Abstract
The lipase production ability of a newly isolated Acinetobacter sp. in submerged (SmF) and solid-state (SSF) fermentations was evaluated. The results demonstrated this strain as one of the rare bacterium, which is able to grow and produce lipase in SSF even more than SmF. Coconut oil cake as a cheap agroindustrial residue was employed as the solid substrate. The lipase production was optimized in both media using artificial neural network. Multilayer normal and full feed forward backpropagation networks were selected to build predictive models to optimize the culture parameters for lipase production in SmF and SSF systems, respectively. The produced models for both systems showed high predictive accuracy where the obtained conditions were close together. The produced enzyme was characterized as a thermotolerant lipase, although the organism was mesophile. The optimum temperature for the enzyme activity was 45°C where 63% of its activity remained at 70°C after 2 h. This lipase remained active after 24 h in a broad range of pH (6–11). The lipase demonstrated strong solvent and detergent tolerance potentials. Therefore, this inexpensive lipase production for such a potent and industrially valuable lipase is promising and of considerable commercial interest for biotechnological applications.
1. Introduction
Lipolytic enzymes catalyse hydrolysis and synthesis of either long-chain (lipases) or short-chain (esterases) acylglycerides [1]. Nowadays, there is an increasing interest in the study of thermostable enzymes not only due to their more thermostability but often due to more resistance to other extreme conditions than their mesophilic homologues [2, 3]. Although thermostable enzymes usually are produced by thermophilic microorganisms, limited mesophilic microorganisms can also produce thermostable or thermotolerant lipases. Microbial extracellular enzymes are of considerable commercial interest for biotechnological applications as they can be produced at low cost [4].
Among the various bacterial lipases being exploited, those from the genera Acinetobacter, Pseudomonas, and Burkholderia have shown activity at a wide range of pH and temperature, and the unique properties of chemo-, regio-, and enantioselectivity have made them the catalysts of choice by most organic chemists and pharmacologists [5]. Thus, extensive research has been focused on lipases from mentioned species in multitude directions like isolation of novel strains, optimization of enzyme production, lipase gene cloning and expression, improvement of enzyme properties by conventional mutagenesis, and emerging approaches like directed evolution [6, 7]. Acinetobacter that is ubiquitous in geographical distribution is a strictly aerobic, gram-negative coccobacillus [8, 9]. Interests in Acinetobacter lipases has increased recently, with the growth of the enzyme industry and the concomitant widening search for novel enzymes and applications [9].
Generally, production of enzymes has been carried out using submerged fermentation (SmF); however, solid-state fermentation (SSF) systems appear promising due to the natural potential and advantages they offer [10]. Coconut oil cake (COC) is an inexpensive, abundant agroindustrial residue which can be used as an attractive substrate for industrialy production of lipase in SSF. COC provides not only nutrients but also good surface area for proper growth and aeration [10, 11].
The engineering of culture condition is an effective and economical mode to improve the enzyme production for particular biotechnological applications [12]. Furthermore, predictive models have been accepted as informative tools for rapid and cost-effective study of microbial growth, their products development, risk assessment and scientific purposes [13]. In the last decade, artificial neural networks (ANNs), adaptive computational data processing systems, have emerged as attractive tools for developing nonlinear empirical models and optimizing the multifactor, nonlinear, and timevariant bioprocess [14–16].
In the present work, a mesophilic thermotolerant lipase-producing bacterium was isolated from oily food waste in Malaysia and identified as Acinetobacter sp. Lipase production by this strain was evaluated in both SmF and SSF systems, and the effective culture parameters were optimized using ANN. Subsequently, the produced enzyme was characterized with emphasis on its biotechnological importance.
2. Materials and Methods
2.1. Bacterial Strain
The bacterial strain used in this study was isolated from oily food waste in Serdang, Selangor, Malaysia and identified as Acinetobacter sp. by the German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany. This strain showed excellent fluorescent-forming ability under UV-light on Rhodamine B agar plate (nutrient agar contained; 1% (v/v) olive oil (as lipase substrate) and 0.001% (w/v) Rhodamine B (as lipase activity indicator)) [17]. The new strain was preserved in sterile 16% (v/v) glycerol in Tryptic Soy Broth (TSB) at −80°C. For the seed culture, it was inoculated into 50 mL of TSB in a 250 mL blue cap bottle and incubated at 29°C under 150 rpm agitation. After 8 h it was harvested and diluted with sterile phosphate buffer (50 mM, pH 7.0) to reach OD600 = 0.5 (inoculum size).
2.2. Lipase Production in SmF
The selected SmF lipase production medium was composed of (% w/v): peptone (5), yeast extract (1), NaCl (0.05), CaCl2 (0.05), lactose (1); and coconut oil (1% v/v). The medium was sterilized for 20 min at 121°C. The SmF cultures were performed in 250 mL blue cap bottles in a rotary incubator shaker (0–250 rpm). The agitation, inoculum size, initial pH, temperature, and time were adjusted according to the central composite rotatable design (CCRD). After lipase production, the cell-free supernatant was obtained by centrifugation at 12,000 × g, 4°C for 10 min prior to lipase assay.
2.3. Substrate Preparation and Lipase Production in SSF
Coconut oil cake (COC) was used as substrate. It was obtained from a local coconut oil mill shop in Serdang, Selangor, Malaysia. Ten grams of dried COC was mixed with distilled water to adjust the required moisture level (1) into a 250 mL Erlenmeyer flask and autoclaved at 121°C for 20 min [18]. The moisture content of the COC was estimated by drying 10 grams of COC at 105°C. To adjust the initial moisture content of the solid medium, COC was soaked with the desired quantity of water. After soaking, the sample was again dried as described above and percentage of moisture content was calculated [19] as follows:
| (1) |
Solid-state fermentations were carried out in 250 mL flasks, containing 10 g COC. The initial pH, moisture content and coconut oil percentage were fixed according to the CCRD design and sterilized by autoclaving at 121°C for 20 min. Each flask was then inoculated with 2% inoculum size per gram solid substrate from the seed culture. Samples were incubated with temperature control and taken according to the time given by the design (CCRD).
Triplicate flasks were processed for each treatment. The enzyme was extracted by adding phosphate buffer (5 mL/g; 50 mM, pH 7.0) in to each flask containing the fermented solid in a rotary shaker at 30°C, 200 rpm for 1 h. Afterwards, samples were pressed through cheese cloths to separate the solid-liquid phases, followed by centrifugation at 2000 × g for 2 min [20]. The supernatant was used for lipase activity assay.
