Genes belonging to the AG subfamily regulate conserved functions in reproductive organ identity determination and meristem determinacy in eudicots. This work describes a functional analysis of all four AG subfamily members of rice, revealing that their function in reproductive organ development and meristem determinacy is conserved in monocots.
Abstract
Reproductive organ development is one of the most important steps in the life cycle of plants. Studies using core eudicot species like thale cress (Arabidopsis thaliana) and snapdragon (Antirrhinum majus) have shown that MADS domain transcription factors belonging to the AGAMOUS (AG) subfamily regulate the identity of stamens, carpels, and ovules and that they are important for floral meristem determinacy. Here, we investigate the genetic interactions between the four rice (Oryza sativa) AG subfamily members, MADS3, MADS13, MADS21, and MADS58. Our data show that, in contrast with previous reports, MADS3 and MADS58 determine stamen and carpel identity and, together with MADS13, are important for floral meristem determinacy. In the mads3 mads58 double mutant, we observed a complete loss of reproductive organ identity and massive accumulation of lodicules in the third and fourth floral whorls. MADS21 is an AGL11 lineage gene whose expression is not restricted to ovules. Instead, its expression profile is similar to those of class C genes. However, our genetic analysis shows that MADS21 has no function in stamen, carpel, or ovule identity determination.
INTRODUCTION
Twenty years ago, Coen and Meyerowitz (1991) proposed, based on the analysis of thale cress (Arabidopsis thaliana) and snapdragon (Antirrhinum majus) floral homeotic mutants, the genetic ABC model. This model explains how three classes of floral homeotic genes, termed class A, B, and C genes, determine the identity of the four floral organ types: sepals, petals, stamens, and carpels. C-function genes regulate stamen and carpel identity in whorl 3 and 4. Furthermore, they are important to prevent meristem indeterminacy in whorl 4. The Arabidopsis agamous (ag) mutant has normal sepals and petals, whereas the stamens in whorl 3 are homeotically transformed into petals and in whorl 4 instead of a pistil, which in Arabidopsis is composed of two fused carpels, a new ag flower develops (Bowman et al., 1989). AG was the first class C gene cloned and encodes a MIKC-type MADS domain transcription factor (Yanofsky et al., 1990; Parenicová et al., 2003). AG expression starts at stage 3 of flower development in the domains of the floral meristem (FM) where stamen and carpel primordia develop, and its expression remains during all stages of stamen and carpel development. AG is also expressed during ovule development and plays a role in ovule identity determination (Pinyopich et al., 2003; Brambilla et al., 2007).
In Arabidopsis, four genes, AG, SHATTERPROOF1 (SHP1), SHP2, and SEEDSTICK (STK; previously known as AGL11), form the AG monophyletic subfamily within the MADS box gene phylogeny (Parenicová et al., 2003; Kramer et al., 2004; Zahn et al., 2006). Analysis of the evolutionary history of gene duplications showed that the SHP1 and SHP2 genes are the products of a recent duplication event, which probably occurred after the divergence of rosid and asterid eudicots (Vision et al., 2000). These two Arabidopsis genes are expressed in developing pistils, fruit, and ovules. They are redundantly involved in stigma, style, and medial tissue development and specify the dehiscence zone, which is important for the dispersal of seeds (Liljegren et al., 2000; Colombo et al., 2010). SHP1 and SHP2 are also redundant with STK in determining ovule identity, since in the stk shp1 shp2 triple mutant, ovules are converted into carpel-like structures (Pinyopich et al., 2003; Brambilla et al., 2007). STK expression is restricted to the placenta and developing ovules, and the stk single mutant develops ovules with a longer funiculus, which upon seed maturation does not abscise from the seeds (Rounsley et al., 1995; Favaro et al., 2003; Pinyopich et al., 2003).
The duplication event that gave rise to the AG (euAG clade) and the SHP genes (PLENA clade) took place early in the history of the core eudicots (Kramer et al., 2004; Zahn et al., 2006), whereas a more ancient duplication event occurred early in angiosperm evolution, marking the origin of the AG and AGL11 lineages (Kramer et al., 2004; Zahn et al., 2006).
In rice (Oryza sativa), 75 MADS box genes have been identified (Arora et al., 2007) of which four genes, MADS3, MADS58, MADS13, and MADS21, belong to the AG subfamily. MADS3 and MADS58 fall into the AG lineage, whereas MADS13 and MADS21 are AGL11 lineage genes (Kramer et al., 2004; Yamaguchi et al., 2006; Dreni et al., 2007). Although Zahn et al. (2006) placed MADS13 in the AG lineage, other phylogenetic analyses, gene structure, and specific amino acid residues strongly suggest that MADS13 belongs to the AGL11 lineage (Kramer et al., 2004; Dreni et al., 2007).
In rice, the basal unit of the inflorescence is the spikelet, which bears one single floret. The two outer basal bracts, named glumes, are extremely reduced in rice and are called rudimentary glumes. Subsequently, there are two empty glumes, or sterile lemmas, that are homologous to lemmas but fail to initiate florets. The floret is composed of a lemma and palea, which are considered the first-whorl organs and enclose the other floral organs: two lodicules (second whorl), six stamens (third whorl), and a carpel containing a single ovule (fourth whorl).
Recently, rice MADS3 and MADS58, which are considered to be paralogous genes, were characterized (Yamaguchi et al., 2006). Disruption of MADS3 by a T-DNA insertion caused homeotic transformations of stamens into lodicules and ectopic development of lodicules in the second whorl near the palea, whereas carpels developed almost normally. The silencing of MADS58 by an RNA interference (RNAi) approach resulted in carpel developmental defects and FM indeterminacy. From these observations, the authors concluded that during rice evolution subfunctionalization occurred between these two genes. The subfunctionalization of AG clade genes might be a more common feature of grasses. In maize (Zea mays), there are three AG lineage genes, namely, ZAG1 (ortholog of rice MADS58) and ZMM2 and ZMM23 (both orthologs of rice MADS3). ZAG1 and ZMM2 transcripts are detectable in the ear and tassel reproductive organs; however, ZAG1 transcript levels are higher in carpels, whereas those of ZMM2 are more abundant in stamens (Schmidt et al., 1993; Mena et al., 1996). A transposon-induced mutation in ZAG1 resulted in the loss of female FM determinacy, but carpel identity was not affected and male flowers developed normally (Mena et al., 1996). Functional analysis of ZMM2 has not been reported yet, but its expression profile suggests a role in reproductive organ development. Functional and expression analyses of ZMM23 (Münster et al., 2002) have not been reported yet.
Interestingly, in none of the rice AG gene mutants, including the mads3 mads58-RNAi double mutant, was carpel identity lost, suggesting that other genes might regulate carpel identity. This would be in contrast with core eudicot plants where carpel identity has shown to be determined by the AG lineage genes. DROOPING LEAF (DL), a gene belonging to the YABBY transcription factor family, has been suggested as a candidate to determine carpel identity in rice (Nagasawa et al., 2003; Yamaguchi et al., 2006). DL is essential for the development of carpels since in the dl mutant, carpels are homeotically transformed into multiple stamens (Nagasawa et al., 2003). DL is expressed in the carpel anlagen and subsequently during carpel development (Yamaguchi et al., 2004). In the mads58-RNAi and the mads3 mads58-RNAi double mutant lines, DL was still expressed in the carpels, indicating that its expression is independent of MADS3 and MADS58 activity.
