven Genes for Muscular Strength study (LGfMS). These analyses focused on chromosomal regions harboring genes involved in the myostatin signaling pathway, as described elsewhere (23, 26). Single and multipoint microsatellite marker-based linkage analyses revealed that the chromosomal regions 12q12–14 [logarithm of the odds (LOD) 3.4, P = 0.00004], 12q22-23 (LOD 2.6, P = 0.0002) and 13q14.2 (LOD 2.74, P = 0.0002) showed significant or suggestive linkage to knee muscle strength (23, 26). It can, however, not be excluded that genes other than the original myostatin pathway candidate genes cause these linkage findings and therefore further fine mapping of these regions is warranted. In a recent report we followed up upon the chr12q12-14 region and identified the activin receptor 1 B (ACVR1B) gene as a strength determining gene using a comprehensive two-staged fine-mapping strategy (60).
Here we present the results of a similar two-staged fine mapping of the 12q22-23 region containing 143 known genes. The purpose of this study was to prioritize these genes and identify genes and polymorphisms that are associated with knee muscle strength characteristics. Therefore the study focused on 51 prioritized candidate genes and identified the NUAK1 and ATP2A2/PPP1CC loci to be related to variation in muscle strength.
MATERIAL AND METHODS
Study Design
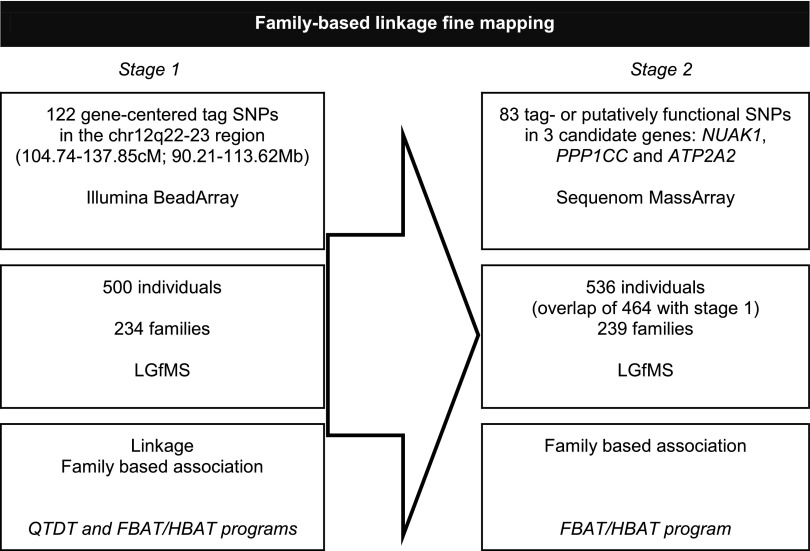
A two-staged approach was designed to fine map the chr12q22-23 linkage region, as shown in Fig. 1.
Fig. 1.
Study design. The 2 stages of the study are displayed. Sample description, genotyping details, and statistical analyses are presented from top to bottom. LGfMS, Leuven Genes for Muscular Strength study; SNP, single nucleotide polymorphism; QTDT, quantitative transmission disequilibrium testing; FBAT, family-based association test; HBAT, haplotype-based family-based association test.
Fine mapping stage 1.
Single nucleotide polymorphisms (SNPs) were selected under the 1-LOD confidence interval of a previously defined multipoint microsatellite-based linkage peak (26). Thereto, an empirical two-step fine-mapping strategy, in which candidate genes are prioritized using a bioinformatics approach and the top genes are chosen for further SNP selection with a linkage disequilibrium-based method, was applied. A detailed description of this method and its application to another linkage region can be found elsewhere (60, 61). In brief, the 143 genes within the 1-LOD confidence interval were ranked according to their similarity to genes known to be involved in (the regulation of) muscle strength using Endeavour software (3). These known genes include candidate genes from the original myostatin pathway (23, 26), structural elements of muscle (actin and myosin related genes, together with troponin, titin, and nebulin), and genes identified based on Gene Ontology (GO) terms “contraction,” “muscle development,” and “regulation (negative and positive) of contraction.” Rankings were made for each of these five reference sets and combined into one global ranking.
We considered the top 20% of the genes according to every reference set for further analyses as well as those genes ranked within the top 20% in the global ranking. Of these, genes without evidence for skeletal muscle expression in publicly available databases were excluded. A total of 51 genes (36% of the original 143 genes in this region) were selected for further SNP analysis. Large-scale validation experiments in the original Endeavour article (3) revealed that the gene of interest, in this particular case the true strength determining gene, is ranked among the top 36% in ∼90% of the cases. Since we were able to discard 64% of the genes with a 90% certitude of still including the gene of interest, the use of Endeavour software is clearly beneficial for our study.
Within these selected 51 genes tagging SNPs were determined based on CEPH genotypes available from the HapMap (47) using Tagger (13), implemented in Haploview (7). Aggressive tagging and a r2 threshold of 0.8 were applied on the SNPs with a minor allele frequency (MAF) >0.05. A list of the candidate genes and corresponding polymorphisms is provided as supplemental material (Supplemental Table S1, available online).1 Linkage analyses and family-based association analyses were performed to test association of these polymorphisms with knee strength measurements (cfr. Statistical Analyses below).
Fine mapping stage 2.
An extended sample was genotyped for the stage 1 SNPs as well as additional SNPs in the most relevant genes (i.e., the genes with the highest significance and/or significance over different strength measurements) from the family-based association tests (ATP2A2, PPP1CC and NUAK1). These additional polymorphisms included other tagSNPs and polymorphisms likely to have functional consequences such as exonic SNPs, SNPs located in intron/exon boundaries, SNPs in (putative) transcription factor binding sites or in highly conserved regions. Assessment of functionality was based on queries from SNPselector (63), SNPseek, and publicly available genetic databases (Entrez Gene, UCSC genome browser, Ensembl). A list of these SNPs can be found in Table 1.
Table 1.