2.4. Lipase Activity Assay and Protein Content Measurement
Lipase activity was assayed according to Kwon and Rhee method [21] using olive oil as substrate. The reaction mixture, consisting of 1 mL crude enzyme (culture filtrate using cellulose acetate filter, pore size of 0.22 μm, Sartorius), 2.5 mL olive oil emulsion (properly mixed of an equal volume olive oil with sodium phosphate buffer, 50 mM, pH 7.0), and 0.02 mL of 20 mM CaCl2, was incubated in a water bath shaker for 30 min at 30°C under 200 rpm agitation. The enzyme reaction in the emulsion system was stopped by adding HCl (1 mL, 6 M) and isooctane (5 mL), followed by properly mixing for 1 min. The upper isooctane layer (4 mL) containing the free fatty acid was transferred to a test tube and properly mixed with 1 mL copper reagent. The reagent was prepared by adjusting the solution of 5% (w/v) copper (II) acetate-1-hydrate to pH 6.1 with pyridine. The free fatty acid dissolved in isooctane was determined by measuring the absorbance of the upper layer at 715 nm after mixture settlement. Lipase activity was determined by measuring the amount of free fatty acid released based on the standard curve of oleic acid in isooctane. One unit of lipase activity was defined as 1.0 μmol of free fatty acid liberated min−1 and reported as Uml−1.
Protein concentration was determined according to the Bradford method using the Bio-Rad assay reagent (catalog number 500-0006) and bovine serum albumin as standard, according to the manufacturer's instructions.
2.5. Experimental Design
A five-level-five-variable CCRD was employed in this study, to reduce the number of experiments and solve the ANN problem which need high amount of training data [22]. According to CCRD, 26 experimental runs were necessary, 5 of them were replicates in centre point. Since in ANN modeling, the replicates do not improve the prediction capability of the network [23], only average of center points was used instead of center points. The experimental data were divided into three sets: training, testing, and validating sets [24], and all tests were performed in triplicate.
The variables and their selected levels, after the preliminary study, for the lipase production optimization in SmF were incubation temperature (27–45°C); initial pH (6–9); inoculum size (1–5%); agitation rate (0–200 rpm); incubation period (24–96 h). The variables and their levels in SSF were: incubation temperature (27–45°C); initial pH (6–9); moisture content (60–100%); olive oil (0–20%); incubation period (72–168 h).
2.6. Artificial Neural Network Analysis
A commercial ANN software, NeuralPower version 2.5 (CPC-X Software) was used for the lipase production optimization in this study. In order to predict the lipase production, multilayer normal feedforward and multilayer full feedforward neural networks were employed separately. Different learning algorithms (incremental back propagation, IBP; batch back propagation, BBP; quickprob, QP; genetic algorithm, GA; Levenberg-Marquardt algorithm, LM) were used for training the networks. Each ANN was trained until the network root of mean square error (RMSE) was lower than 0.001, and average correlation coefficient (R) and average determination coefficient (DC) were equal to 1. Other ANN parameters were chosen as the default values of the software [22]. Three remaining points (3 out of 22) were used to test the obtained network. Finally, experimental values of predicted optimal conditions (Table 4) were used as validating set.
Table 4.
Optimum conditions predicted by ANNs in SmF and SSF systems.
| T (°C) | pH | IS (%) | t (h) | Agt (rpm) | Predicted Lipase activity (U/mg protein) | Actual Lipase activity (U/mg protein) | |
|---|---|---|---|---|---|---|---|
| SmF | 29 | 6.0 | 1 | 24 | 200 | 31.0 | 32.2 |
| T (°C) | pH | M (%) | t (h) | Oo (%) | Predicted Lipase activity (U/mg protein) | Actual Lipase activity (U/mg protein) | |
| SSF | 30 | 6.5 | 90 | 81.6 | 5 | 79.0 | 75.4 |
AAD for SmF: 0.04%; AAD for SSF: 0.05%.
T: growth temperature (°C), pH: initial pH, IS: inoculum size (%), t: incubation period (h), Agt: agitation rate (rpm), Oo: olive oil (%), M: moisture content (%).
2.7. Validation of the Optimized Condition
In order to test the reliability of the estimation capabilities of the employed technique for both SmF and SSF systems, the predicted responses obtained from ANN analysis were compared with the experimental values in triplicate. The coefficient of determination (R2) and absolute average deviation (AAD) were determined, and these values were used together to evaluate the ANN model. The AAD and R2 were calculated by (2) and (3), respectively,
| (2) |
| (3) |
where yi,exp and yi,cal were the experimental and calculated responses, respectively, and p was the number of the experimental runs. n was the number of experimental data. R2 is a measure of the amount of the reduction in the variability of response obtained by using the repressor variables in the model. Since R2 alone is not a measure of the model's accuracy, it is necessary to use absolute average deviation (AAD) analysis, which is a direct method for describing the deviations. Evaluation of R2 and AAD values together would be better to check the accuracy of the model [23]. R2 must be close to 1.0, and the AAD between the predicted and observed data must be as small as possible. The acceptable values of R2 and AAD values mean that the model equation defines the true behavior of the system, and it can be used for interpolation in the experimental domain [23].
2.8. Partial Characterization of Acinetobacter sp. Lipase
2.8.1. Effect of pH and Temperature on the Lipase Activity and Stability
The optimum pH for Acinetobacter sp. lipase activity was determined by carrying out the enzyme assay at different pH (6–9) and our standard assay temperature (30°C). The pH stability of the lipase was tested by incubating the enzyme at different pH (4–11) for 24 h, following by standard enzyme assay [21] and reported as residual activity. The optimum temperature for lipase activity was determined by carrying out the enzyme assay at different temperatures (25–70°C) at the optimum pH obtained. The lipase thermostability was determined by incubating the enzyme solution at different temperatures (50–70°C) for 120 min. The relative activity was determined under standard assay method. All tests were performed in triplicate.
2.8.2. Effect of Organic Solvents on Lipase Activity
To study the effect of organic solvents, aliquots of the enzyme were incubated with the solvents (30% v/v in phosphate buffer, 50 mM, pH 7.0) for 30 min at 30°C under 150 rpm shaking. The solvents were selected based on their different log P values (values in the parenthesis) as follows: methanol (−0.76), acetonitrile (−0.394), ethanol (−0.235), acetone (−0.208), 2-propanol (0.074), ethyl acetate (0.7), and hexane (3.6). The lipolytic activity was measured relative to the control (without solvent) at optimum temperature obtained for lipase activity (45°C).
2.8.3. Effects of Surfactants on Lipase Activity
Effect of various surfactants on the lipase activity was investigated by pre-incubating the enzyme for 30 min at 30°C in phosphate buffer (50 mM, pH 7.0) containing following surfactants (nonionic surfactants; 0.1 and 1% (v/v)): Tween 20, Tween 80, Triton X-100, Span 20; SDS as ionic surfactant (1 and 5 mM) a commercial domestic dish washing detergent (Glo manufactured by colgate-palmolive sdn. Bhd., Malaysia). The lipolytic activity was measured relative to the control (without surfactant) at optimum temperature obtained for lipase activity (45°C).