Of the rice AGL11 lineage genes, MADS13 has been characterized in most detail. Its expression is, like STK of Arabidopsis, restricted to the ovule primordium and carpel inner epidermis, and its expression remains during all phases of ovule development (Lopez-Dee et al., 1999). In the mads13 mutant, ovules are homeotically transformed into carpelloid structures (Dreni et al., 2007). Based on this phenotype, MADS13 was classified as a class D homeotic gene (Colombo et al., 1995). In the mads13 mutant, a “carpels inside carpels” phenotype was often observed, indicating a loss of FM determinacy. This phenotype was not observed in the Arabidopsis stk shp1 shp2 triple mutant, which also shows homeotic conversions of ovules into carpels (Pinyopich et al., 2003). This might be due to the fact that the ovule in the rice floret develops directly from the FM, whereas in Arabidopsis the developing carpels completely use up the FM, and ovules develop later from meristematic protrusions arising from the placenta (Dreni et al., 2007; Colombo et al., 2008).
MADS13 and MADS21 are paralogous genes; however, MADS21 expression is not restricted to ovules. Its expression is also observed in stamens and carpel tissues, although at low levels (Arora et al., 2007; Dreni et al., 2007). The mads21 single mutant did not show any aberrant phenotype, and when combined with mads13, it did not enhance the ovule phenotype, indicating that this gene does not have a role in ovule identity determination.
Here, we report a detailed characterization of the rice AG subfamily. For our analysis, we used insertion mutants for all four genes and our results provide important new insights into the function of these genes in reproductive organ development in rice. We show, in contrast with Yamaguchi et al. (2006), that mads58 mutant flowers do not have significant developmental defects. However, when the mads58 mutant was combined with the mads3 mutant, reproductive organ identity was completely lost, very similar to what has been described for the ag mutant in Arabidopsis. This confirms that the C-function is also conserved in monocotyledonous plants. Furthermore, generating the mads3 mads13 and mads13 mads58 double mutants confirmed an important role for MADS13 in FM determinacy since an astonishing proliferation of carpels was observed in the center of these double mutant flowers. Interestingly, when the mads21 mutant was combined with these double mutants, none of the higher order mutant combinations showed an enhanced phenotype, which suggests that MADS21 does not contribute to the determination of reproductive organ identity.
RESULTS
Expression Analysis of Rice AG Subfamily Genes
By in situ hybridization, we analyzed in detail the expression of the four rice AG subfamily genes during flower development. MADS3 expression was first detected in the third-whorl founder cells positioned laterally to the FM and remained expressed in the emerging stamen primordia (Figures 1A to 1C). By contrast, MADS58 expression was observed uniformly throughout the FM; in the stamen primordia, it seemed to be preferentially expressed in the epidermal cells (Figures 1F and 1G). During early stages of stamen development and differentiation, MADS3 and MADS58 were expressed in the filament and in the anther wall (Figures 1D and 1H). In the microspore mother cells, we observed only a weak MADS3 expression, whereas MADS58 transcripts were not detected.
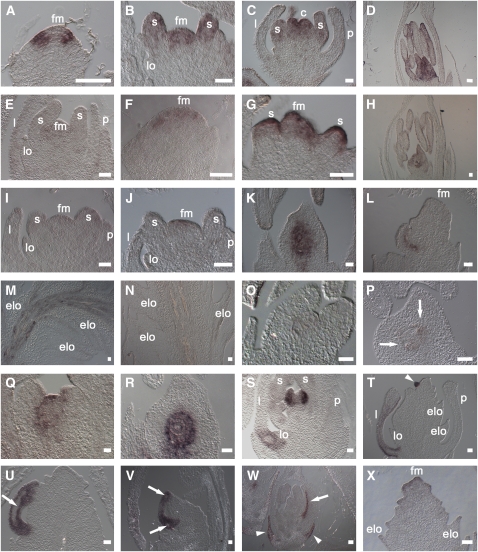
Figure 1.
Expression Analysis of AG Family MADS Box Genes.
(A) to (E) Expression profile of MADS3 in wild-type floral buds ([A] to [D]) and in the mads3-3 mutant as negative control (E).
(F) to (I) Expression profile of MADS58 in wild-type floral buds ([F] to [H]) and in the mads58 mutant as negative control (I).
(J) to (N) MADS13 activation in the wild-type FM (J) and its expression in wild-type ovules (K), at the adaxial side of a mads3-3 mads58 double mutant palea-like primordium (L) and at a later developmental stage of the same mutant in the central vasculature parenchyma (M). As expected, no signal is visible in the mads3-3 mads13 mads58 triple mutant vasculature (N).
(O) to (R) Comparative expression of MADS21 at similar stages of wild-type ([O] and [P]) and pMADS13:MADS21 ([Q] and [R]) ovule primordia. Arrows in (P) indicate the integument primordia.
(S) to (U) DL expression in the wild-type carpel (S) and in the mads3-3 mads58 double mutant palea-like primordia ([T] and [U]). In (S) and (T), the expression is clearly visible also in the lemma midrib. The early palea-like primordium (T) and the palea-like vasculature (U) are indicated with an arrowhead and an arrow, respectively. There is no obvious expression in the vasculature.
(V) DL expression in the mads3-3 mads13 mads58 triple mutant palea-like primordium; no expression is observed in the differentiating vascular region (arrows).
(W) and (X) G1 expression profile.
(W) Expression in the wild-type sterile glumes (arrowheads) and palea (arrow).
(X) Expression in the mads3-3 mads58 double mutant floral apex during the formation of the fourth whorl.
c, carpel; elo, ectopic lodicule; fm, floral meristem; l, lemma; lo, lodicule; p, palea; s, stamen. Bars = 50 μm.
As soon as the stamen primordia emerged, both MADS3 and MADS58 were expressed in the FM and remained stably expressed in the carpel and ovule primordia with a very similar expression pattern (Figures 1B to 1D and 1G to 1H). In general, MADS3 and MADS58 have a very similar expression profile, which is reminiscent of AG expression in Arabidopsis (Drews et al., 1991). Yamaguchi et al. (2006) reported a significantly different expression profile for MADS3, since they found that its expression was only transient during initial stages of stamen, carpel, and ovule primordia formation. For our experiments we used specific probes spanning the K-box and C-terminal region of MADS3 and MADS58 (see Supplemental Figure 1 online). To completely exclude cross-hybridization of our probes, we also performed in situ hybridizations using the mads3-3 and mads58 knockout mutants (see below), which confirmed specificity of the probes for each of the two genes (Figures 1E and 1I). The MADS3 expression profile was also confirmed using another antisense RNA probe targeting a region between the C terminus and 3′ untranslated region, which has no significant sequence similarity with MADS58 (see Supplemental Figures 1 and 2M online). These expression profiles are in agreement with data from Arora et al. (2007), who observed by microarray analysis that both MADS3 and MADS58 were continuously and increasingly expressed during panicle development. Furthermore, the presence of MADS3/RAG transcripts in stamen primordia was also reported by Hu et al. (2011), Ikeda et al. (2005), Kyozuka et al. (2000), and Kang et al. (1995).
The expression profiles of the two STK orthologs, MADS13 and MADS21, have been reported previously (Lopez-Dee et al., 1999; Dreni et al., 2007). The ovule identity gene MADS13 starts to be expressed in the apical part of the FM just before the differentiation of the carpel and ovule primordia. Subsequently, its expression continues in the ovule primordium and in the inner layer of the developing carpel wall (Figures 1J and 1K). During panicle and flower development, MADS21 is by far the lowest expressed AG subfamily gene of rice (Arora et al., 2007; Dreni et al., 2007). Although MADS21 belongs to the AGL11 lineage, its expression profile is not ovule specific and is reminiscent of a typical C-function gene (Dreni et al., 2007). MADS21 expression is observed in developing stamens, carpels, and ovules. Expression in stamens appeared very weak, and the strongest signal was detected in ovule integument primordia, but not in the developing nucellus (Figure 1P).
Analysis of the expression of the four rice AG subfamily genes shows that during the differentiation of the FM into the ovule primordium, the expression of all four genes overlap in this domain.
Analysis of mads3 and mads58 Single Mutants
To characterize the function of MADS3, we analyzed plants homozygous for the mads3-3 or mads3-4 mutant alleles (Yamaguchi et al., 2006; Hu et al., 2011).