Polymorphisms selected in NUAK1, ATP2A2, and PPP1CC and putative function
| SNP | No. | Coordinate* | Minor allele | Minor Allele Frequency | Putative Function |
|---|---|---|---|---|---|
| NUAK1 | |||||
| rs10861553 | 1 | 104976944 | C | 0.48 | Tagging SNP, genotyped stage 1 |
| rs6539247 | 2 | 104984032 | T | 0.44 | Conserved |
| rs3741883 | 3 | 104985068 | C | 0.22 | Tagging SNP, genotyped stage 1 |
| rs55774704 | 4 | 104985440 | A | 0.0 | Nonsynonymous [G/D] |
| rs3741885† | 5 | 104985987 | Conserved | ||
| rs3741886 | 6 | 104990989 | C | 0.071 | Tagging SNP, genotyped stage 1 |
| rs934085† | 7 | 104993301 | Conserved | ||
| rs17038085† | 8 | 104996331 | Conserved | ||
| rs17038089 | 9 | 104998855 | T | 0.041 | Conserved |
| rs7133815 | 10 | 105000270 | C | 0.19 | Conserved |
| rs12146713 | 11 | 105000935 | C | 0.065 | Conserved |
| rs1560757 | 12 | 105001319 | G | 0.20 | Tagging SNP |
| rs11112856 | 13 | 105002687 | A | 0.16 | Conserved, located in transcription factor binding site (TFBS) |
| rs34594928† | 14 | 105004761 | Frameshift mutation | ||
| rs12582194 | 15 | 105006063 | A | 0.25 | Tagging SNP, genotyped stage 1 |
| rs1366041 | 16 | 105008642 | G | 0.066 | Conserved |
| rs967872 | 17 | 105016425 | G | 0.45 | Tagging SNP, genotyped stage 1 |
| rs17038111 | 18 | 105017017 | C | 0.030 | Conserved |
| rs1560000 | 19 | 105017277 | C | 0.41 | Conserved |
| rs11112868 | 20 | 105022585 | G | 0.46 | Conserved |
| rs1427785 | 21 | 105024538 | C | 0.42 | Conserved |
| rs4964439† | 22 | 105025580 | Conserved | ||
| rs10861556† | 23 | 105027775 | Conserved | ||
| rs12297025 | 24 | 105030377 | A | 0.11 | Tagging SNP, genotyped stage 1 |
| rs10492351 | 25 | 105032945 | G | 0.082 | Conserved |
| rs1215604 | 26 | 105041118 | G | 0.49 | Tagging SNP |
| rs2434081 | 27 | 105052584 | C | 0.38 | Conserved, located in TFBS |
| rs2559602 | 28 | 105054291 | C | 0.48 | Conserved |
| rs1215597 | 29 | 105056202 | A | 0.49 | conserved |
| rs55663911 | 30 | 105056410 | G | 0.0 | Nonsynonymous [L/S] |
| ATP2A2 | |||||
| rs7304243 | 1 | 109146710 | C | 0.32 | Conserved |
| rs3026421 | 2 | 109169412 | T | 0.0 | Located in (TFBS) |
| rs3026432 | 3 | 109203486 | T | 0.0 | Conserved, located in CpG island |
| rs3026433† | 4 | 109203575 | Located in CpG island | ||
| rs3026434 | 5 | 109204058 | G | 0.00096 | Synonymous SNP [E/E], conserved, located in CpG island |
| rs3026436 | 6 | 109205386 | G | 0.0 | Conserved |
| rs3026452 | 7 | 109212232 | G | 0.038 | Tagging SNP |
| rs929518 | 8 | 109220198 | A | 0.0010 | Tagging SNP |
| rs4630352 | 9 | 109231802 | A | 0.28 | Tagging SNP |
| rs3026468 | 10 | 109242464 | A | 0.075 | Tagging SNP, genotyped stage 1 |
| rs1803737 | 11 | 109250126 | C | 0.0 | Nonsynonymous [A/V], located in TFBS |
| rs3026473 | 12 | 109254251 | A | 0.0 | Conserved |
| rs35616357 | 13 | 109262096 | C | 0.0 | Located in TFBS |
| rs2302372 | 14 | 109263086 | T | 0.0 | Synonymous SNP [D/D], conserved, located in TFBS |
| rs12312588 | 15 | 109264520 | A | 0.0 | Synonymous SNP [L/l] |
| rs1860561 | 16 | 109267624 | A | 0.18 | Tagging SNP |
| rs12297171 | 17 | 109268602 | T | 0.0030 | Synonymous SNP [L/l], conserved |
| rs1042649 | 18 | 109268757 | 0.0 | Conserved | |
| rs1042650 | 19 | 109268768 | A | 0.0 | Conserved |
| rs1042653 | 20 | 109268818 | A | 0.0 | Conserved, located in TFBS |
| rs1803736 | 21 | 109268904 | T | 0.0 | Conserved, located in TFBS |
| rs3026489 | 22 | 109268961 | C | 0.025 | Tagging SNP |
| rs1803738 | 23 | 109269281 | C | 0.0 | Conserved, located in TFBS |
| rs1803735 | 24 | 109269449 | T | 0.0 | Conserved |
| rs3211481 | 25 | 109269506 | T | 0.0 | Conserved, located in TFBS |
| rs3026490 | 26 | 109269892 | A | 0.0 | Conserved |
| rs12638 | 27 | 109273087 | T | 0.0 | Conserved, located in TFBS |
| rs3415 | 28 | 109273140 | C | 0.0 | Conserved |
| rs9540 | 29 | 109273238 | G | 0.011 | Conserved, located in TFBS |
| PPP1CC | |||||
| rs7132423 | 1 | 109609452 | C | 0.0 | Located in regulatory region |
| rs35343396 | 2 | 109610578 | A | 0.0 | Intergenic region |
| rs12826018 | 3 | 109636538 | C | 0.0 | Located in TFBS |
| rs11065703 | 4 | 109637378 | G | 0.0 | Intergenic region |
| rs17682482 | 5 | 109639171 | G | 0.064 | Tagging SNP |
| rs3191028 | 6 | 109642234 | T | 0.0 | Located in 3′ UTR, conserved |
| rs1050587 | 7 | 109642544 | G | 0.076 | Located in 3′ UTR, exonic splice enhancer |
| rs11558236 | 8 | 109642611 | G | 0.0010 | Located in 3′ UTR, conserved |
| rs11558238 | 9 | 109643266 | A | 0.0 | Nonsynonymous [K/*], conserved |
| rs11558239 | 10 | 109643272 | G | 0.0 | Nonsynonymous [K/E], conserved |
| rs1973505 | 11 | 109644386 | A | 0.12 | Tagging SNP, synonymous [C/C], genotyped stage 1 |
| rs28702860 | 12 | 109644818 | C | 0.0 | Nonsynonymous [N/D], conserved |
| rs11558237† | 13 | 109646916 | Nonsynonymous [F/S], conserved | ||
| rs7960761 | 14 | 109648798 | G | 0.076 | Tagging SNP, genotyped Stage 1 |
| rs11558235 | 15 | 109652822 | 0.0 | Nonsynonymous [C/R], conserved | |
| rs11558234† | 16 | 109654001 | Synonymous [S/S], conserved | ||
| rs11558240 | 17 | 109654064 | 0.0 | Synonymous [N/N], conserved | |
| rs7301769 | 18 | 109655409 | G | 0.078 | Tagging SNP, genotyped stage 1 |
| rs1476470 | 19 | 109751414 | T | 0.45 | Tagging SNP, conserved |
| rs11065729 | 20 | 109761871 | C | 0.14 | Intergenic region, conserved |
| rs12312903† | 21 | 109764018 | Intergenic region, conserved | ||
| rs12312907† | 22 | 109764023 | Intergenic region, conserved | ||
| rs12303008 | 23 | 109769214 | G | 0.028 | Intergenic region |
| rs2339635 | 24 | 109769255 | G | 0.34 | Intergenic region |
Based on National Center for Biotechnology Information build 36;
failed assay. Informative single nucleotide polymorphisms (SNPs) are indicated in boldface.
A two-staged fine-mapping strategy similar to the one described for this study has already been successfully used in the fine mapping of a linkage region on chr12q12-14 and identified ACVR1B as a muscle strength-related gene (60).
Study Sample
Siblings analyzed in stage 1 and stage 2 of this study were selected from the LGfMS project (748 men, aged 17–36 yr) based on family size and DNA availability. The recruitment protocol and subject characteristics have been described elsewhere (23, 26). The medical and ethical committee of the Katholieke Universiteit Leuven approved the study, and all participants gave their written informed consent.