2.8.4. Effect of Calcium on the Lipase Activity
To determine the effect of calcium ions on the lipase activity, various concentrations of Ca2+ (CaCl2; 10–100 mM) were used in assay condition. The control contained no calcium ions (5 mM ethylene diamine tetraacetic acid (EDTA) as di- and trivalent metal ions chelating agent was employed to remove the calcium ions). The results were reported as relative activity under standard assay method. All tests were performed in triplicate.
3. Results and Discussion
3.1. Production of Lipase in SmF and SSF Systems
Bacteria mostly have been investigated in submerged fermentation in the case of growth and different enzymes production. One of the rare reports about the bacterial enzyme production in SSF would be Burkholderia cepacia lipase production on corn bran [25]. Among different bacterial isolates in our laboratory, only Acinetobacter sp. was able to grow on SSF (data not shown), and the related solid substrate was coconut oil cake (COC) which is abundant and cheap in Malaysia. COC, a byproduct of the extraction of coconut oil, contains soluble sugars (1.6%), starch (29%), fat (6.6%), and the initial moisture content of 9% [18]. It is produced in large amounts in Malaysia and is normally incorporated into pet food. We cultivated the local isolated Acinetobacter sp. in both SmF and SSF systems in order to produce lipase. Lipase production was achieved in SmF production medium and surprisingly on SSF without any additional nutrients. This inexpensive lipase production for such a popular Acinetobacterial lipase was promising and made it a possible candidate for future industrial applications. The average lipase activities were 10.7 U/mg protein in SmF and 15.1 U/mg protein in SSF. Therefore, we tried to enhance the lipase production in both conditions by optimization of effective physical parameters using ANN. The only nutritional factor employed was oil as lipase inducer. It has been reported that oil contents higher than 1.5% led to serious oxygen transfer limitations [26], therefore a fixed amount of 1% coconut oil in SmF. Since the COC lipid content is relatively low, we managed to investigate the influence of additional commercial coconut oil (concentration of 0–20%) as lipase production inducer, in SSF. According to the preliminary studies, the effective parameters for SmF were incubation temperature, initial pH, inoculum size, agitation rate, and incubation period (Table 1). In addition, the effective parameters for SSF were incubation temperature, initial pH, coconut oil content, moisture content, and incubation period (Table 2).
Table 1.
Experimental design used in ANN studies by using five independent variables showing observed values of lipase activity in SmF.
| Run | Temperature (°C) | pH | Inoculum (%) | Time (h) | Agitation (rpm) | Lipase activity (U/mg protein) |
|---|---|---|---|---|---|---|
| 1 | 40.9 | 8.3 | 1.9 | 79.8 | 45.1 | 8.0 |
| 2 | 40.9 | 6.7 | 4.1 | 79.8 | 45.1 | 3.6 |
| 3 | 31.1 | 8.3 | 4.1 | 40.2 | 154.9 | 10.6 |
| 4 | 40.9 | 8.3 | 4.1 | 40.2 | 45.1 | 5.5 |
| 5 | 40.9 | 8.3 | 1.9 | 40.2 | 154.9 | 4.8 |
| 6 | 40.9 | 6.7 | 1.9 | 79.8 | 154.9 | 2.4 |
| 7 | 31.1 | 6.7 | 4.1 | 79.8 | 154.9 | 7.2 |
| 8 | 31.1 | 8.3 | 1.9 | 79.8 | 154.9 | 8.5 |
| 9 | 40.9 | 6.7 | 4.1 | 40.2 | 154.9 | 5.6 |
| 10 | 31.1 | 8.3 | 4.1 | 79.8 | 45.1 | 5.2 |
| 11 | 31.1 | 6.7 | 1.9 | 40.2 | 45.1 | 8.3 |
| 12 | 27.0 | 7.5 | 3.0 | 60.0 | 100.0 | 11.2 |
| 13 | 45.0 | 7.5 | 3.0 | 60.0 | 100.0 | 9.4 |
| 14 | 36.0 | 6.0 | 3.0 | 60.0 | 100.0 | 9.8 |
| 15 | 36.0 | 9.0 | 3.0 | 60.0 | 100.0 | 15.0 |
| 16 | 36.0 | 7.5 | 1.0 | 60.0 | 100.0 | 9.2 |
| 17 | 36.0 | 7.5 | 5.0 | 60.0 | 100.0 | 7.6 |
| 18 | 36.0 | 7.5 | 3.0 | 24.0 | 100.0 | 9.2 |
| 19 | 36.0 | 7.5 | 3.0 | 96.0 | 100.0 | 8.4 |
| 20 | 36.0 | 7.5 | 3.0 | 60.0 | 0 | 4.2 |
| 21 | 36.0 | 7.5 | 3.0 | 60.0 | 200.0 | 10.6 |
| 22 | 36.0 | 7.5 | 3.0 | 60.0 | 100.0 | 7.2 |
| 23 | 36.0 | 7.5 | 3.0 | 60.0 | 100.0 | 7.6 |
| 24 | 36.0 | 7.5 | 3.0 | 60.0 | 100.0 | 5.9 |
| 25 | 36.0 | 7.5 | 3.0 | 60.0 | 100.0 | 8.1 |
| 26 | 36.0 | 7.5 | 3.0 | 60.0 | 100.0 | 8.6 |
Table 2.
Experimental design used in ANN studies by using five independent variables showing observed values of lipase activity in SSF.