The mads3-3 allele carries a T-DNA insertion in the second intron of MADS3. This intron typically contains conserved regulatory elements important for the proper expression of AG-like genes (Causier et al., 2009). mads3-3 is the only MADS3 full knockout mutant allele that has been reported thus far. However, the original line that we received from Postech (line 1A-19842, Dongjin cultivar background) was not suitable for our genetic studies, since all the plants showed a dwarf phenotype and tiny flowers producing almost no pollen and were almost completely sterile (95%). This phenotype was also observed in wild-type segregants. To eliminate this background effect, we performed backcrosses, using plants heterozygous for the mads3-3 allele as male parent, to Dongjin wild-type plants. By selfing plants across a few generations, we obtained a mads3-3 mutant line in a restored genetic background that did not show this dwarf phenotype. The mads3-4 mutant contains a frame-shift allele caused by a 2-bp deletion in the fifth exon encoding the second K-box α-helix, resulting in a C-terminal truncation of 100 amino acids.
Under our growing conditions and in the restored genetic background, the mads3-3 knockout mutant had a milder phenotype than previously reported by Yamaguchi et al. (2006). Half of the flowers had six floral organs in the third whorl and a single carpel in the fourth whorl (Figure 2C), whereas the other flowers showed a weak loss of FM determinacy, leading to one to three additional floral organs in both the third and fourth whorls (Figure 2D). All of the third-whorl stamens of mads3-3 mutant flowers were partially or totally converted into lodicule-like structures. When the homeotic conversion was incomplete, we observed the formation of chimeric lodicule filaments bearing anther-like structures (Figures 2C and 2D). Furthermore, these mutant anthers did not develop pollen grains. In comparison to wild-type flowers, the gynoecium was often altered in shape, showing enlarged or elongated ovaries and multiple stigmas. Consistently with the MADS3 expression profile and with MADS3 being an AG subfamily member, we never observed defects in second-whorl lodicules, which developed normally at the lemma side of the mads3-3 mutant flowers. This is in contrast with the previous observations made by Yamaguchi et al. (2006), which reported the formation of ectopic lodicules at the palea side of the second whorl. We also observed a fully developed third lodicule at the palea side of most mads3-3 flowers; however, this ectopic organ clearly developed instead of a third whorl stamen and not from the second whorl (Figure 2C). Interestingly, identity determination of this palea side stamen seems to be particularly sensitive to a reduction in MADS3 activity. This became especially clear from the analysis of the milder mads3-4 mutant allele, which shows a less severe conversion of stamens into lodicules, a narrow ovary, and usually no loss of FM determinacy. In each flower, a variable number of anthers were able to form a normal amount of pollen grains (Hu et al., 2011), although these pollen grains failed to reach maturity. Though the mads3-4 mutant shows a milder phenotype, the identity of the palea-side stamen was much more affected relative to the other stamens, but it was never completely converted into a lodicule (Figure 2B). This partial conversion of the palea-side stamen nicely confirms that the extra lodicule, which is usually observed in the more severe mads3-3 mutant, derives from the homeotic conversion of the third-whorl stamen and not from the second whorl. Despite the fact that the second-whorl lodicules do not seem to be affected in these mutants, anthesis never occurred in mads3-3 and mads3-4 mutant flowers.
Figure 2.
Rice AG Family Gene Mutant Phenotypes.
(A) Wild-type mature rice flower.
(B) Mads3-4 mutant flower. The arrow indicates the more severely affected palea-side stamen.
(C) Mild mads3-3 mutant phenotype. The arrow indicates the third-whorl ectopic lodicule replacing the palea-side stamen.
(D) Severe mads3-3 mutant phenotype.
(E) Mads3-3/+ mads21 mads58 mutant flower showing no phenotype.
(F) Mads3-3 mads58/+ flower.
(G) Mads3-3 mads58 double mutant. A palea-like organ developed in place of the carpel toward the lemma side (arrow).
(H) Mads3-4 mads58 double mutant. Rarely, like in this picture, the palea-like organ does not develop at the lemma side (arrow).
(I) Mads3-3 mads13 mads58 triple mutant showing an increased development of the palea-like organ.
(J) and (K) Mads3-3 mads13 (J) and mads13 mads58 (K) double mutants.
(L) Mads3-3 mads13/+ mads58/+ flower. After producing a few ectopic carpelloid structures, the indeterminate FM switched to the differentiation of ectopic lodicule primordia (arrow).
To show the inner whorls, lemma and palea were partially or completely removed. In all of the pictures, the asterisk marks the second whorl lodicule, whereas the arrowheads in (G), (J), and (L) indicate the visible indeterminate FM. Bars = 100 μm.
To characterize MADS58 functionally, we obtained a mads58 insertion mutant carrying a dSpm element in the second intron. Quantitative RT-PCR analysis, using RNA extracted from 2-cm-long panicles of the mads58 homozygous mutant, showed that MADS58 mRNA levels were strongly reduced with respect to the wild type during floral organogenesis (nearly 35-fold reduction) (see Supplemental Figure 3A online). Yamaguchi et al. (2006) reported dramatic phenotypes in both strong and weak MADS58 RNAi silencing lines, resembling those of intermediate Arabidopsis ag mutants (Mizukami and Ma, 1995; Sieburth et al., 1995). Surprisingly, despite the drastic reduction in mRNA levels observed in our mads58 knockout mutant, we did not detect any of the previous reported phenotypes. Plants were highly fertile and flowers developed normally, apart from 5% of the flowers in which one or both stigmas were bifurcated (see Supplemental Figure 2B online). This ratio increased to 20% when plants were simultaneously heterozygous for the mads3-3 allele.
MADS3 and MADS58 Redundantly Regulate Reproductive Organ Identity
The mads3 single mutant phenotype shows the importance of MADS3 for correct stamen development, although stamen identity was only partially lost. Yamaguchi et al. (2006) assessed the phenotypic effects when combining the mads3 mutant with a MADS58 knockdown construct. However, for this analysis, they used the mild mads3-2 mutant, and based on our mads58 knockout mutant data, it is difficult to judge the specificity of their mads58-RNAi construct. Therefore, we decided to analyze functional redundancy between MADS3 and MADS58 using the stable mads3-3 and mads58 knockout alleles. Plants homozygous for mads3-3 and heterozygous for mads58 already showed a strong increase in the mads3-3 mutant phenotype (i.e., loss of stamen identity, FM indeterminacy, and alterations in carpel morphology) (Figure 2F). The phenotype of mads3-3 mads58 homozygous double mutant flowers was very dramatic. They showed a complete loss of sexual organ identity and FM determinacy (Figure 2G). In addition, the size of the FM was strongly increased (Figure 3A), and the ortholog of the Arabidopsis CLV3 gene FON2/FON4 (Chu et al., 2006; Suzaki et al., 2006) remained stably expressed in the indeterminate FM (see Supplemental Figure 2D online). The combination of these features resulted in a striking enlargement of the third whorl, which frequently consisted of even more than 20 ectopic lodicules arranged in a spiral phyllotaxis (Figure 3A). During early stages of development, the second-whorl lodicules were forced against the lemma wall due to the development of this ectopic mass, and as a consequence they obtained an enlarged and flattened appearance (Figure 2G). In 52% of the flowers (n = 71 spikelets), mostly at the lemma side of the floral axis, a small green lemma/palea-like structure replaced the carpel in the fourth whorl (Figures 2G and 3A). Furthermore, the FM continued to produce lodicule-like organs, and frequently we observed branching of the FM (see Supplemental Figure 2E online). Twenty percent of the flowers had two to three lemma/palea-like organs in and after the fourth whorl, whereas the remaining 28% produced only lodicules. The phenotype of the mads3-4 mads58 double mutant (n = 61 spikelets) was nearly identical (Figure 2H), with indeterminate flowers showing one (64%) or two to three (8%) lemma/palea-like organs in place of the carpel, or only lodicules (28%).