For stage 1, data from 500 brothers of 234 Caucasian families (26 singles, 160 duos, 38 trios, and 10 quads) were included in the statistical analyses, resulting in an overlap of 169 subjects with the preceding multipoint microsatellite-based linkage study (26). For stage 2, subjects for whom genotyping failed in stage 1 were excluded and additional subjects for whom new DNA was collected between the two stages were included. As a result, the stage 2 sample consisted of 536 siblings (239 families, 7 singles, 176 duos, 47 trios, and 9 quads) with an overlap of 464 individuals with stage 1.
Strength Measurements
Knee flexion and extension strength tests were performed on a Cybex NORM dynamometer (Lumex, Ronkonkoma, NY) and are described in detail elsewhere (23, 26). Maximal isometric knee strength and dynamic knee strength at 60°/s, 120°/s and 240°/s were retained for analyses in this paper. Torque was measured at specific angles, i.e., 60° for knee extension (quadriceps) and 30° for knee flexion (hamstrings) (0° = full extension of the knee) as the force-length relationship of a muscle predicts that optimal strength is generated at longer muscle length.
SNP Genotyping
Genomic DNA was extracted either from EDTA whole blood by a standard salting-out method or from saliva collected in Oragene DNA Self-Collection Kits (Oragene, Kanata, Canada) using the protocol provided by the manufacturer.
In stage 1 a total of 122 SNPs was genotyped on an Illumina Bead Array platform (Illumina, San Diego, CA). Genotypes were determined using GenCall 5.2.0 software and visually checked. The overall sample success rate, locus success rate, and genotype call rate were 99.8% (499/500), 97.5% (119/122), and 98.9% (58,722/59,381), respectively. These SNPs capture 48% of all available alleles at r2 > 0.8. Per gene, coverage is highly variable because the number of SNPs per gene was set relative to the Endeavour ranking so that for the top genes more polymorphisms were selected than for the 20–30th gene. For example for MYBPC1 which ranked first in the global ranking eight tagSNPs were selected, resulting in a coverage of 57% of the alleles with MAF >0.05. In contrast, for UBE3B that ranked 30th, only one tagSNP was selected and only 17% of alleles with MAF >0.05 was captured.
In the second stage of the study, genotyping of 83 SNPs was performed using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry iPLEX Gold platform (Sequenom, San Diego, CA). Genotypes determined by Sequenom's Typer 4.0 software were visually checked prior to statistical analysis. Overall sample success rate, locus success rate, and genotype call rate were 98.9% (530/536), 84.3% (70/83), and 94.9% (35,209/37,100), respectively.
Statistical Analyses
Pedstats v0.6.4 (57) was used to check pedigrees for Mendelian (in)consistency and to test Hardy-Weinberg equilibrium. Unlikely genotypes were identified and discarded using the error-checking option of Merlin v1.0.1.(1).
Haploview (7) was used to examine the linkage disequilibrium (LD) structure across the region of interest and to calculate r2 between all pairs of SNPs within a candidate region.
Linkage analyses were performed using Merlin v1.2.1.(1). We conducted family-based association tests (FBAT v1.7.3)(21, 30) and used the haplotype version of this test (HBAT)(22) to obtain empirical P values by means of the Monte-Carlo permutation procedures implemented in HBAT (10,000 permutations were performed). Trait offsets were specified as the sample mean of the trait.
Covariates could not be included directly in the FBAT/HBAT analyses. Therefore a multiple regression of the strength measurements on age, stature, and fat-free mass was performed using SAS v9.1.3 and v9.2 (SAS Institute, Cary, NC), and the resulting residuals were used as input phenotypes for FBAT/HBAT. SAS 9.2 software was also used to calculate descriptive statistics.
RESULTS
Descriptive Statistics
In this two-staged genetic fine-mapping study 500 and 536 individuals from the LGfMS were included in stage 1 and stage 2, respectively, with an overlap of 464 individuals between both stages. Somatic and strength characteristics of these individuals are given in Table 2. Study samples for the two stages did not differ significantly for any of these traits.
Table 2.
Somatic characteristics and knee strength statistics
|
Stage 1 |
Stage 2 |
|||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| n | 500 | 536 | ||
| Age, yr | 24.2 | 4.4 | 24.2 | 4.5 |
| Weight, kg | 74.1 | 9.6 | 74.0 | 9.8 |
| Stature, cm | 180.7 | 6.3 | 180.6 | 6.4 |
| Flexion | ||||
| Isometric, Nm | 148.7 | 26.8 | 149.0 | 27.4 |
| Torque at 60°/s, Nm | 121.3 | 22.2 | 121.7 | 22.6 |
| Torque at 120°/s, Nm | 105.6 | 18.9 | 105.6 | 18.9 |
| Torque at 240°/s, Nm | 74.9 | 16.7 | 75.0 | 16.5 |
| Extension | ||||
| Isometric, Nm | 261.3 | 48.3 | 260.9 | 48.7 |
| Torque at 60°/s, Nm | 196.7 | 34.5 | 196.8 | 34.9 |
| Torque at 120°/s, Nm | 159.1 | 25.5 | 159.5 | 26.0 |
| Torque at 240°/s, Nm | 107.3 | 19.2 | 107.6 | 19.8 |
All flexion and extension measurements were taken at an angle of 30 and 60°, respectively.
Stage 1 Genotyping
To reduce the number of genes and SNPs for the fine-mapping analyses, an empirical two-step selection approach was used. First, the 143 genes in the 12q22-23 region were ranked according to their similarity to five different reference sets of genes known to be involved in (the regulation of) muscle strength. Selection of the top 20% genes for each of these reference sets and exclusion of genes that are not expressed in skeletal muscle tissue resulted in the selection of 51 candidate genes. Second, tagging SNPs were determined on CEPH genotypes downloaded from the HapMap (47) website (MAF > 0.05, r2 > 0.8, aggressive tagging). The most informative SNPs per gene (i.e., tagging the most other SNPs) were selected, resulting in the selection of 117 SNPs in 40 candidate genes. For the 11 remaining genes no polymorphisms meeting our criteria [Illumina designability rank = 1 (i.e., high success rate of assay design); MAF > 0.05] were present. Five additional SNPs in the gaps in between the 43 genes were selected (61). A list of these SNPs can be found as supplemental material (Supplemental Table S1, available online).
Of these 122 SNPs, 119 were successfully genotyped, one of which was excluded due to Hardy-Weinberg disequilibrium (rs3817552 in MYBPC1, P < 0.001).