| Run | Temperature (°C) | pH | Moisture content (%) | Time (h) | Olive oil (%) | Lipase activity (U/mg protein) |
|---|---|---|---|---|---|---|
| 1 | 40.9 | 8.3 | 69.0 | 146.4 | 4.5 | 15.0 |
| 2 | 40.9 | 6.7 | 91.0 | 146.4 | 4.5 | 17.0 |
| 3 | 31.1 | 8.3 | 91.0 | 93.6 | 15.5 | 27.5 |
| 4 | 40.9 | 8.3 | 91.0 | 93.6 | 4.5 | 15.5 |
| 5 | 40.9 | 8.3 | 69.0 | 93.6 | 15.5 | 17.0 |
| 6 | 40.9 | 6.7 | 69.0 | 146.4 | 15.5 | 0 |
| 7 | 31.1 | 6.7 | 91.0 | 146.4 | 15.5 | 10.0 |
| 8 | 31.1 | 8.3 | 69.0 | 146.4 | 15.5 | 0 |
| 9 | 40.9 | 6.7 | 91.0 | 93.6 | 15.5 | 16.0 |
| 10 | 31.1 | 8.3 | 91.0 | 146.4 | 4.5 | 27.5 |
| 11 | 31.1 | 6.7 | 69.0 | 93.6 | 4.5 | 14.0 |
| 12 | 27.0 | 7.5 | 80.0 | 120.0 | 10.0 | 0 |
| 13 | 45.0 | 7.5 | 80.0 | 120.0 | 10.0 | 0.5 |
| 14 | 36.0 | 6.0 | 80.0 | 120.0 | 10.0 | 8.5 |
| 15 | 36.0 | 9.0 | 80.0 | 120.0 | 10.0 | 7.0 |
| 16 | 36.0 | 7.5 | 60.0 | 120.0 | 10.0 | 20.0 |
| 17 | 36.0 | 7.5 | 100 | 120.0 | 10.0 | 8.5 |
| 18 | 36.0 | 7.5 | 80.0 | 72.0 | 10.0 | 11.5 |
| 19 | 36.0 | 7.5 | 80.0 | 168.0 | 10.0 | 30.5 |
| 20 | 36.0 | 7.5 | 80.0 | 120.0 | 0 | 24.5 |
| 21 | 36.0 | 7.5 | 80.0 | 120.0 | 20.0 | 0.5 |
| 22 | 36.0 | 7.5 | 80.0 | 120.0 | 10.0 | 9.5 |
| 23 | 36.0 | 7.5 | 80.0 | 120.0 | 10.0 | 13.0 |
| 24 | 36.0 | 7.5 | 80.0 | 120.0 | 10.0 | 11.5 |
| 25 | 36.0 | 7.5 | 80.0 | 120.0 | 10.0 | 10.0 |
| 26 | 36.0 | 7.5 | 80.0 | 120.0 | 10.0 | 12.5 |
3.2. Artificial Neural Network Analysis and Modeling
The best selected ANN model for lipase production in SmF system was a multilayer normal feed forward incremental backpropagation network, while the best model for lipase production in SSF system was a multilayer full feed forward incremental backpropagation network. The optimized values of learning rate and momentum for both fermentation system networks were 0.15 and 0.8, respectively. The best topology was Gaussian transfer function consisted of a 5-15-1 (inputs-hidden layer-output neurons) for both SmF and SSF systems. These models architectures and topologies were very close to model obtained by Ebrahimpour et al. [22] and Khoramnia et al. [27] which indicates the usefulness of this kind of network for microbial lipase production studies. The learning was accomplished in RMSE < 0.001, R = 1, and DC = 1. In the case of SmF selected network, R2 was 0.998, and related AADs were 1.2% (for training) and 1.7% (for testing). In the case of SSF, R2 was 0.997, and related AADs were 0.9% (for training) and 2.0% (for testing) (Table 3).
Table 3.
Actual and ANN predicted lipase activities in SmF and SSF systems along with R2 and AAD.
| SmF | SSF | ||
|---|---|---|---|
| Lipase activity (U/mg protein) | Lipase activity (U/mg protein) | ||
| Actual | Predicted | Actual | Predicted |
| 8.0 | 7.9 | 15.0 | 16.5 |
| 3.6 | 3.7 | 17.0 | 18.5 |
| 10.6 | 7.0 | 27.5 | 29.5 |
| 5.5 | 7.0 | 15.5 | 14.0 |
| 4.8 | 4.1 | 17.0 | 19.5 |
| 2.4 | 1.1 | 0 | 0 |
| 7.2 | 7.5 | 10.0 | 11.0 |
| 8.5 | 6.2 | 0 | 0.5 |
| 5.6 | 6.3 | 16.0 | 15.5 |
| 5.2 | 5.6 | 27.5 | 28.0 |
| 8.3 | 8.6 | 14.0 | 15.0 |
| 11.2 | 10.2 | 0 | 0 |
| 9.4 | 10.4 | 0.5 | 0.5 |
| 9.8 | 9.9 | 8.5 | 8.0 |
| 15.0 | 13.4 | 7.0 | 7.0 |
| 9.2 | 9.5 | 20.0 | 21.5 |
| 7.6 | 3.4 | 8.5 | 8.5 |
| 9.2 | 11.9 | 11.5 | 10.5 |
| 8.4 | 8.3 | 30.5 | 32.0 |
| 4.2 | 5.8 | 24.5 | 21.5 |
| 10.6 | 9.8 | 0.5 | 0 |
| 7.2 | 10.0 | 9.5 | 7.0 |
SmF: AAD ANN (training) = 1.2%, AAD ANN (testing, italic) = 1.7%, R2 = 0.998,
SSF: AAD ANN (training) = 0.9%, AAD ANN (testing, italic) = 2.0%, R2 = 0.997
3.3. Optimum Conditions and Verification Study
The optimum conditions for the lipase production in SmF and SSF systems were predicted by each corresponding best fitted model of ANN (Table 4). The optimum lipase production of Acinetobacter sp. in SmF was 32.2 U/mg protein (3-fold increase) under the condition of growth temperature (29°C), inoculum size (1%), agitation rate (200 rpm), incubation period (24 h), and initial pH (6). The optimum lipase production condition for SSF was temperature (30°C), olive oil (5%), incubation period (81.5 h), moisture content (90%), and initial pH (6.5) which revealed 75.4 U/mg protein (5 times increase). In order to confirm these results, lipase production was carried out under optimum conditions for both fermentation processes in triplicate. As it has been shown in Table 4, the actual values of both SmF and SSF lipase activities were very close to the predicted values. The well correlation between predicted and experimental values justified the validity of the ANN models and the existence of optimum points. According to the obtained values of R2 and AAD for all data sets, it can be concluded that ANN optimization system has enough capability to predict and generalize both known (training) and unknown (testing and validating) data sets. Optimization by ANNs has also drastically enhanced the lipase production in both fermentation systems. These results confirmed the previous findings, which indicated the superiority of ANNs in comparison to other modeling methods in biological systems [15, 16, 22, 23, 27, 28].
3.4. Main Effects and Interactions between Parameters in SmF and SSF Lipase Production
Three dimensional response surface curves were plotted to study the optimum levels and interaction effects of the parameters on lipase production in both SmF and SSF systems. Each three-dimensional plot represents the effect of two independent variables on the lipase production while the other factors were fixed on their optimum points.
Figure 1(a) shows the interaction between time and temperature in SmF, it is obvious that the lipase production of Acinetobacter sp. decreased remarkably as time and temperature increased. The optimum points obtained for time and temperature in SmF were 24 h and 29°C, which is in accordance with the graph behavior. Therefore, to maximize the lipase production, both variables must be kept at their lowest tested levels. Lipases are produced throughout the bacterial growth, with optimum production at late exponential growth phase [29]. Thus, depending on environmental conditions and characteristics of the microorganism itself, the optimum incubation time is based on duration of log phase where limitation of growth elements occurs, and this can be an inducer for the production of some enzymes [22].
Figure 1.