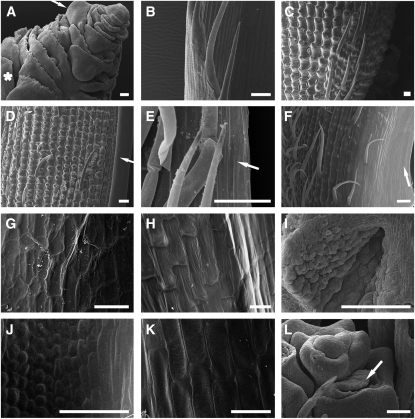
Figure 3.
Scanning Electron Microscopy Analysis of Mutant Flowers.
(A) Inner organs of a mads3-3 mads58 double mutant floral bud showing a second-whorl lodicule (asterisk), a fourth-whorl lemma-side palea-like primordium (arrow), several ectopic lodicules, and the enlarged FM.
(B) Wild-type sterile lemma with the abaxial surface oriented on the left.
(C) to (F) Abaxial surface of wild-type lemma (C), wild-type palea (D), mads3-3 mads58 double mutant palea-like organ (E), and mads3-3 mads13 mads58 triple mutant palea-like organ (F). The palea and palea-like marginal regions are shown in (D) to (F) (arrows).
(G) to (K) Adaxial surface of wild-type lemma (G), wild-type palea (H), mads3-3 mads58 double mutant palea-like organ ([I] and [J]), and mads3-3 mads13 mads58 triple mutant palea-like organ (K).
(L) Mads3-3 mads13 double mutant indeterminate FM surrounded by several developing carpel primordia and also an ectopic lodicule primordium (arrow).
Bars = 100 μm in (D) and 50 μm in the other pictures.
In the most indeterminate flowers of the mads3-3 mads58 double mutant plants, the FM remained strongly active for even more than 1 month after heading. In these flowers, the ectopic mass of hundreds of lodicules forced the lemma and palea away and they protruded outside the flower (see Supplemental Figure 2E online). At these late stages, the FM sometimes produced a number of small, well-developed sterile lemma- and lemma/palea-like structures, from which axillary indeterminate FMs developed that produced again only lodicules (see Supplemental Figure 2F online). These mads3 mads58 double mutant phenotypes revealed that a double knockout of MADS3 and MADS58 produces a rice mutant flower in which reproductive organ identity is completely lost and in which the FM becomes indeterminate, which is very similar to the ag mutant phenotype in Arabidopsis (Bowman et al., 1989).
As described above, a lemma/palea-like organ often replaced the gynoecium in the mads3-3 mads58 double mutant. We carefully analyzed the cellular and molecular identity of this ectopic organ by means of microscopy and by in situ RNA hybridization experiments.
Lemma and palea have a similar epidermal cell morphology, with rounded projections and trichomes on the abaxial surface and smooth, wide, rectangular cells on the adaxial surface. However, the palea has a distinctive marginal tissue that is smooth and trichome-less, which is similar to the outer glume epidermis (Prasad et al., 2001; Prasad and Vijayraghavan, 2003; Figures 3B to 3D). The glume-like organs that developed in place of carpels in mads3-3 mads58 double mutant flowers showed rounded projections and trichomes on the abaxial surface and smooth trichome-less marginal tissue, which strongly supports palea identity for this organ (Figure 3E). However, in situ hybridization analysis showed that DL was expressed in these palea-like structures. DL expression is typical for the fourth-whorl carpel tissue, and it seems that its expression remained unaffected in this ectopic organ, except for the marginal tissues and the midvein where no DL expression was observed (Figures 1S to 1U; see Supplemental Figure 2J online). In the first-whorl organs, DL expression occurs only in the lemma rudimentary midrib but never in the palea (Yamaguchi et al., 2004; Figures 1S and 1T). As expected, the B-class gene MADS2 was expressed in the ectopic lodicules but not in the palea-like primordium where DL remained expressed (see Supplemental Figure 2L online). Using in situ hybridization experiments, we also investigated the expression of G1/ELE, which is a specific marker for the sterile lemmas and palea that is not expressed in the lemma (Yoshida et al., 2009; Hong et al., 2010). G1 is initially highly expressed in the sterile lemmas and palea primordia, and subsequently its expression gradually fades during the development of these floral organs (Figure 1W). Even if we observed G1 activation in the mads3-3 mads58 indeterminate floral apices, its expression did not clearly mark the palea-like primordia (Figure 1X).
Thus, in the rice mads3 mads58 double mutant, the carpel is converted into an organ with a palea-like appearance, which, however, markedly differs from the first-whorl palea at the molecular level and retains expression of genes normally expressed in the fourth whorl.
Genetic Interactions between MADS3, MADS13, and MADS58
Our previous analysis of the mads13 mutant revealed a carpel-in-a-carpel phenotype, which points to indeterminacy of the FM (Dreni et al., 2007). However, this phenotype could also be a consequence of the homeotic conversion of the ovule into a carpel that itself tries to make an ovule, which in turn is transformed in a carpel again. Since our data and those of Yamaguchi et al. (2006) clearly showed that MADS3 and MADS58 redundantly regulate FM determinacy, we investigated whether MADS13 shares this role with these genes in the center of the fourth whorl where they all are coexpressed during the transition of the FM into the ovule primordium. Therefore, we analyzed double and triple mutant combinations involving these three AG subfamily genes.
The mads3-3 mads13 double mutant showed a complete loss of FM determinacy inside the fourth whorl (Figure 2J). As expected, the stamen phenotype remained the same as in the mads3-3 single mutant, since MADS13 is not expressed in the third whorl. The carpel was mostly unclosed and contained a reiteration of similar ectopic carpels; often this ectopic mass was able to force the lemma and palea away from each other within a few weeks after heading (see Supplemental Figure 2G online). The FM often branched (Figure 2J), but its size was smaller than that observed in the mads3-3 mads58 double mutant (Figure 3L), and it produced a minor number of ectopic organs.
The mads13 mads58 double mutant flowers showed a similar but milder phenotype, and carpel morphology was less affected (Figure 2K). Curiously, in both the mads3-3 mads13 and the mads13 mads58 double mutants, sometimes the first ectopic organs arising from the carpel were two small stamens, with a subsequent reiteration of ectopic carpels (see Supplemental Figures 2H and 2I online). Scanning electron microscopy analysis revealed that, in rare events, the mads3-3 mads13 double mutant formed ectopic lodicules between the ectopic carpels (Figure 3L).
As described above, in plants homozygous for mads3-3 and heterozygous for mads58, FM determinacy was partially lost in the inner two whorls (Figure 2F). However, ovule-like structures usually developed inside of the ovary. This effect was greatly enhanced in plants homozygous for mads3-3 and heterozygous for both mads58 and mads13. In this genetic background, the FM remained active inside the gynoecium, producing a reiteration of ectopic carpels and often also lodicules, indicating a partial ectopic expression of B-function genes (Figure 2L).