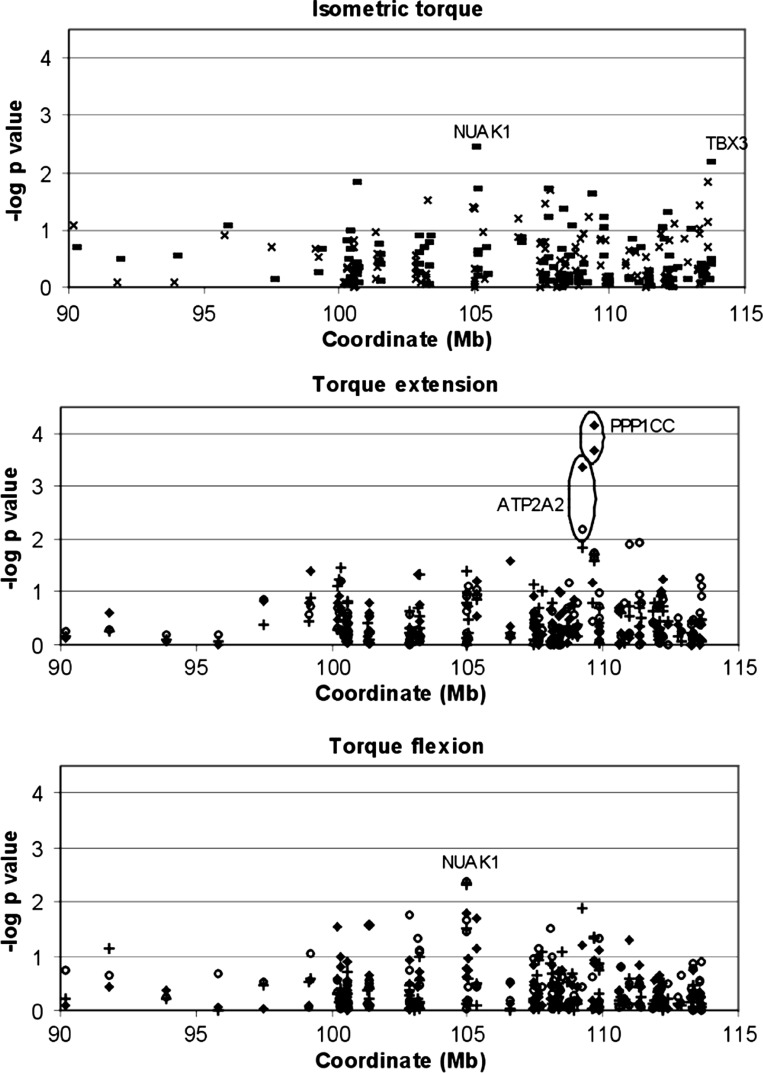
Figure 2 shows the empirical P values for family-based association for all of the 118 polymorphisms. Three genes were repeatedly highly suggestive (P < 0.01, −log P value >2) over different strength measurements: NUAK1 (NUAK family, SNF1-like kinase, 1), PPP1CC (protein phosphatase 1, catalytic subunit, gamma isoform), and ATP2A2 (ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2) and were therefore selected for further follow-up.
Fig. 2.
Family-based association results using single SNP markers. Empirical P values from HBAT analyses were calculated after 10,000 permutations. Isometric strength for knee extension (×) and flexion (bold line) and dynamic strength at 60°/s (+), at 120°/s (circle) and at 240°/s (♦) are shown. Significant results (P < 0.01, −log P value > 2.0) were marked with associated genes. All flexion and extension measurements were taken at an angle of 30 and 60°, respectively.
The effect of age, stature, and estimated fat-free mass on muscle strength was also evaluated. None of the polymorphisms significantly associated with muscle strength were associated with age, stature, or fat-free mass (data not shown). FBAT analyses of the strength characteristics corrected for these confounding factors showed a reduced significance for ATP2A2, PPP1CC and NUAK1 and increased significance of TXNRD1 (thioredoxin reductase 1) located next to NUAK1 compared with the uncorrected P values. These results are available as Supplemental Fig. S1.
Stage 2 Genotyping
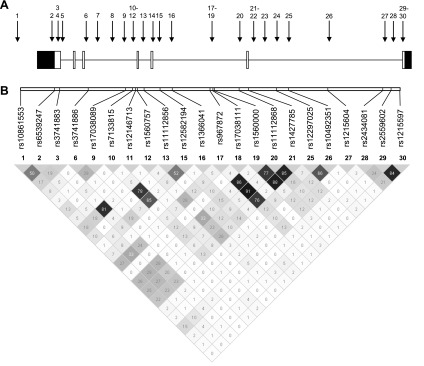
Table 1 and Fig. 3 show the putative function and the location of the 30 SNPs selected in NUAK1. Six assays failed during analyses and two SNPs were 100% homozygous, resulting in 22 informative SNPs, all in Hardy-Weinberg equilibrium (P > 0.001). Three SNPs (rs10861553, rs3741886, and rs2434081) showed significant associations with all flexion strength measurements (P values ranging from 0.040 to 0.00092) (Table 3). Including age, stature, and fat-free mass as covariates resulted in P values ranging between 0.09 and 0.006 for the associations between the latter three SNPs and the strength measurements as shown in Supplemental Table S2.
Fig. 3.
Location of the selected SNPs in NUAK1. A: the locations of the SNPs are indicated by the arrows with numbers (cfr. Table 1). Exons and introns are depicted as boxes and lines, respectively. White boxes are exons and black boxes are untranslated regions. B: pairwise r2 was estimated and is displayed by a gray-shaded square with the hundredths of r2 value inside (Haploview). Darker shades of gray indicate higher degrees of linkage disequilibrium (LD).
Table 3.
Empirical P values for family-based association analyses for NUAK1
| rs10861553 | rs6539247 | rs3741883 | rs3741886 | rs17038089 | rs7133815 | rs12146713 | rs1560757 | rs11112856 | rs12582194 | rs1366041 | rs967872 | rs17038111 | rs1560000 | rs11112868 | rs1427785 | rs12297025 | rs10492351 | rs1215604 | rs2434081 | rs2559602 | rs1215597 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Knee extensors | ||||||||||||||||||||||
| Isometric, Nm | 0.061 | 0.22 | 0.99 | 0.013 | 0.064 | 0.16 | 0.45 | 0.72 | 0.43 | 0.97 | 0.091 | 0.81 | 0.34 | 0.39 | 0.86 | 0.58 | 0.17 | 0.66 | 0.30 | 0.42 | 0.85 | 0.60 |
| Concentric, 60°/s, Nm | 0.15 | 0.50 | 0.98 | 0.032 | 0.74 | 0.053 | 0.50 | 0.43 | 0.19 | 0.45 | 0.14 | 0.67 | 0.14 | 0.10 | 0.67 | 0.058 | 0.45 | 0.34 | 0.86 | 0.17 | 0.99 | 0.65 |
| Concentric, 120°/s, Nm | 0.30 | 0.57 | 0.93 | 0.22 | 0.30 | 0.082 | 0.82 | 0.42 | 0.37 | 0.31 | 0.70 | 0.77 | 0.046 | 0.27 | 0.96 | 0.083 | 0.45 | 0.077 | 0.84 | 0.17 | 0.34 | 0.65 |
| Concentric, 240°/s, Nm | 0.16 | 0.20 | 0.74 | 0.13 | 0.83 | 0.40 | 0.11 | 0.56 | 0.80 | 0.86 | 0.61 | 0.75 | 0.085 | 0.59 | 0.44 | 0.44 | 0.54 | 0.35 | 0.53 | 0.88 | 0.040 | 0.41 |
| Knee flexors | ||||||||||||||||||||||
| Isometric, Nm | 0.00092 | 0.026 | 0.29 | 0.0065 | 0.11 | 0.35 | 0.079 | 0.77 | 0.71 | 0.61 | 0.21 | 0.33 | 0.16 | 0.76 | 0.19 | 0.42 | 0.10 | 0.64 | 0.17 | 0.018 | 0.71 | 0.92 |
| Concentric, 60°/s, Nm | 0.00228 | 0.59 | 0.75 | 0.0049 | 0.12 | 0.52 | 0.11 | 0.74 | 0.65 | 0.34 | 0.53 | 0.34 | 0.67 | 0.95 | 0.34 | 0.58 | 0.44 | 0.57 | 0.15 | 0.0059 | 0.14 | 0.69 |
| Concentric, 120°/s, Nm | 0.00132 | 0.13 | 0.97 | 0.0038 | 0.23 | 0.99 | 0.20 | 0.74 | 0.66 | 0.21 | 0.55 | 0.38 | 0.10 | 0.50 | 0.17 | 0.60 | 0.20 | 0.87 | 0.28 | 0.030 | 0.52 | 0.82 |
| Concentric, 240°/s, Nm | 0.11 | 0.50 | 0.73 | 0.023 | 0.41 | 0.99 | 0.15 | 0.38 | 0.38 | 0.37 | 0.54 | 0.071 | 0.20 | 0.32 | 0.055 | 0.16 | 0.14 | 0.091 | 0.57 | 0.39 | 0.49 | 0.96 |
All flexion and extension measurements were taken at an angle of 30 and 60°, respectively. Boldface indicates significant associations at P < 0.05.