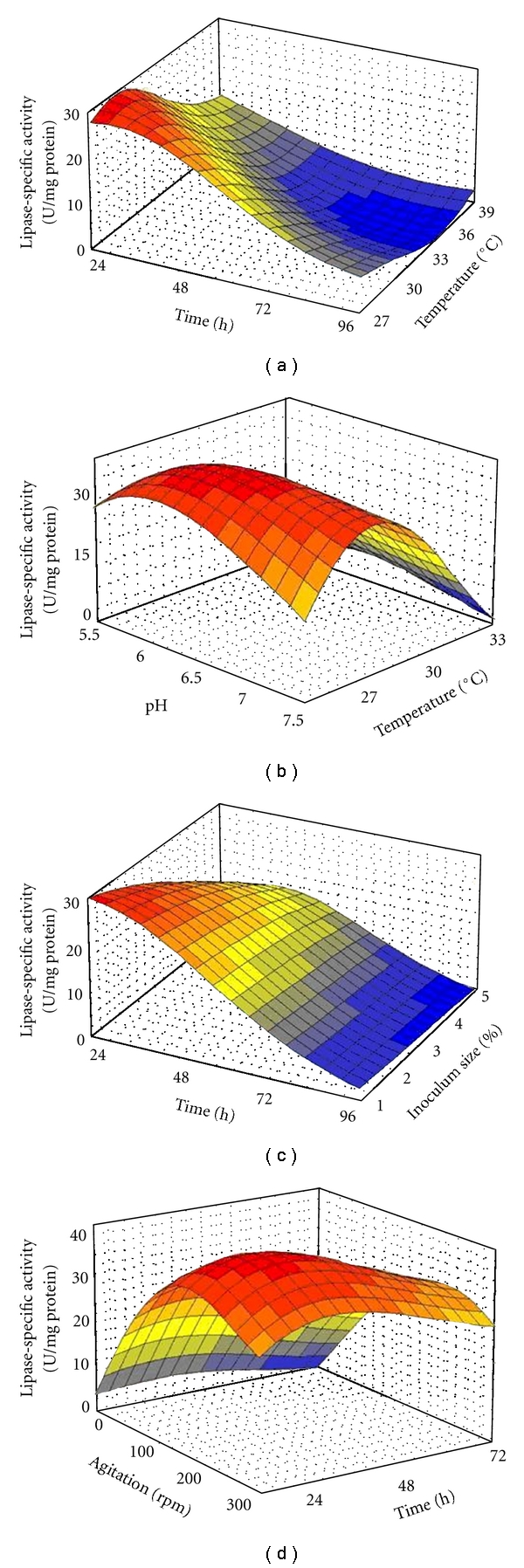
Three dimensional plots for the interaction effect of: (a) time and temperature; (b) pH and temperature; (c) time and inoculum size; (d) agitation and time on Acinetobacter sp. lipase production in SmF. The colors from blue to gray, green, yellow, orange, and red, respectively, show increasing the lipase-specific activity.
Figure 1(b) shows the interaction between pH and temperature in SmF. As it has been shown in this figure, although the amount of temperature significantly changed the graph from minimum to maximum, the pH amount had no significant effect on the lipase production of Acinetobacter sp. Similar behaviors were also observed in the case of temperature-pH interaction (Figure 2(b)) and olive oil-pH interaction (Figure 2(d)) in SSF. The lipase production changed drastically with different temperatures (Figure 2(b)) and decreased with increasing of olive oil concentration (Figure 2(d)), but never changed significantly with different pH values in both graphs. Therefore, in both SmF and SSF lipase productions of Acinetobacter sp., parameter of pH seems not to play an important role in process optimization. This fact is also revealed in Figure 3 that shows the percentage of important parameters released from ANN analysis.
Figure 2.

Three dimensional plots for the interaction effect of: (a) olive oil and temperature; (b) pH and temperature; (c) moisture content and temperature and (d) olive oil and pH, on Acinetobacter sp. lipase production in SSF. The colors from blue to gray, green, yellow, orange, and red, respectively, show increasing the lipase-specific activity.
Figure 3.

Importance of effective parameters on lipase production in SmF (a) and SSF (b) systems.
Figure 1(c) shows the interaction between time and inoculum size in SmF, where the highest lipase production was at the lowest levels of these parameters. As it has been reported by Ebrahimpour et al. [22], in order to enhance the bacterial growth and lipase production, inoculum size must be at suitable amount. The suitable inoculum size is important for bacterium to reach the sufficient nutrient and oxygen levels [22]. Figure 3(a) reveals that inoculum size had the lowest importance for the lipase production by Acinetobacter sp. in SmF compared to other factors. Figure 1(c) also clearly shows that the lipase production was stable in higher values of incubation time in different amount of inoculum size whilst in the optimum incubation time (24 h), inoculum size significantly influenced the amount of the lipase production.
Figure 1(d) shows the time and agitation interaction effects on the lipase production of Acinetobacter sp. where the optimum amount of agitation for lipase activity was 200 rpm. Therefore, to maximize the lipase production, agitation must be kept at the highest tested level but time at the lowest level. Generally, suitable agitation leads to increase of nutrient uptake and dissolve of oxygen in the media [30]. Figures 2(a) and 2(c) represent the olive oil-temperature and moisture content-temperature interactions in SSF, respectively. These plots reveal that the optimum lipase production was achieved at 5% of olive oil and 90% of moisture content. Further increase or decrease in these amounts led to the decrease in the enzyme production.
3.5. Comparison of Lipase Production by Acinetobacter sp. in Different Fermentation Systems
Acinetobacter sp. lipase was produced via SmF and SSF cell cultures. Maximum lipase production by this strain was achieved almost in the same temperature in both SmF and SSF systems. According to the optimum values for Acinetobacter sp. lipase production, pH 6 and 6.5 were the best in SmF and SSF systems, respectively. It was concluded that the bacterium needs slightly acidic or neutral pH values for maximum lipase production. In accordance, it has been reported that the optimal temperature and pH for lipase fermentation of Acinetobacter radioresistens were 30°C and 7, respectively [31]. Unlike the optimum temperature and pH values, which were technically similar in SmF and SSF systems, the amount of lipase inducer (coconut oil) showed a big difference between the systems. Based on preliminary studies, oil was not selected as an effective parameter for lipase production in SmF system. Therefore, a fixed amount of coconut oil (1%) was used in all combinations. In contrast, the importance of coconut oil percentage was attended as one of the most significant parameters in process optimization of lipase production in SSF (Figure 3(b)). The highest predicted yield of lipase production in SSF was achieved with a medium containing 5% of coconut oil. It has been also reported by Fernandes et al. [25] that the maximum lipase activity was achieved when 5% (v/w) of corn oil was added to corn bran, whereas the initial oil concentrations above 5% (v/w) caused a very sticky consistency on the corn bran solids, making them unsuitable for SSF. This amount of coconut oil (5%) in SSF cannot be used in SmF as it has been well established that microorganisms growth and lipase production decrease due to oxygen transfer limitation when large amounts of oil present in the liquid culture media [26]. In this context, it has been reported that olive oil contents higher than 1.5% led to serious oxygen transfer limitations [32]. In the case of incubation time, the big difference between SmF (24 h) and SSF (81.5 h) systems might be due to the lower amount of free water in SSF that may increase the time of bacterial growth and enzyme production.