Since reproductive floral organs completely disappeared in the mads3-3 mads58 double mutant, we expected that MADS13 expression would be completely absent from these double mutant flowers. To test this hypothesis, we performed quantitative RT-PCR (qRT-PCR) experiments using RNA extracted from pools of mads3-3 mads58 double mutant and from wild-type developing panicles of 1 to 14 cm long. Surprisingly, MADS13 expression in the double mutant showed only a reduction of nearly fourfold, whereas MADS21, which is already weakly expressed in wild-type panicles, was reduced by 23-fold (see Supplemental Figure 3B online). To localize the residual MADS13 expression in the mads3-3 mads58 double mutant, we performed in situ RNA hybridization experiments. Surprisingly, MADS13 mRNAs were not detected in the indeterminate FM during fourth whorl formation and also not during later developmental stages, but transcripts were detected only in the adaxial epidermis of the ectopic palea-like lateral organ primordium, exactly as in the corresponding epidermis of the fourth-whorl carpel in wild-type plants (Figure 1L). At later stages, we observed an irregular spotted expression in the parenchyma cells of the central vascular bundle developing at the center of the elongating floral axis, indicating that this ectopic vascular system might correspond to the ovule dorsal vascular bundle, where MADS13 is normally highly expressed in the wild type (Figure 1M). In agreement with this expression pattern, the mads3-3 mads13 mads58 triple mutant differed from the mads3-3 mads58 double mutant only in the palea-like organ, which was more enlarged (Figure 2I). Scanning electron microscopy analysis revealed that at the basal part of the adaxial surface of the mads3-3 mads58 double mutant palea-like organ, often the epidermal layer did not develop, thus exposing the subepidermal cells (Figure 3I). In the rest of the adaxial surface, some of the cells were converted into palea epidermis cells, but often patches of smaller polyhedral cells still resembling those of carpels were visible (Figure 3J). By contrast, in the mads3-3 mads13 mads58 triple mutant, the ectopic palea adaxial side was more regular and completely converted into a first-whorl palea epidermis (Figure 3K). This means that MADS3, MADS13, and MADS58 redundantly specify the identity of the carpel inner epidermis, since in the mads13 single mutant, the identity of these cells was not lost.
The DL expression pattern in the palea-like organs did not markedly differ between the mads3-3 mads58 double and the mads3-3 mads13 mads58 triple mutants (Figures 1T to 1V). Also, the G1 activation in the indeterminate floral apex was similar between these two mutants (see Supplemental Figure 2O online).
Analysis of the Role of MADS21 in Reproductive Organ Development
Since the AGL11 lineage gene MADS21 has a similar expression profile as MADS3 and MADS58, we tested the hypothesis that this gene also contributes to the C-function in rice. Since the mads3-3 mads21 mads58 triple mutant is not informative (because MADS21 expression is almost completely suppressed in mads3-3 mads58 flowers [see Supplemental Figure 3B online] and because reproductive organ identity is already completely lost in this double mutant), we analyzed the consequences of the loss of MADS21 activity in a reduced C-function background. Flowers of the mads21 mutant were completely normal, in agreement with our previous observations (Dreni et al., 2007), and the mads3-3 and mads58 phenotypes were not enhanced in mads3-3 mads21 and mads21 mads58 double mutants, respectively. Furthermore, mads3-3 mads58/+ and mads3-3 mads21 mads58/+ plants had the same phenotype. Most mads3-3/+ mads21 mads58 flowers were normal (Figure 2E), and the frequency of bifurcated stigmas did not increase when compared with mads3-3/+ mads58 mutant plants.
Similarly, the mads13 mads21 mads58 triple mutant flowers phenocopied those of the mads13 mads58 double mutant. Taken together, these data strongly suggest that MADS21 does not play a crucial role in the formation of reproductive organs and FM determinacy.
MADS21 Retains Ovule Identity Determination Activity
The higher order mutants described above show that MADS21 does not contribute to the C-function. We previously reported that the mads13 phenotype was not enhanced in the mads13 mads21 double mutant (Dreni et al., 2007). This suggests that the AGL11 lineage gene MADS21 has no class C activity and also does not determine ovule identity (class D activity). This is curious considering its phylogenetic classification. A possible explanation for the absence of class D gene activity might be its low expression level. To test this, we made a gene construct in which the MADS21 gene was expressed under the control of the MADS13 upstream region. Five independent transgenic rice lines were obtained, and in none of these lines did MADS21 upregulation cause a phenotypic effect. One of these lines showed an ~10-fold increase in MADS21 expression in developing panicles (see Supplemental Figure 3C online), and this line was used for crosses with the mads13 mutant. In situ RNA hybridization confirmed that the MADS13 upstream region is able to drive strong expression in developing ovules (Figures 1Q and 1R), although the expression seemed variable between different flowers. In contrast with wild-type samples, a strong MADS21 signal was visible even in the developing nucellus (Figure 1R). In the F2 generation, we analyzed ovule development in mads13 mutant plants carrying the pMADS13:MADS21 construct. In the mads13 mutant, amorphic or normal appearing carpels replace ovules (Dreni et al., 2007; Figures 4C and 4F). In the mads13/pMADS13:MADS21 plants, 37% of the flowers (n = 35 spikelets) showed partial complementation of the mads13 phenotype. Nucellus and integument-like structures or naked nucella without embryo sacs were observed. Some ovaries developed complete ovule-like structures enveloped by integuments, in which surprisingly the megaspore mother cell was replaced by a short tracheary element (Figures 4B and 4E). This preliminary result shows that higher expression levels of MADS21 can partially compensate for the loss of MADS13 function, although ovule development was not completely restored, suggesting that the protein might have lost some specific characteristics for correct ovule development. This hypothesis will be tested in the future by the analysis of more independent pMADS13:MADS21 transformants and using transgenic pMADS13:MADS13 control lines in the mads13 mutant background.
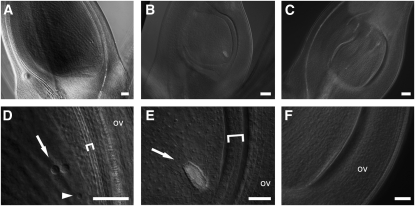
Figure 4.
Whole-Mount Tissue Clearing Analysis.
(A) and (D) Wild-type mature ovule showing details (D) of the central cell nuclei (arrow), the egg cell nucleus (arrowhead), and the inner integument, which is laterally ~4 μm thick (bracket).
(B) and (E) mads13/pMADS13:MADS21 mutant ovule showing details (E) of the tracheary element replacing the embryo sac (arrow) and the inner integument, which is laterally ~16 μm thick (bracket).
(C) and (F) A mads13 mutant ovary that is filled with carpelloid cells.
ov, ovary wall. Bars = 50 μm for (A) to (C) and 20 μm for (D) to (F).
DISCUSSION
The C-Function
Our analysis using the model plant rice clearly shows that the C-function is highly conserved between core eudicot and monocot plants. Similarly, rice DEF/GLO orthologs have been shown to determine the identity of second and third whorl floral organs (B-function), whereas SQUA-like MADS box genes are more likely required to influence FM identity rather than having a true A function, although further analysis will be required (reviewed in Kater et al., 2006). Recessive A-function mutants, clearly showing identity defects in first- and second-floral whorl organs, have thus far been reported only in Arabidopsis, and probably only a BC model can be applied to other model core eudicots (Schwarz-Sommer et al., 2003; Causier et al., 2010).
Recently, the functional conservation of AG-like genes in mediating the C-function, which means the determination of stamen and carpel identity, and the regulation of FM determinacy has also been demonstrated by virus-induced gene silencing experiments in the basal eudicots California poppy (Eschscholzia californica) and opium poppy (Papaver somniferum), both belonging to the Papaveraceae family (Hands et al., 2011; Yellina et al., 2010). Interestingly, in both these basal eudicots and in the asterid A. majus, the inhibition of AG-like gene function led to the homeotic conversion of the gynoecium into petal-like organs, rather than into sepal-like organs as would be predicted by the classic ABC model, suggesting an involvement of AG-like genes in the repression of B-function genes in the fourth whorl of these species (Davies et al., 1999; Yellina et al., 2010; Hands et al., 2011).