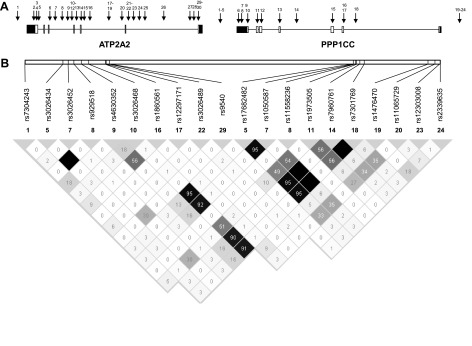
The functionality and position of the 24 and 29 polymorphisms selected in PPP1CC and ATP2A2, respectively, are depicted in Table 1 and Fig. 4. Five assays failed during analyses and 28 appeared to be 100% homozygous, giving a total of 20 informative SNPs, of which one was not in Hardy-Weinberg equilibrium (rs4630352). Results from the family-based association analyses can be found in Table 4. Significant associations and trends toward significance over several strength measurements were present for rs3026468 in ATP2A2 and for rs1050587, rs7960761, and rs7301769 in PPP1CC. However, as the LD between rs3026468 and the significant PPP1CC SNPs is rather high (r2 values between 0.91 and 0.95), it is possible that they all point toward the same “strength increasing allele.”
Fig. 4.
Location of the selected SNPs in ATP2A2 and PPP1CC. A: the locations of the SNPs are indicated by the arrows with numbers (cfr. Table 1). Exons and introns are depicted as boxes and lines, respectively. White boxes are exons and black boxes are untranslated regions. B: pairwise r2 was estimated and is displayed by a gray-shaded square with the hundredths of r2 value inside (Haploview). Darker shades of gray indicate higher degrees of LD.
Table 4.
Empirical P values for family-based association analyses for ATP2A2 and PPP1CC
|
ATP2A2 |
PPP1CC |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7304243 | rs3026434 | rs3026452 | rs929518 | rs4630352 | rs3026468 | rs1860561 | rs12297171 | rs3026489 | rs9540 | rs17682482 | rs1050587 | rs11558236 | rs1973505 | rs7960761 | rs7301769 | rs1476470 | rs11065729 | rs12303008 | rs2339635 | |
| Knee extensors | ||||||||||||||||||||
| Isometric, Nm | 0.81 | 0.49 | 0.95 | 0.54 | 0.54 | 0.016 | 0.020 | 0.11 | 0.92 | 0.34 | 0.14 | 0.023 | 0.49 | 0.14 | 0.055 | 0.042 | 0.20 | 0.84 | 0.69 | 0.66 |
| Concentric, 60°/s, Nm | 0.99 | 0.51 | 0.98 | 0.49 | 0.80 | 0.022 | 0.36 | 0.78 | 0.67 | 0.68 | 0.075 | 0.081 | 0.53 | 0.21 | 0.091 | 0.079 | 0.39 | 0.71 | 0.80 | 0.72 |
| Concentric, 120°/s, Nm | 0.20 | 0.49 | 0.45 | 0.45 | 0.36 | 0.017 | 0.16 | 0.55 | 0.58 | 0.58 | 0.17 | 0.14 | 0.51 | 0.68 | 0.16 | 0.14 | 0.48 | 0.46 | 0.35 | 0.32 |
| Concentric, 240°/s, Nm | 0.28 | 0.52 | 0.25 | 0.50 | 0.625 | 0.0015 | 0.21 | 0.48 | 0.63 | 0.84 | 0.017 | 0.0091 | 0.47 | 0.17 | 0.0091 | 0.0098 | 0.68 | 0.42 | 0.55 | 0.76 |
| Knee flexors | ||||||||||||||||||||
| Isometric, Nm | 0.094 | 0.51 | 0.86 | 0.52 | 0.64 | 0.0044 | 0.42 | 0.13 | 0.79 | 0.56 | 0.13 | 0.012 | 0.47 | 0.23 | 0.025 | 0.028 | 0.010 | 0.88 | 0.39 | 0.11 |
| Concentric, 60°/s, Nm | 0.16 | 0.46 | 0.19 | 0.50 | 0.17 | 0.019 | 0.40 | 0.51 | 0.36 | 0.24 | 0.39 | 0.058 | 0.49 | 0.93 | 0.079 | 0.091 | 0.15 | 0.81 | 0.86 | 0.99 |
| Concentric, 120°/s, Nm | 0.38 | 0.52 | 0.051 | 0.53 | 0.54 | 0.27 | 0.49 | 0.12 | 0.61 | 0.64 | 0.98 | 0.56 | 0.50 | 0.36 | 0.75 | 0.73 | 0.54 | 0.091 | 0.73 | 0.40 |
| Concentric, 240°/s, Nm | 0.26 | 0.51 | 0.11 | 0.50 | 0.58 | 0.17 | 0.85 | 0.66 | 0.69 | 0.70 | 0.69 | 0.46 | 0.52 | 0.44 | 0.53 | 0.53 | 0.99 | 0.72 | 0.74 | 0.79 |
All flexion and extension measurements were taken at an angle of 30 and 60°, respectively. Bold face indicates significant difference at P < 0.05.
Inclusion of age, stature, and fat-free mass as covariates reduced the evidence for association between PPP1CC and the muscle strength characteristics (Supplemental Table S3). Three SNPs show significant associations with a single strength phenotype (rs17682482, extensor strength at 240°/s, P = 0.048; rs1050587 - isometric flexor strength, P = 0.047 and rs11065729, flexor strength at 120°/s, P = 0.050). In ATP2A2, significant associations remain present between rs3026468 and extensor strength (0°/s and 240°/s, P values of 0.045 and 0.033, respectively). Associations between rs7304243, rs3026452, and rs1860561 and a single strength phenotype are also observed.
DISCUSSION
This paper described a two-staged fine mapping of a previously identified linkage peak on chr12q22-23 (23, 26). FBATs showed an association between knee muscle torque production and rs10861553 and rs3741886 in NUAK1, rs3026468 located in ATP2A2, and rs1050587 and rs7301769 in PPP1CC. Additional putatively functional polymorphisms were not associated with knee muscle strength.