Eventually, the crude Acinetobacter sp. lipase in SSF showed superior productivities than SmF. As COC is an abundant and inexpensive industrial residue in Malaysia, lipase production by this newly isolated Acinetobacter sp. would be promising for large scale lipase production with significantly reduced cost.
3.6. Partial Characterization of Acinetobacter sp. Lipase
3.6.1. Effects of pH and Temperature on the Lipase Activity and Stability
Temperature and pH are effective parameters, not only on lipase production but also for lipase activity. Therefore, effects of temperature and pH on the newly isolated Acinetobacter sp. lipase activity were studied. The results showed the optimum pH value of 6.0 for lipase activity. The slightly acidic pH is in contrast with previous studies that reported the alkaline lipases from Acinetobacter [9, 33–35]. On the other hand, this lipase was stable at a broad range of pH values between 6 and 11 after 24 h incubation, which makes it applicable in different industrial purposes. The optimal temperature of 45°C (Figure 4) was higher than Chen et al. [33] report, which was 37°C, and lower than Ahmed et al. [34] and Uttatree et al. [35] reports, which were 60°C. Our newly isolated Acinetobacter sp. lipase was stable against thermal denaturation where it remained 63% of its original activity at 70°C after 120 min. Since thermostable lipases, which are active and stable in acidic and alkaline media, are very attractive and have a great potential for different industrial applications, this locally isolated Acinetobacter sp. lipase would be a potent and valuable enzyme for further applications.
Figure 4.

Acinetobacter sp. lipase thermostability (a) and effect of temperature on its rate of olive oil hydrolysis (b).
3.6.2. Effect of Organic Solvents on Lipase Activity
Stability in organic solvents is desirable in enzymatic synthesis reactions. Organic solvents shift the reaction equilibrium toward completion of the reverse reaction of hydrolysis. Moreover, these solvents are able to enhance the solubility of nonpolar substrates, facilitate the nonpolar product recovery, and increase the enzyme thermostability [36]. However, application of enzymes is usually hampered by denaturation and inactivation in the presence of organic solvents [37].
Among different organic solvents (log P values: −0.76 to 3.6) tested in this study, the lipase activity in presence of almost all of them increased as follows: methanol (114.7%), acetonitrile (105.9%), ethanol (153.4%), acetone (133.9%), 2-propanol (135.9%), ethyl acetate (111.4%), and hexane (114.0%) (Figure 5). These results revealed that the lipase not only was stable in the presence of water miscible and water immiscible solvents tested but also most of the solvents tested could even enhance the enzyme activity, where ethanol showed the highest activation (153.4%). As a conclusion, there was no clear correlation between the log P value of an organic solvent and the stability of the lipase in its presence (Figure 5). It can be suggested that the water miscibility is not the only critical factor of solvents affecting enzyme stability. Other factors such as the solvents molecular structures and their functional groups as well as enzyme structure and the type of surface amino acids may also play their roles [38].
Figure 5.
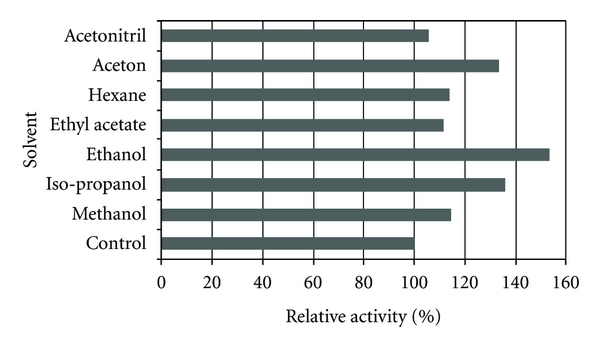
Effects of various organic solvents on Acinetobacter sp. lipase activity. The enzyme was preincubated for 30 min at 30°C with 30% v/v of solvents with 150 rpm agitation. The lipolytic activity was measured at 65°C relative to the control (without solvent), using olive oil (emulsified, 1 : 1 v/v, in 50 mM Tris-HCl buffer, pH 8.0) as substrate.
Review of the literature reveals that microbial lipases are generally stable in organic solvents but they possess different sensitivity to the solvents. Although there is a general belief that polar water miscible solvents are more destabilizing than water immiscible solvents [39, 40], lipases are diverse in their sensitivity to organic solvents [40].
In accordance to our results, Ebrahimpour et al. [38] reported that the activity of lipase from Geobacillus sp. strain ARM in presence of following organic solvents was increased: 2,3-butanediol (100.4%), methanol (107.3%), ethanol (116.4%), benzene (142.0%), toluene (164.9%), 1-octanol (137.3%), o-xylene (136.1%), hexane (139.3%), heptane (149.6%), iso-octane (128.6%), 1-dodecanol (149.6%), n-tetradecane (115.8%), n-pentadecane (128.5%), and heptadecane (142.5%). Hun et al. [41] and Lin [42] found that the lipase activities of Bacillus sphaericus 205y and Pseudomonas pseudoalcaligenes F-111 were enhanced in n-hexane by 3.5 and 2.5 fold, respectively. Lin [42] reported that isooctane enhanced the lipase activity of Pseudomonas pseudoalcaligenes F-111.
Ethanol and methanol enhanced the lipase activity of B. thermocatenulatus [43] and AG-8 lipase [44]. Enhancement of lipase activity in benzene and n-hexane has been reported by Eltaweel et al. [45] for Bacillus sp. strain 42 and Nawani et al. [40] for Bacillus J33. In addition, Hun et al. [41] have reported the enhancement of lipase activity of Bacillus sphaericus 205y by n-hexane and p-xylene.
3.6.3. Effects of Surfactants on Lipase Activity
It is well known that surfactants may affect the structure and function of different enzymes including lipases. The effects of various surfactants on the lipase activity are shown in Figure 6. This enzyme showed an obvious stability in presence of not only nonionic surfactants including Tween 20, Tween 80, Triton X-100, Span 20, but also SDS as ionic surfactant (1 and 5 mM) as well as a commercial domestic dish washing detergent (Glo manufactured by colgate-palmolive sdn. Bhd., Malaysia). In addition, some of surfactants tested were able to increase the lipase activity (Figure 6). The highest increase of relative activity (around 30%) was achieved in the presence of 0.1% (v/v) Triton X-100 followed by 0.1% (v/v) of Tween 20 and 1 mM of SDS. Reported thermophilic lipases have shown variable responses to the presence of surfactants. Stimulating effect of surfactants on enzymatic hydrolysis has been reported several times [46]. Ebrahimpour et al. [38] and Castro-Ochoa et al. [47] found that lipase activity of Bacillus sp. was enhanced in the presence of Triton X-100. The Lip-SBRN2 exhibited a high level of activity in the presence of SDS [48].
Figure 6.