Based on our mutant analysis, the two rice AG lineage genes MADS3 and MADS58 redundantly mediate the C-function (Figure 5). Although the mads58 mutant might not be a complete knockout since some transcript was detected by real-time PCR analysis, the contribution of MADS3 seems to be more important since strong mads3 single mutants already showed some defects, whereas most of the mads58 flowers were indistinguishable from wild-type flowers. In particular, the mads3 knockout causes a partial loss of stamen identity, suggesting a major role in third-whorl organ identity specification when compared with MADS58, as already reported by Yamaguchi et al. (2006). This functional difference might be explained by protein subfunctionalization. Analysis of the amino acid sequences highlighted interesting differences between MADS3 and MADS58. For example, the AG motif II (Kramer et al., 2004) is poorly conserved in the grass MADS58 subclade. However, the level of gene expression and the differences in timing of the onset of gene expression might also contribute to this partial subfunctionalization. MADS3 is strongly and specifically activated in the stamen anlagen of the floral apex, whereas at the same stage, MADS58 is activated at a lower level uniformly throughout the FM. After the third whorl is differentiated, MADS3 and MADS58 show a very similar expression profile at the center of the FM and in the deriving gynoecium. Consistent with this, FM determinacy and carpel morphogenesis are only weakly affected in the mads3 knockout mutant, and there is no loss in carpel identity. Interestingly, although MADS21 is weakly expressed in the developing reproductive organs, it does not seem to contribute to the C-function, which is consistent with its classification as an AGL11 lineage gene.
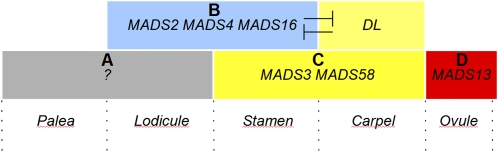
Figure 5.
Genetic Model for Floral Organ Identity Determination in Rice.
The existence of a true rice A function determining the identity of the first- and second-whorl organs has, like in most flowering plants, not been identified yet. The homeotic conversion of the carpel into a palea-like organ in the mads3 mads58 double mutant suggests that only the palea, and not the lemma, can be considered as a first-whorl organ. The class B genes regulate lodicule identity in the second whorl, and together with class C genes, they determine stamen identity in the third whorl. In the fourth whorl, DL represses the expression of class B genes. The main class C genes of rice are MADS3 and MADS58. Since the ovule develops directly from the FM, it can be considered a fifth-whorl organ. Based on current data, MADS13 seems to be the only AG subfamily gene regulating ovule identity in rice. Among the four AG subfamily genes of rice, MADS3, MADS13, and MADS58 regulate also FM determinacy, whereas MADS21 seems not to play important functions during flower development. The MADS domain proteins shown in this model might interact with SEP-like and/or AGL6-like MADS domain proteins providing E-function (Favaro et al., 2002; Kater et al., 2006; Ohmori et al., 2009; Cui et al., 2010; Li et al., 2010; Li et al., 2011a), which for simplicity are not shown in this scheme.
Although we show that the rice mads3 mads58 double mutant largely corresponds to the ag mutant of Arabidopsis, there are also important differences between their phenotypes. mads3 mads58 double mutant flowers displayed an enlargement of the third whorl, bearing even more than 20 ectopic lodicules arranged in a spiral phyllotaxis. Fifty percent of the mutant flowers developed a single palea-like structure in place of the carpel, mostly from the lemma side of the floral axis (Figure 2G), meaning that the position of the fourth-whorl primordium was maintained. Subsequently, the rest of the FM was consumed by the production of a large number of ectopic lodicules. The other 50% of the mads3 mads58 double mutant flowers developed zero or two to three palea-like structures instead of the carpel, and again a large number of ectopic lodicules developed in the center of the flower. Thus, there is no regular alternation of first- and second-whorl floral organs like in the ag mutant. Furthermore, in the ag mutant, the conversion of the carpel into a new ag flower is complete, whereas in the rice mads3 mads13 mads58 triple mutant, the ectopic palea-like structure remains much smaller than the first-whorl palea, and even though its morphology and cell shapes resemble those of a first-whorl palea, it is still not a true palea from a molecular point of view. In particular, in both the mads3 mads58 double and the mads3 mads13 mads58 triple mutants, DL expression remains unaffected in this palea-like organ. Furthermore, G1 appears irregularly activated in the region of the mutant floral apex, but its expression does not clearly mark the arising palea-like primordium as normally occurs in the wild-type palea.
Interestingly, DL expression in the palea-like organ provides new insights about its true functions during carpel development. It seems that DL cannot specify carpel identity without MADS3 and MADS58, and it is possible that it has no C-function at all. DL is required as negative regulator of class B gene expression in the fourth whorl (Nagasawa et al., 2003). This function is enough to explain the dl mutant phenotype, since the ABC model predicts that the simultaneous expression of B and C genes leads to stamen identity. From this point of view, the dl mutant is not a true class C mutant; otherwise, a conversion of the carpel into sterile perianth organs like a palea (or a lodicule due to class B ectopic expression) should be expected. In this context, it might be better to consider DL as a fourth-whorl marker rather than a carpel identity marker, of which its transcriptional regulation is at least in part independent of the true C-function genes MADS3 and MADS58. Based on the observation that in dl mutant flowers the carpel is replaced by two to seven stamens, a direct function of DL in FM determinacy has been proposed (Nagasawa et al., 2003; Yamaguchi et al., 2004). However, this partial loss of FM determinacy might be an indirect effect. In agreement with this hypothesis, it has recently been shown that MADS13, which regulates FM determinacy, is not expressed in dl mutant flowers (Li et al., 2011b). Furthermore, the dl phenotype did not change in the dl mads13 double mutant, whereas the loss of FM determinacy seen in dl flowers was strongly enhanced in the dl mads3 double mutant (Li et al., 2011b), most likely due to the simultaneous repression of MADS3 and MADS13. These data therefore suggest that the main function of DL in carpel development is to repress the B-function genes (Figure 5), a function that its Arabidopsis ortholog CRABS CLAW (CRC), and its ortholog in the basal eudicot E. californica, EcCRC, do not have (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Orashakova et al., 2009). However, these studies clearly indicate that in basal and core eudicots, the CRC-like genes regulate important aspects of carpel morphogenesis. Based on current data, it is not possible to exclude a similar function also for DL in rice.
FM Determinacy
Among the four Arabidopsis AG subfamily genes, only AG seems to regulate FM determinacy. We previously suggested that in rice the ovule identity gene MADS13 also participates in mediating FM determinacy (Dreni et al., 2007). On the contrary, Yamaki et al. (2011) proposed that MADS13, like its orthologs in Arabidopsis and petunia (Petunia hybrida), might be simply a master regulator of ovule identity and not involved in determining FM activity. Here, we show that in rice, three of the four AG subfamily genes (MADS3, MADS13, and MADS58) redundantly regulate FM determinacy. Like in Arabidopsis (Mizukami and Ma, 1995), FM determinacy in rice seems to be sensitive to the available amount of AG-like protein(s), since all the three possible double mutant combinations (mads3 mads58, mads3 mads13, and mads13 mads58) resulted in an enhanced FM indeterminacy. Based on our double and triple mutant combinations, it seems that the contribution of MADS21 to FM determinacy is not significant. MADS3 and MADS13 appear to be the most important genes because FM determinacy is already partially lost in the single knockout mutants.
The Role of AG Subfamily Genes in Ovule Development
Rice possesses two AGL11 lineage genes, namely, MADS13 and MADS21. Interestingly, the determination of ovule identity seems to be an exclusive function of MADS13. This is in contrast with Arabidopsis, where all four AG subfamily genes are largely redundant in carrying out this function (Pinyopich et al., 2003; Brambilla et al., 2007). When expressing MADS21 at high levels inside the carpel under the control of the putative MADS13 promoter, partial complementation of the mads13 mutant phenotype was observed. This shows that MADS21 is potentially able to induce the pathway that leads to the formation of ovules but that this activity normally remains unobserved due to its low expression in the ovule. Since the complementation of the mads13 mutant phenotype was only partial, it is possible that MADS21 has limited D-function activity. However, a higher number of independent transformants and the necessary controls will be needed to confirm this hypothesis. The expression of MADS21 strongly increases in developing kernels (Arora et al., 2007), suggesting that this gene might have an important function only after fertilization.