After stage 1, the ATP2A2, PPP1CC, and NUAK1 genes were selected as candidate genes for follow-up. The ATP2A2 gene encodes the sarcoplasmic reticulum calcium ATPase 2. It is expressed predominantly in cardiac and slow-twitch skeletal muscle (34, 64) and is involved in the calcium handling necessary for the muscle contraction-relaxation cycle (40). Darier's disease patients, who have an altered ATP2A2 expression level due to mutations in the ATP2A2 gene, have a prolonged contraction time and half-relaxation time of the adductor pollicis muscle (36).
NUAK1 and PPP1CC both play a role in glycogen metabolism. NUAK1, an AMP-activated protein kinase, is expressed in skeletal muscle. It could have both a structural and a regulatory role in the reaction of the muscle on muscle contraction. Hoppe et al. (20) suggested that unc82 (the Caenorhabditis elegans homolog of NUAK1) can regulate some aspects of thick filament organization during changes in muscle length. Fisher et al. (18) indicated that NUAK1 could play a regulatory role in muscle metabolism and be active in glucose transport-related pathways that are controlled similarly by muscle contraction and insulin.
PPP1CC (also known as PPP1G) is a subunit of the protein phosphatase 1 and is universal in skeletal muscle (37). Glycogen-associated phosphatases such as the protein phosphatase 1 are responsible for the dephosphorylation and subsequent inactivation of glycogen synthase. PPP1CC is not required for insulin-stimulated glycogen synthesis in skeletal muscle but appears to be a component of the response to contractile action (6, 38).
Since we did not a priori select our candidate genes based on their specific function, it is surprising that two genes from complementary pathways in glycogen metabolism were identified in our study.
In the second stage of the study, we genotyped additional tagging SNPs and SNPs likely to influence gene function in an extended sample. In NUAK1, two tagSNPs and one conserved SNP in a transcription factor binding site showed associations with multiple muscle strength measurements. For ATP2A2 and PPP1CC 3 tagSNPs and one SNP in a splice enhancer showed association.
Because muscle strength can be influenced by factors such as age, stature, and fat free mass, these factors were included as covariates in the analyses. As FBAT/HBAT analyses cannot directly incorporate confounding factors in the analyses, we performed a multiple regression of the strength measurements on age, weight, and fat-free mass. The resulting residuals were then used as input phenotypes for the FBAT/HBAT analyses. For both stage 1 and stage 2 analyses, inclusion of the covariates resulted in reduced significance of the association findings. For NUAK1 evidence for association remained present for rs10861553 (TagSNP), rs2434081, and rs2559602 (two conserved SNPs), while a trend toward association became apparent for rs12146713, a conserved SNP. For the ATP2A2/PPP1CC locus, only rs3026468, a tagSNP, remained significant over different strength measurements.
As most associated SNPs are intronic tagSNPs direct effects on protein function are not expected. However, it is possible that these tagSNPs are located in a currently unknown transcriptional regulatory element or in a regulatory miRNA, often found in intronic sequences (9, 32, 39, 46, 55). Alternatively, these SNPs could be in LD with neighboring, yet untyped, polymorphisms. These polymorphisms can be currently unknown polymorphisms within the NUAK1 or ATP2A2 genes or polymorphisms located in neighboring genes. The latter seems surprising given the low levels of intragenic LD. However, several studies show that even though strong nonrandom association between intragenic polymorphisms is present, a nonnegligible proportion of intragenic SNPs is in weak LD with each other (4, 5, 45, 54). Moreover, LD has been reported between quite distant markers (2, 12, 45). Screening of the LD structure around rs10864553 showed that no other genes are located within this region. The specific source of the association therefore remains unclear. For the ATP2A2/PPP1CC locus, it is possible that one of the hypothetical proteins in the region (HSU79274, FLJ21127, or MGC15619) has a yet unknown function in muscle strength.
In an effort to strengthen our association results with gene expression data, we genotyped the associated SNPs in a sample (n = 23) for which knee strength data and muscle biopsies were available (56). Unfortunately, due to the small sample size, no significant SNP effects on muscle strength or ATP2A2/PPP1CC/NUAK1 expression could be detected (data not shown).
Another explanation of this limited set of significant association findings is the expected number and effect size of associated SNPs with the current sample size. Depending on the approach taken in power analyses [family-based, in PBAT, with a model including MAF, mode of inheritance, LD between marker and trait locus and effect size, or based on r2 of the SNP effect, without the influence of the MAF in Genetic Power Calculator (43)], different outcomes on the power of this study are found. To address this issue, we applied similar assumptions to the power calculations as reported in Hagberg et al. (19). Given that 122 SNPs were tested in the first phase and 83 SNPs in the second phase, the alpha-level was adapted to 0.0004 and 0.0006 respectively (Bonferroni correction: 0.05/122 = 0.0004 - 0.05/83 = 0.0006). With a sample size of 500 (and 536) subjects, this study has 83% (and 87%) power to detect a SNP with an effect size of 4%. This probably represents an effect size that can be expected for SNP variants at the higher end of the effect-size distribution for complex traits and corresponds well to the top four listed SNP effects in genome-wide association (GWA)-detected variants for training-induced responses in VO2max in the HERITAGE family study, which was also based on a similar sample size of 473 whites (10). Power calculations in PBAT for continuous traits show less optimal power as the stage 2 sample could detect a locus with an effect of 15% explained variance with 80% power, if the locus MAF exceeds 0.08. This estimate corresponds well to the lower bound of MAF for the SNPs that were associated at P < 1.5 × 10−4 with change in VO2max in Bouchard et al. (10). Given that the chromosomal region was identified by linkage analysis and SNPs have been selected in muscle-related prioritized genes, we might hypothesize the detection of association effects of larger size (e.g., in the 4% range).
In addition, the availability of numerous genes and environmental factors that possibly influence the phenotypes under study and the presence of gene-gene and gene-environment interactions will also affect the power of the study. However, a strategy similar to the one described here has successfully identified the ACVR1B gene in the chr12q12-14 linkage region (23, 26) as a strength-determining gene (60).
It can be argued that the currently applied fine-mapping strategy is biased toward known genes. Indeed, we did not apply a true hypothesis-free approach for this genomic region as is done in a GWA. In the first stage a prioritizing bioinformatics tool, Endeavour (3), was used to select a set of genes based on their ranked position as candidate genes. Endeavour uses multiple heterogeneous data sources [literature, functional annotation (GO), microarray expression, expressed sequence tag expression, protein domains, protein-protein interactions, pathway membership, cis-regulatory modules, transcriptional motif, and sequence similarity] based on a set of reference genes. We applied five ranking procedures using different sets of reference genes and therefore feel strongly that the selected genes were well chosen and allowed us to apply a cost-effective strategy for SNP selection in those genes. However, the outcome of the in silico-based gene selection needs to be placed within the specific available database information at the time of selection.
In addition, it should be noted that not all genetic variation was captured using our stage 1 SNP selection method because we focused on SNPs with a MAF >5%. It is therefore plausible that genes and/or polymorphisms other than the ones we investigated are involved in the linkage signal. Moreover, genes for which no significant associations were found should not be considered negatively associated as not all SNPs were covered (e.g., rare alleles). However, we were aware of these limitations from the beginning of our study (as stated in Ref. 60) and acknowledge that additional research will be necessary to determine which other genes and polymorphisms are responsible for the linkage signal. The analyses should also take into account that since the onset of this study a considerable amount of new information about human genetic variants has become available (e.g., 1,000 genomes project).