Effects of various surfactants on Acinetobacter sp. lipase activity. The enzyme was preincubated for 30 min at 30°C with 0.1 and 1% (v/v) of each nonionic surfactants and commercial detergent; 1 and 5 mM SDS (ionic surfactant) in phosphate buffer (50 mM, pH 7.0). The lipolytic activity was measured at 65°C relative to the control (without surfactant), using olive oil (emulsified, 1 : 1 v/v, in 50 mM Tris-HCl buffer, pH 8.0) as substrate.
Surfactants may affect the lipase activity by altering the lipase conformation and/or the interfacial property. Explanations for the surfactant effect include enhancement of enzyme stability, improvement of oil substrate solubility in water, and increasing accessibility of the substrate [46, 48].
3.6.4. Effect of Calcium on the Lipase Activity
Metal cations, particularly Ca2+, play important roles in the structure and function of enzymes, and some of the lipases are strictly calcium dependent [49]. In this study it was shown that different concentrations of Ca2+, including concentration zero (the reaction without Ca2+), did not affect the Acinetobacter sp. lipase activity. Therefore, this lipase was not Ca2+ dependent. In accordance with this result, it has been reported that Ca2+ ion was not essential for the enzymatic activity of Acinetobacter baylyi lipase [35]. However, it has been mentioned the positive effect of Ca2+ on enzyme stabilization and activity as a universal property of Acinetobacter lipases [9]. Khoramnia et al. [27] reported that the enzymatic activity of Staphylococcus xylosus lipase was stimulated by Ca2+, but this ion does not seem to be necessary for the lipase activity. In contrast, refolding of Pseudomonas aeruginosa lipase was strictly dependent on calcium [50]. Moreover, it has been demonstrated that the activity of Pseudomonal and Staphylococcal lipases depended on the presence of Ca2+ ions [49, 51].
Acknowledgment
The financial support by Universiti Putra Malaysia is gratefully acknowledged.
References
- 1.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochemical Journal. 1999;343(1):177–183. [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeger KE, Eggert T. Lipases for biotechnology. Current Opinion in Biotechnology. 2002;13(4):390–397. doi: 10.1016/s0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- 3.Sharma R, Soni SK, Vohra RM, Jolly RS, Gupta LK, Gupta JK. Production of extracellular alkaline lipase from a Bacillus sp. RSJ1 and its application in ester hydrolysis. Indian Journal of Microbiology. 2002;42(1):49–54. [Google Scholar]
- 4.Schmidt-Dannert C, Rúa ML, Atomi H, Schmid RD. Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochimica et Biophysica Acta. 1996;1301(1-2):105–114. doi: 10.1016/0005-2760(96)00027-6. [DOI] [PubMed] [Google Scholar]
- 5.Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409(6817):258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 6.Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annual Review of Microbiology. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Guo D, Yan Y. Cloning, expression and characterization of a novel thermal stable and short-chain alcohol tolerant lipase from Burkholderia cepacia strain G63. Journal of Molecular Catalysis B. 2007;45(3-4):91–96. [Google Scholar]
- 8.Nobel WC. Hospital epidemiology of Acinetobacter infection. In: Towner K, Bergogne-Bérézin E, Fewson CA, editors. The Biology of Acinetobacter. New York, NY, USA: Plenum; 1991. [Google Scholar]
- 9.Snellman EA, Colwell RR. Acinetobacter lipases: molecular biology, biochemical properties and biotechnological potential. Journal of Industrial Microbiology and Biotechnology. 2004;31(9):391–400. doi: 10.1007/s10295-004-0167-0. [DOI] [PubMed] [Google Scholar]
- 10.Benjamin S, Pandey A. Coconut cake—A potent substrate for the production of lipase by Candida rugosa in solid-state fermentation. Acta Biotechnologica. 1997;17(3):241–251. [Google Scholar]
- 11.Benjamin S, Pandey A. Mixed-solid substrate fermentation. A novel process for enhanced lipase production by Candida rugosa. Acta Biotechnologica. 1998;18(4):315–324. [Google Scholar]
- 12.Hsu KH, Lee GC, Shaw JF. Promoter analysis and differential expression of the Candida rugosa lipase gene family in response to culture conditions. Journal of Agricultural and Food Chemistry. 2008;56(6):1992–1998. doi: 10.1021/jf073076o. [DOI] [PubMed] [Google Scholar]
- 13.Ross T. Predictive Food Microbiology Models in the Meat Industry. Sydney, Australia: Meat and livestock Australia; 1999. [Google Scholar]
- 14.Liu CH, Lin YH, Chen CY, Chang JS. Characterization of Burkholderia lipase immobilized on celite carriers. Journal of the Taiwan Institute of Chemical Engineers. 2009;40(4):359–363. [Google Scholar]
- 15.Liu L, Sun J, Zhang D, Du G, Chen J, Xu W. Culture conditions optimization of hyaluronic acid production by Streptococcus zooepidemicus based on radial basis function neural network and quantum-behaved particle swarm optimization algorithm. Enzyme and Microbial Technology. 2009;44(1):24–32. doi: 10.1002/btpr.278. [DOI] [PubMed] [Google Scholar]
- 16.Patnaik PR. Synthesizing cellular intelligence and artificial intelligence for bioprocesses. Biotechnology Advances. 2006;24(2):129–133. doi: 10.1016/j.biotechadv.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Kouker G, Jaeger KE. Specific and sensitive plate assay for bacterial lipases. Applied and Environmental Microbiology. 1987;53(1):211–213. doi: 10.1128/aem.53.1.211-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey A, Ashakumary L, Selvakumar P. Copra waste—A novel substrate for solid-state fermentation. Bioresource Technology. 1995;51(2-3):217–220. [Google Scholar]
- 19.Adinarayana K, Prabhakar T, Srinivasulu V, Anitha Rao M, Jhansi Lakshmi P, Ellaiah P. Optimization of process parameters for cephalosporin C production under solid state fermentation from Acremonium chrysogenum. Process Biochemistry. 2003;39(2):171–177. [Google Scholar]
- 20.Gombert AK, Pinto AL, Castilho LR, Freire DMG. Lipase production by Penicillium restrictum in solid-state fermentation using babassu oil cake as substrate. Process Biochemistry. 1999;35(1-2):85–90. [Google Scholar]
- 21.Kwon DY, Rhee JS. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. Journal of the American Oil Chemists’ Society. 1986;63(1):89–92. [Google Scholar]