By contrast, MADS3 and MADS58 are both strongly expressed at all stages of ovule development; thus, they are likely required for ovule development, possibly in MADS13-independent pathways. It could be that they are needed to establish a genetic background necessary for MADS13 functions. Interestingly, MADS13 expression in the mads3 mads58 double mutant flowers was restricted to the palea-like fourth-whorl organ and the central vascular bundle of the floral axis, which suggests that MADS13 expression is only dependent on class C gene activity for its early activation in the FM.
In the wild type, MADS13 is expressed in developing ovules and in the ovary inner epidermis; however, no developmental defects are evident in the epidermal layer in the mads13 mutant. Indeed, when comparing the mads3 mads58 double mutant with the mads3 mads13 mads58 triple mutant, we demonstrated that these three genes redundantly determine the identity of the ovary adaxial epidermis. Therefore, the mads3 mads58 mutant will provide a good genetic background in which to study MADS13 functions in the development of the ovary adaxial epidermis, since in this way these functions will not be masked by redundancy.
Conservation of Expression of AG and AGL11 Lineage Genes
Since both AG and AGL11 lineage genes have been isolated in the main Angiosperm taxa, it is thought that these two lineages of the AG subfamily arose from a gene duplication event that occurred in the most recent common ancestor of flowering plants (Kramer et al., 2004). Many dicot and monocot AG lineage genes are expressed in stamen, carpel, and ovule, although a restriction of the expression domain is not uncommon, especially in the core eudicot PLENA subclade. It is thus likely that the AG lineage ancestor gene had an expression profile similar to AG of Arabidopsis (see Supplemental Figure 4 online) (Zahn et al., 2006).
In core eudicots, AGL11 lineage genes are specifically expressed in developing ovules and annexed ovary tissues with only a few exceptions, so very likely this expression profile has been inherited and conserved from the early eudicot plants (Zahn et al., 2006). In monocots, a similar expression profile has been reported for the rice gene MADS13, its maize ortholog ZAG2, the lily (Lilium longiflorum) gene LMADS2, and MADS1 from hyacinth (Hyacinthus orientalis). However, expression in stamens has also been shown for monocot AGL11 lineage genes, for example, MADS21 in rice (Dreni et al., 2007) and AVAG2 in asparagus (Asparagus officinalis; Yun et al., 2004). Thus, it seems plausible that the angiosperm AGL11 lineage ancestral gene had an expression pattern already restricted to the ovules and annexed ovary tissues and that the conservation of this expression domain, and the related D-function, is less strict in monocots when compared with core eudicots. However, this assumption can be confirmed only by expression analysis of AGL11 lineage genes in basal angiosperms like Amborella and Nymphaea, in basal eudicots, and basal monocots, whose data are still completely missing.
METHODS
Plant Material and Growth Conditions
We used plants of rice (Oryza sativa ssp japonica) of the following lines: cultivars Nipponbare and Dongjin, mads13 and mads21 mutants (Dreni et al., 2007); mads3-4 mutant (Hu et al., 2011); mads3-3 mutant (line 1A-19842, with a T-DNA insertion 4656 bp after the 5′-splice site of the second intron, from the mutant population of Pohang University of Science and Technology, Republic of Korea; Jeon et al., 2000; Jeong et al., 2006); mads58 mutant (line RdSpm2035B_3.1, with a dSpm insertion 117 bp after the 5′-splice site of the second intron, from the Nipponbare mutant population of the Sundaresan Lab, Department of Plant Biology, University of California, Davis, CA).
Plants were grown in a phytotron under 28°C during the light cycle and under 24°C during the dark cycle.
Primers Used for PCR Genotyping of Mutant Plants
The following primers were used: mads3-3 mutant, OsP297F and OsP298R for the wild-type allele and OsP95/T-DNALB and OsP298R for the mutant allele; mads3-4 mutant, OsP270F and OsP272R for the wild-type allele and OsP271F forward and Osp272R for the mutant allele (alternatively, plant genotyping was performed by PCR amplification and sequencing of the fifth exon); mads58 mutant, OsP201F and OsP202R for the wild-type allele and Osp201F and OsP196/Spm3′-3 for the mutant allele. The PCR genotyping of mads13 and mads21 mutants has been described previously (Dreni et al., 2007).
Plasmid Construction and Plant Transformation
A 3435-bp fragment of the MADS13 upstream region (pMADS13), comprising the 5′ untranslated region and leader intron sequences, but not the start codon, was amplified with primers OsP131F-SacI and OsP132R-SpeI, using Phusion High-Fidelity DNA polymerase (Finnzymes). The PCR product was digested with SacI (partial digestion) and SpeI restriction enzymes. The binary vector pK2GW7 (Karimi et al., 2002), carrying the NptII selectable marker gene for in planta selection, was digested with the same enzymes to excise the 35S promoter. The pMADS13 fragment was then ligated into the digested plasmid to generate the NOB1186 vector. The MADS21 coding sequence was amplified from Nipponbare panicle cDNA using the primers OsP48-attB1 and OsP49-attB2 and recombined into NOB1186 using Gateway Technology (Invitrogen). Finally, this pMADS13:MADS21 vector was introduced in the Agrobacterium tumefaciens strain LBA 4404. Rice transformation was performed using geneticin (G418) selection and the protocol of Hiei et al. (1994).
cDNA Preparation and qRT-PCR Analysis
Total RNA from three biological replicates was extracted with the LiCl method, and its integrity was checked on agarose gels. Total RNA was then treated with recombinant DNase I, RNase-free (Roche Diagnostics) in 1× TAB II buffer, after which ~0.5 μg of RNA was reverse transcribed with the iScript cDNA synthesis kit (Bio-Rad). Negative controls were performed without the addition of reverse transcriptase into the mix. Tenfold dilutions of cDNA were tested in RT-PCR and qRT-PCR experiments using reference genes. No amplification was observed in negative controls. For qRT-PCR analyses, cDNA templates (10-fold dilutions) and primers (OsP274F and OL121R for MADS13; OsP20F and OsP57R for MADS21; OsP226F and OsP222R for MADS58) were added to iQ SYBR Green Supermix (Bio-Rad). qRT-PCR reactions were performed with three technical replicates/samples using a Bio-Rad CFX96 real-time PCR detection system, and data were managed using the CFX Manager software based on the delta-delta Ct method. Data were normalized using the two reference genes LOC_Os06g11170.1 (coding for a putative nucleic acid binding protein) and LOC_Os06g48970.1 (coding for a putative protein kinase) with the same primers published by Narsai et al. (2010) (see Supplemental Table 1 online). Before performing expression analysis, the primers’ efficiency was estimated through a five-point standard curve (10-, 40-, 160-, 640-, and 2560- fold dilutions of cDNA). We obtained the following amplification efficiencies: 81% for MADS13, 95% for MADS21, 96% for MADS58, 92% for LOC_Os06g11170.1, and 95% for LOC_Os06g48970.1.
Scanning Electron Microscopy Analysis
Samples were prepared and analyzed as described previously (Favaro et al., 2003).
In Situ RNA Hybridization Experiments and Whole-Mount Tissue Clearing
In situ hybridization experiments were performed essentially as described previously (Dreni et al., 2007) with the only difference that sections were pretreated following the method described by Jackson (1991). The preparation of MADS13, MADS21, DL, and FON4 digoxygenin-labeled antisense RNA probes was described previously (Chu et al., 2006; Dreni et al., 2007). The cDNA fragments for the in vitro transcription of the other RNA probes were amplified with the following primers: OsP16F and OsP17R-T7 for MADS3 (first probe), OsP72F and OsP73R-T7 for MADS3 (second probe), OsP63F OsP64R-T7 for MADS58, OsP307F and OsP308R-T7 for G1, and MADS2F and MADS2R for MADS2.