Even though dense SNP panels (current study, 122 SNPs, average marker distance 0.28 cM) are shown to induce higher genetic informativeness than sparser microsatellite markers (original study, 11 microsatellites, average marker distance 4.9 cM) (17, 59), we could not confirm the original linkage peak using our SNP selection. This difference could be related to sample size differences or regional informativeness of closely chosen markers. Therefore we reran the analyses on the subset of individuals that had both microsatellite and SNP data available (169 siblings from 57 families). In this subset, a clear linkage peak is present in all analyses. An example of these analyses can be found in the supplemental material (Supplemental Fig. S1, available online). In addition, this model was compared with a model including both linkage and association, and results support an association between rs10861553 and rs3741886 in NUAK1 and strength of the knee flexors (data not shown).
In the original linkage study (26) insulin-like growth factor 1 (IGF1) and myosin binding protein C, slow type (MYBPC1) were suggested as possible candidate genes as IGF1 has a role in muscle growth, repair, and hypertrophy (8), and MYBPC1 is involved in muscle contraction (29). However, for none of the eight SNPs in MYBPC1 and the five SNPs in IGF1 associations reached our significance threshold.
In summary, we conducted a genetic fine-mapping study to identify and prioritize a small number of genes within a list of muscle-related prioritized genes underlying a previously reported linkage peak for knee muscle strength on chr12q22-23. TagSNPs in the NUAK1, ATP2A2, and PPP1CC genes were associated with knee muscle strength. Despite detailed analyses of putatively functional polymorphisms in these genes, no additional associations could be detected. Further in-depth analyses are necessary to elucidate the exact role of these loci in human muscle strength and to determine which (functional) polymorphisms are at the base of the linkage and association results.
GRANTS
A. Windelinckx and W. Huygens were funded by the Research Fund of the K. U. Leuven (OT/04/44 and OT/98/39, respectively). G. De Mars was funded by Research Foundation Flanders (FWO) Grant G.0496.05; he passed away on February 28th 2010. Maarten Peeters is a postdoctoral researcher for the Research Foundation Flanders. The fine mapping phase of the Leuven Genes for Muscular Strength Study is funded by OT/04/44 and the FWO (G.0496.05).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Ivo Salden for support during the preparation of DNA samples; Ruben van 't Slot, Bart Claes, and Gilian Peuteman for the genotyping; Monique Ramaekers and Els Van den Eede for assistance in the biopsy study; and Karolina Szlufcik for support during the mRNA expression analyses.
Present address of C. Wijmenga: Dept. Genetics, University Medical Center Groningen and University of Groningen, Groningen, Nl-9713 GZ, The Netherlands.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101, 2002. [DOI] [PubMed] [Google Scholar]
- 2.Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WO. Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet 68: 191–197, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat Biotechnol 24: 537–544, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet 69: 582–589, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3: 299–309, 2002. [DOI] [PubMed] [Google Scholar]
- 6.Arsic N, Zacchigna S, Zentilin L, Ramirez-Correa G, Pattarini L, Salvi A, Sinagra G, Giacca M. Vascular endothelial growth factor stimulates skeletal muscle regeneration in Vivo. Mol Ther 10: 844–854, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Barton ER. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl Physiol Nutr Metab 31: 791–797, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Beohar N, Kawamoto S. Transcriptional regulation of the human nonmuscle myosin II heavy chain-A gene. Identification of three clustered cis-elements in intron-1 which modulate transcription in a cell type- and differentiation state-dependent manner. J Biol Chem 273: 9168–9178, 1998. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard C, Sarzynski M, Rice TK, Kraus WE, Church TS, Sung YJ, Rao DC, Rankinen T. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J Appl Physiol 110: 1160–1170, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc 41: 35–73, 2009. [DOI] [PubMed] [Google Scholar]
- 12.Collins A, Lonjou C, Morton NE. Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci USA 96: 15173–15177, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 37: 1217–1223, 2005. [DOI] [PubMed] [Google Scholar]
- 14.De Mars G, Windelinckx A, Huygens W, Peeters MW, Beunen GP, Aerssens J, Vlietinck R, Thomis MAI. Genome-wide linkage scan for maximum and length-dependent knee muscle strength in young men: significant evidence for linkage at chromosome 14q24.3. J Med Genet 45: 275–283, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Mars G, Windelinckx A, Huygens W, Peeters MW, Beunen GP, Aerssens J, Vlietinck R, Thomis MA. Genome-wide linkage scan for contraction velocity characteristics of knee musculature in the Leuven Genes for Muscular Strength Study. Physiol Genomics 35: 36–44, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvigneaud N, Matton L, Wijndaele K, Deriemaeker P, Lefevre J, Philippaerts R, Thomis M, Delecluse C, Duquet W. Relationship of obesity with physical activity, aerobic fitness and muscle strength in Flemish adults. J Sports Med Phys Fitness 48: 201–210, 2008. [PubMed] [Google Scholar]
- 17.Evans DM, Cardon LR. Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet 75: 687–692, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher JS, Ju JS, Oppelt PJ, Smith JL, Suzuki A, Esumi H. Muscle contractions, AICAR, and insulin cause phosphorylation of an AMPK-related kinase. Am J Physiol Endocrinol Metab 289: E986–E992, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagberg JM, Rankinen T, Loos R, Perusse L, Roth SM, Wolfarth B, Bouchard C. Advances in exercise, fitness, and performance genomics in 2010. Med Sci Sports Exerc 43: 743–752, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppe PE, Chau J, Flanagan KA, Reedy AR, Schriefer LA. C. elegans unc-82 encodes a serine/threonine kinase important for myosin filament organization in muscle during growth. Genetics 184: 79–90, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet 9: 301–306, 2001. [DOI] [PubMed] [Google Scholar]
- 22.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 26: 61–69, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck R, Beunen G. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics 17: 264–270, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck RF, Beunen G. A quantitative trait locus on 13q14.2 for trunk strength. Twin Res 7: 603–606, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Can J Appl Physiol 29: 186–200, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Huygens W, Thomis MAI, Peeters MW, Aerssens J, Vlietinck R, Beunen GP. Quantitative trait loci for human muscle strength: linkage analysis of myostatin pathway genes. Physiol Genomics 22: 390–397, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Karasik D, Kiel DP. Genetics of the musculoskeletal system: a pleiotropic approach. J Bone Miner Res 23: 788–802, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet 75: 1143–1148, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RHA. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res 86: 51–58, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol 19, Suppl 1: S36–S42, 2000. [DOI] [PubMed] [Google Scholar]