- 22.Ebrahimpour A, Rahman RNZRA, Ean Ch’ng DH, Basri M, Salleh AB. A modeling study by response surface methodology and artificial neural network on culture parameters optimization for thermostable lipase production from a newly isolated thermophilic Geobacillus sp. strain ARM. BMC Biotechnology. 2008;8, article 96 doi: 10.1186/1472-6750-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bas D, Boyaci IH. Modeling and optimization I: usability of response surface methodology. Journal of Food Engineering. 2007;78(3):836–845. [Google Scholar]
- 24.Cohran WG, Cox GM. Experimental Design. New York, NY, USA: John Wiley & Sons; 2002. [Google Scholar]
- 25.Fernandes MLM, Saad EB, Meira JA, Ramos LP, Mitchell DA, Krieger N. Esterification and transesterification reactions catalysed by addition of fermented solids to organic reaction media. Journal of Molecular Catalysis B. 2007;44(1):8–13. [Google Scholar]
- 26.Lima VMG, Krieger N, Sarquis MIM, Mitchell DA, Ramos LP, Fontana JD. Effect of nitrogen and carbon sources on lipase production by Penicillium aurantiogriseum. Food Technology and Biotechnology. 2003;41(2):105–110. [Google Scholar]
- 27.Khoramnia A, Lai OM, Ebrahimpour A, Tanduba CJ, Tan SV, Mukhlis S. Thermostable lipase from a newly isolated staphylococcus xylosus strain; process optimization and characterization using RSM and ANN. Electronic Journal of Biotechnology. 2010;13(5):22 pages. [Google Scholar]
- 28.Dutta JR, Dutta PK, Banerjee R. Optimization of culture parameters for extracellular protease production from a newly isolated Pseudomonas sp. using response surface and artificial neural network models. Process Biochemistry. 2004;39(12):2193–2198. [Google Scholar]
- 29.Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology. 2004;64(6):763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- 30.Kumar CG, Takagi H. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnology Advances. 1999;17(7):561–594. doi: 10.1016/s0734-9750(99)00027-0. [DOI] [PubMed] [Google Scholar]
- 31.Chen SJ, Cheng CY, Chen TL. Production of an alkaline lipase by Acinetobacter radioresistens. Journal of Fermentation and Bioengineering. 1998;86(3):308–312. [Google Scholar]
- 32.Mateos Diaz JC, Rodríguez JA, Roussos S, et al. Lipase from the thermotolerant fungus Rhizopus homothallicus is more thermostable when produced using solid state fermentation than liquid fermentation procedures. Enzyme and Microbial Technology. 2006;39(5):1042–1050. [Google Scholar]
- 33.Li CY, Cheng CY, Chen TL. Fed-batch production of lipase by Acinetobacter radioresistens using Tween 80 as the carbon source. Biochemical Engineering Journal. 2004;19(1):25–31. [Google Scholar]
- 34.Ahmed EH, Raghavendra T, Madamwar D. An alkaline lipase from organic solvent tolerant Acinetobacter sp. EH28: application for ethyl caprylate synthesis. Bioresource Technology. 2010;101(10):3628–3634. doi: 10.1016/j.biortech.2009.12.107. [DOI] [PubMed] [Google Scholar]
- 35.Uttatree S, Winayanuwattikun P, Charoenpanich J. Isolation and characterization of a novel thermophilic-organic solvent stable lipase from Acinetobacter baylyi. Applied Biochemistry and Biotechnology. 2010;162(5):1362–1376. doi: 10.1007/s12010-010-8928-x. [DOI] [PubMed] [Google Scholar]
- 36.Ogino H. Organic solvent-stable enzymes. In: Thomas KS, Thomas T, editors. Protein Adaptation in Extremophiles. Huntington, NY, USA: Nova Science Publishers; 2008. [Google Scholar]
- 37.Ogino H, Yasui K, Shiotani T, Ishihara T, Ishikawa H. Organic solvent-tolerant bacterium which secretes an organic solvent- stable proteolytic enzyme. Applied and Environmental Microbiology. 1995;61(12):4258–4262. doi: 10.1128/aem.61.12.4258-4262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebrahimpour A, Rahman RNZRA, Basri M, Salleh AB. High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresource Technology. 2011;102(13):6972–6981. doi: 10.1016/j.biortech.2011.03.083. [DOI] [PubMed] [Google Scholar]
- 39.Kamini NR, Iefuji H. Lipase catalyzed methanolysis of vegetable oils in aqueous medium by cryptococcus spp. S-2. Process Biochemistry. 2001;37(4):405–410. [Google Scholar]
- 40.Nawani N, Dosanjh NS, Kaur J. A novel thermostable lipase from a thermophilic Bacillus sp.: characterization and esterification studies. Biotechnology Letters. 1998;20(10):997–1000. [Google Scholar]
- 41.Hun CJ, Rahman RNZA, Salleh AB, Basri M. A newly isolated organic solvent tolerant Bacillus sphaericus 205y producing organic solvent-stable lipase. Biochemical Engineering Journal. 2003;15(2):147–151. [Google Scholar]
- 42.Lin SF. Production and stabilization of a solvent-tolerant alkaline lipase from Pseudomonas pseudoalcaligenes F-111. Journal of Fermentation and Bioengineering. 1996;82(5):448–451. [Google Scholar]
- 43.Schmidt-Dannert C, Sztajer H, Stocklein W, Menge U, Schmid RD. Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenulatus. Biochimica et Biophysica Acta. 1994;1214(1):43–53. doi: 10.1016/0005-2760(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 44.Sharma AK, Tiwari RP, Hoondal GS. Properties of a thermostable and solvent stable extracellular lipase from a Pseudomonas sp. AG-8. Journal of Basic Microbiology. 2001;41(6):363–366. doi: 10.1002/1521-4028(200112)41:6<363::AID-JOBM363>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 45.Eltaweel MA, Rahman RNZRA, Salleh AB, Basri M. An organic solvent-stable lipase from Bacillus sp. strain 42. Annals of Microbiology. 2005;55(3):187–192. [Google Scholar]
- 46.Kristensen JB, Börjesson J, Bruun MH, Tjerneld F, Jørgensen H. Use of surface active additives in enzymatic hydrolysis of wheat straw lignocellulose. Enzyme and Microbial Technology. 2007;40(4):888–895. [Google Scholar]
- 47.Castro-Ochoa LD, Rodríguez-Gómez C, Valerio-Alfaro G, Oliart Ros R. Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme and Microbial Technology. 2005;37(6):648–654. [Google Scholar]
- 48.Kanjanavas P, Khuchareontaworn S, Khawsak P, et al. Purification and characterization of organic solvent and detergent tolerant lipase from thermotolerant Bacillus sp. RN2. International Journal of Molecular Sciences. 2010;11(10):3783–3792. doi: 10.3390/ijms11103783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Khattabi M, Van Gelder P, Bitter W, Tommassen J. Role of the calcium ion and the disulfide bond in the Burkholderia glumae lipase. Journal of Molecular Catalysis B. 2003;22(5-6):329–338. [Google Scholar]
- 50.Shibata H, Kato H, Oda J. Calcium ion-dependent reactivation of a Pseudomonas lipase by its specific modulating protein, LipB. Journal of Biochemistry. 1998;123(1):136–141. doi: 10.1093/oxfordjournals.jbchem.a021900. [DOI] [PubMed] [Google Scholar]
- 51.Rosenstein R, Götz F. Staphylococcal lipases: biochemical and molecular characterization. Biochimie. 2000;82(11):1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]