For clearing of ovaries, a 2-d treatment with Herr’s 41/2 clearing solution (Herr 1971), followed by 2 d in lactic acid saturated with chloral hydrate, was used. Samples where then transferred and mounted in chloral hydrate/glycerol/water (8:1:2, w/v/v) to improve the contrast.
Samples were subsequently observed using a Zeiss Axiophot D1 microscope equipped with differential interface contrast optics (Carl Zeiss MicroImaging). Images were captured on a Zeiss Axiocam MRc5 camera using AxioVision software.
Primers Used in This Work
The sequences are listed in Supplemental Table 1 online.
Images
The multifocus pictures were made using CombineZP software (http://combinezp.software.informer.com/). Figures and panels were assembled and processed using Adobe Photoshop CS3.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: MADS3 (L37528), MADS13 (AF151693), MADS21 (AY551913), and MADS58 (AB232157).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of MADS3 and MADS58 Coding Sequences.
Supplemental Figure 2. Morphological and Molecular Analyses.
Supplemental Figure 3. Quantitative RT-PCR Expression Analysis.
Supplemental Figure 4. Scheme of the Ancient Gene Duplication That Led to the Formation of the AG and AGL11 Lineages.
Supplemental Table 1. Sequence of Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Venkatesan Sundaresan and Patrick E. McGuire for providing the mads58 mutant and Gynheung An for the mads3-3 mutant. This work was supported by Ministero dell’Istruzione dell’Università e della Ricerca, Programmi di Ricerca di Rilevante Interesse Nazionale 2007, and by Ministero dell’Istruzione dell’Università e della Ricerca, Fondo per gli Investimenti della Ricerca di Base in the frame of the European ERA-Net Plant Genomics network “Seeds for Growth” 2007–2010.
AUTHOR CONTRIBUTIONS
L.D. performed genetic, histological, and molecular experiments and wrote the manuscript. A. Pilatone and S.E. performed genotyping and phenotyping experiments. D.Y. and D.Z. provided the mads3-4 single and mads3-4 mads58 double mutants and rice MADS2 in situ data. A. Pajoro did rice transformation experiments. E.C. did the scanning electron microscopy analysis. M.M.K. coordinated the research and wrote the manuscript.
References
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 [DOI] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S., Kater M.M., Colombo L. (2007). Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Bradley D., Cook H., Davies B. (2009). Conserved intragenic elements were critical for the evolution of the floral C-function. Plant J. 58: 41–52 [DOI] [PubMed] [Google Scholar]
- Causier B., Schwarz-Sommer Z., Davies B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21: 73–79 [DOI] [PubMed] [Google Scholar]
- Chu H., Qian Q., Liang W., Yin C., Tan H., Yao X., Yuan Z., Yang J., Huang H., Luo D., Ma H., Zhang D. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142: 1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Colombo L., Battaglia R., Kater M.M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13: 444–450 [DOI] [PubMed] [Google Scholar]
- Colombo L., Franken J., Koetje E., van Went J., Dons H.J., Angenent G.C., van Tunen A.J. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Brambilla V., Marcheselli R., Caporali E., Kater M.M., Colombo L. (2010). A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev. Biol. 337: 294–302 [DOI] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., Xu Q., Chong K., Theissen G., Meng Z. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Davies B., Motte P., Keck E., Saedler H., Sommer H., Schwarz-Sommer Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18: 4023–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P.B., An G., Colombo L., Kater M.M. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Favaro R., Immink R.G., Ferioli V., Bernasconi B., Byzova M., Angenent G.C., Kater M., Colombo L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L., Ditta G., Yanofsky M.F., Kater M.M., Colombo L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hands P., Vosnakis N., Betts D., Irish V.F., Drea S. (2011). Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy. Ann. Bot. 107: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr J.M., Jr (1971). A new clearing-squash technique for the study of ovule development in angiosperms. Am. J. Bot. 58: 785–790 [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hong L., Qian Q., Zhu K., Tang D., Huang Z., Gao L., Li M., Gu M., Cheng Z. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37: 101–115 [DOI] [PubMed] [Google Scholar]
- Hu L., Liang W., Yin C., Cui X., Zong J., Wang X., Hu J., Zhang D. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Nagasawa N., Nagato Y. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282: 349–360 [DOI] [PubMed] [Google Scholar]
- Jackson D. (1991). In-situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach Bowles D.J., Gurr S.J., McPherson M.J., (Oxford, UK: Oxford University Press; ), pp. 163–174 [Google Scholar]
- Jeon J.S., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong D.H., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Kang H.G., Noh Y.S., Chung Y.Y., Costa M.A., An K., An G. (1995). Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Mol. Biol. 29: 1–10 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kater M.M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Jaramillo M.A., Di Stilio V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J., Kobayashi T., Morita M., Shimamoto K. (2000). Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41: 710–718 [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Hu Y., Zhu L., Yin C., Xu J., Dreni L., Kater M.M., Zhang D. (2011a). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang W., Jia R., Yin C., Zong J., Kong H., Zhang D. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Yin C., Zhu L., Zhang D. (2011b). Genetic interaction of OsMADS3, DROOPING LEAF and OsMADS13 in specifying rice floral organs identities and meristem determinacy. Plant Physiol. 156: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lopez-Dee Z.P., Wittich P., Enrico Pè M., Rigola D., Del Buono I., Gorla M.S., Kater M.M., Colombo L. (1999). OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev. Genet. 25: 237–244 [DOI] [PubMed] [Google Scholar]
- Mena M., Ambrose B.A., Meeley R.B., Briggs S.P., Yanofsky M.F., Schmidt R.J. (1996). Diversification of C-function activity in maize flower development. Science 274: 1537–1540 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. (1995). Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol. Biol. 28: 767–784 [DOI] [PubMed] [Google Scholar]
- Münster T., Deleu W., Wingen L.U., Ouzunova M., Cacharrón J., Faigl W., Werth S., Kim J.T.T., Saedler H., Theißen G. (2002). Maize MADS-box genes galore. Maydica 47: 287–301 [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Narsai R., Ivanova A., Ng S., Whelan J. (2010). Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 10: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., Nagato Y., Yoshida H. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orashakova S., Lange M., Lange S., Wege S., Becker A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 58: 682–693 [DOI] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Prasad K., Sriram P., Kumar C.S., Kushalappa K., Vijayraghavan U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Prasad K., Vijayraghavan U. (2003). Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S.D., Ditta G.S., Yanofsky M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.J., Veit B., Mandel M.A., Mena M., Hake S., Yanofsky M.F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5: 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Davies B., Hudson A. (2003). An everlasting pioneer: The story of Antirrhinum research. Nat. Rev. Genet. 4: 657–666 [DOI] [PubMed] [Google Scholar]
- Sieburth L.E., Running M.P., Meyerowitz E.M. (1995). Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7: 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Toriba T., Fujimoto M., Tsutsumi N., Kitano H., Hirano H.Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Vision T.J., Brown D.G., Tanksley S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S., Nagato Y., Kurata N., Nonomura K. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351: 208–216 [DOI] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yellina A.L., Orashakova S., Lange S., Erdmann R., Leebens-Mack J., Becker A. (2010). Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica). Evodevo. 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H.Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 106: 20103–20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun P.Y., Kim S.Y., Ochiai T., Fukuda T., Ito T., Kanno A., Kameya T. (2004). AVAG2 is a putative D-class gene from an ornamental asparagus. Sex. Plant Reprod. 17: 107–116 [Google Scholar]
- Zahn L.M., Leebens-Mack J.H., Arrington J.M., Hu Y., Landherr L.L., dePamphilis C.W., Becker A., Theissen G., Ma H. (2006). Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: Evidence of independent sub- and neofunctionalization events. Evol. Dev. 8: 30–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.