- 31.Landers KA, Hunter GR, Wetzstein CJ, Bamman MM, Weinsier RL. The interrelationship among muscle mass, strength, and the ability to perform physical tasks of daily living in younger and older women. J Gerontol A Biol Sci Med Sci 56: B443–B448, 2001. [DOI] [PubMed] [Google Scholar]
- 32.Lin SL, Miller JD, Ying SY. Intronic MicroRNA (miRNA). J Biomed Biotechnol 2006: 26818, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loos R, Thomis M, Maes HH, Beunen G, Claessens AL, Derom C, Legius E, Derom R, Vlietinck R. Gender-specific regional changes in genetic structure of muscularity in early adolescence. J Appl Physiol 82: 1802–1810, 1997. [DOI] [PubMed] [Google Scholar]
- 34.Lytton J, Zarain-Herzberg A, Periasamy M, MacLennan DH. Molecular cloning of the mammalian smooth muscle sarco(endo)plasmic reticulum Ca2+-ATPase. J Biol Chem 264: 7059–7065, 1989. [PubMed] [Google Scholar]
- 35.Marzetti E, Leeuwenburgh C. Skeletal muscle apoptosis, sarcopenia and frailty at old age. Exp Gerontol 41: 1234–1238, 2006. [DOI] [PubMed] [Google Scholar]
- 36.Marziniak M, Hasselmann D, Tilgen W, Sommer C, Dillmann D. Influence of a mutation of the calcium ATPase 2 (SERCA 2) gene in patients with Darier's disease on the central and peripheral nervous system. Klinische Neurophysiologie 35, 2004. [Google Scholar]
- 37.Newgard CB, Brady MJ, O'Doherty RM, Saltiel AR. Organizing glucose disposal: emerging roles of the glycogen targeting subunits of protein phosphatase-1. Diabetes 49: 1967–1977, 2000. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen JN, Richter EA. Regulation of glycogen synthase in skeletal muscle during exercise. Acta Physiol Scand 178: 309–319, 2003. [DOI] [PubMed] [Google Scholar]
- 39.Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet 32: 650–654, 2002. [DOI] [PubMed] [Google Scholar]
- 40.Periasamy M, Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 35: 430–442, 2007. [DOI] [PubMed] [Google Scholar]
- 41.Phan HM, Alpert JS, Fain M. Frailty, inflammation, and cardiovascular disease: evidence of a connection. Am J Geriatr Cardiol 17: 101–107, 2008. [PubMed] [Google Scholar]
- 42.Phillips SM. Resistance exercise: good for more than just Grandma and Grandpa's muscles. Appl Physiol Nutr Metab 32: 1198–1205, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Cherny SS, Sham P. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150, 2003. [DOI] [PubMed] [Google Scholar]
- 44.Silventoinen K, Magnusson PK, Tynelius P, Kaprio J, Rasmussen F. Heritability of body size and muscle strength in young adulthood: a study of one million Swedish men. Genet Epidemiol 32: 341–349, 2008. [DOI] [PubMed] [Google Scholar]
- 45.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293: 489–493, 2001. [DOI] [PubMed] [Google Scholar]
- 46.Surinya KH, Cox TC, May BK. Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. J Biol Chem 273: 16798–16809, 1998. [DOI] [PubMed] [Google Scholar]
- 47. The International HapMap Consortium. The International HapMap Project. Nature 426: 789–796, 2003. [DOI] [PubMed] [Google Scholar]
- 48.Thomis MA, De Mars G, Windelinckx A, Peeters MW, Huygens W, Aerssens J, Beunen GP. Genome-wide linkage scan for resistance to muscle fatigue. Scand J Med Sci Sports 21: 580–588, 2011. [DOI] [PubMed] [Google Scholar]
- 49.Thomis MA, Van Leemputte M, Maes HH, Blimkie CJR, Claessens AL, Marchal G, Willems E, Vlietinck RF, Beunen GP. Multivariate genetic analysis of maximal isometric muscle force at different elbow angles. J Appl Physiol 82: 959–967, 1997. [DOI] [PubMed] [Google Scholar]
- 50.Thomis MAI, Beunen GP, Van Leemputte M, Maes HH, Blimkie CJ, Claessens AL, Marchal G, Willems E, Vlietinck RF. Inheritance of static and dynamic arm strength and some of its determinants. Acta Physiol Scand 163: 59–71, 1998. [DOI] [PubMed] [Google Scholar]
- 51.Tiainen K, Sipila S, Alen M, Heikkinen E, Kaprio J, Koskenvuo M, Tolvanen A, Pajala S, Rantanen T. Shared genetic and environmental effects on strength and power in older female twins. Med Sci Sports Exerc 37: 72–78, 2005. [DOI] [PubMed] [Google Scholar]
- 52.Tiainen KM, Perola M, Kovanen VM, Sipila S, Tuononen KA, Rikalainen K, Kauppinen MA, Widen EI, Kaprio J, Rantanen T, Kujala UM. Genetics of maximal walking speed and skeletal muscle characteristics in older women. Twin Res Hum Genet 11: 321–334, 2008. [DOI] [PubMed] [Google Scholar]
- 53.Tiainen K, Sipila S, Alen M, Heikkinen E, Kaprio J, Koskenvuo M, Tolvanen A, Pajala S, Rantanen T. Heritability of maximal isometric muscle strength in older female twins. J Appl Physiol 96: 173–180, 2004. [DOI] [PubMed] [Google Scholar]
- 54.Tiret L, Poirier O, Nicaud V, Barbaux S, Herrmann SM, Perret C, Raoux S, Francomme C, Lebard G, Tregouet D, Cambien F. Heterogeneity of linkage disequilibrium in human genes has implications for association studies of common diseases. Hum Mol Genet 11: 419–429, 2002. [DOI] [PubMed] [Google Scholar]
- 55.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, Furukawa H, Nagashima M, Yoshino S, Mabuchi A, Sekine A, Saito S, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 35: 341–348, 2003. [DOI] [PubMed] [Google Scholar]
- 56.Vincent B, De Bock K, Ramaekers M, Van den Eede E, Hespel P, Thomis MA. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol Genomics 32: 58–63, 2007. [DOI] [PubMed] [Google Scholar]
- 57.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics 21: 3445–3447, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Wijndaele K, Duvigneaud N, Matton L, Duquet W, Thomis M, Beunen G, Lefevre J, Philippaerts RM. Muscular strength, aerobic fitness, and metabolic syndrome risk in Flemish adults. Med Sci Sports Exerc 39: 233–240, 2007. [DOI] [PubMed] [Google Scholar]
- 59.Wilcox MA, Pugh EW, Zhang H, Zhong X, Levinson DF, Kennedy GC, Wijsman EM. Comparison of single-nucleotide polymorphisms and microsatellite markers for linkage analysis in the COGA and simulated data sets for Genetic Analysis Workshop 14: Presentation Groups 1, 2, and 3. Genet Epidemiol 29, Suppl 1: S7–S28, 2005. [DOI] [PubMed] [Google Scholar]
- 60.Windelinckx A, De Mars G, Huygens WH, Peeters MW, Vincent B, Wijmenga C, Lambrechts D, Delecluse C, Roth SM, Metter EJ, Ferrucci L, Aerssens J, Vlietinck R, Beunen GP, Thomis MAI. Comprehensive fine mapping of chr12q12–14 and follow-up replication identify activin receptor 1B (ACVR1B) as a muscle strength gene. Eur J Hum Genet 19: 208–215, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Windelinckx A, Vlietinck R, Aerssens J, Beunen G, Thomis MA. Selection of genes and single nucleotide polymorphisms for fine mapping starting from a broad linkage region. Twin Res Hum Genet 10: 871–885, 2007. [DOI] [PubMed] [Google Scholar]
- 62.Wolfe RR. The underappreciated role of muscle in health and disease. Am J Clin Nutr 84: 475–482, 2006. [DOI] [PubMed] [Google Scholar]
- 63.Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics 21: 4181–4186, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zarain-Herzberg A, MacLennan DH, Periasamy M. Characterization of rabbit cardiac sarco(endo)plasmic reticulum Ca2+-ATPase gene. J Biol Chem 265: 4670–4677, 1990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.