Oilseed germination requires gluconeogenesis, the conversion of storage lipids into carbohydrates, to sustain seedling heterotrophic growth. Pyrophosphate (PPi), a byproduct of ATP hydrolysis, is released by active metabolism in imbibed seeds. We report that the removal of PPi by plant vacuolar H+-pyrophosphatase is central for successful gluconeogenesis and resumption of postembryonic growth.
Abstract
Postgerminative growth of seed plants requires specialized metabolism, such as gluconeogenesis, to support heterotrophic growth of seedlings until the functional photosynthetic apparatus is established. Here, we show that the Arabidopsis thaliana fugu5 mutant, which we show to be defective in AVP1 (vacuolar H+-pyrophosphatase), failed to support heterotrophic growth after germination. We found that exogenous supplementation of Suc or the specific removal of the cytosolic pyrophosphate (PPi) by the heterologous expression of the cytosolic inorganic pyrophosphatase1 (IPP1) gene from budding yeast (Saccharomyces cerevisiae) rescued fugu5 phenotypes. Furthermore, compared with the wild-type and AVP1Pro:IPP1 transgenic lines, hypocotyl elongation in the fugu5 mutant was severely compromised in the dark but recovered upon exogenous supply of Suc to the growth media. Measurements revealed that the peroxisomal β-oxidation activity, dry seed contents of storage lipids, and their mobilization were unaffected in fugu5. By contrast, fugu5 mutants contained ~2.5-fold higher PPi and ~50% less Suc than the wild type. Together, these results provide clear evidence that gluconeogenesis is inhibited due to the elevated levels of cytosolic PPi. This study demonstrates that the hydrolysis of cytosolic PPi, rather than vacuolar acidification, is the major function of AVP1/FUGU5 in planta. Plant cells optimize their metabolic function by eliminating PPi in the cytosol for efficient postembryonic heterotrophic growth.
INTRODUCTION
ATP is the main molecule for storage and transfer of biochemical energy in all living organisms, from the simplest to the most complex. However, in almost 200 known biochemical reactions, ATP hydrolysis releases pyrophosphate (PPi), which becomes a metabolic inhibitor at high concentrations in cells and must be hydrolyzed immediately to facilitate the biosynthesis of various macromolecules (Heinonen, 2001). In seed plants, on imbibition, hydrated seeds switch from quiescence to highly active metabolism to cope with rapid postembryonic growth. Early in germination of oilseeds, when they are still unable to perform photosynthesis due to the lack of functionally differentiated chloroplasts, embryos are primarily nourished by the recycling of storage proteins and lipids that provide nitrogen and carbon sources, respectively. Simultaneously, active metabolism, including biosynthetic processes for macromolecules such as proteins, DNA, RNA, and cellulose is initiated along with fatty acid β-oxidation and Suc metabolism. Consequently, many of these biochemical reactions generate PPi as a by-product (Maeshima, 2000; Heinonen, 2001) and increase the PPi concentration in the cytosol.
In Arabidopsis thaliana, the vacuolar H+-translocating pyrophosphatase (V-PPase) uses energy from the hydrolysis of PPi to power active proton transport across the membranes (Martinoia et al., 2007). Generally, the V-PPase activity is high in young tissues characterized with high proliferative activity (Martinoia et al., 2007). The importance of pyrophosphatases has been reported in several other organisms. For example, the ppa gene is essential for growth in Escherichia coli; in fact, when the pyrophosphatase level was decreased, the PPi level increased and growth stopped (Chen et al., 1990). Similarly, in Saccharomyces cerevisiae, the IPP1 protein is essential for the viability of the yeast cell (Lundin et al., 1991). Moreover, a null mutant of pyp-1, a worm (Caenorhabditis elegans) PPase gene, revealed developmental arrest at early larval stages and exhibited gross defects in intestinal morphology and function (Ko et al., 2007). In Arabidopsis, the V-PPase loss-of-function mutant avp1-1 has been reported to have a severely disrupted development of root, shoot, and flowers and to be infertile (Li et al., 2005). These developmental abnormalities of avp1-1 are interpreted as a failure of proton pumping that may eventually cause inappropriate auxin distribution (Li et al., 2005). Nevertheless, the biological roles and effects of PPi in vivo remained largely unknown in these Arabidopsis mutants.
We are interested in mechanisms of organogenesis and have been focusing on leaf morphogenesis. Although leaf developmental dynamism has been the subject of vigorous research (Donnelly et al., 1999; White, 2006; Ferjani et al., 2007; Usami et al., 2009), little is known about leaf size–coordinating mechanisms (Ingram and Waites, 2006; Anastasiou and Lenhard, 2007; Ferjani et al., 2008; Tsukaya, 2008; Micol, 2009; Krizek, 2009). Recently, emerging data indicate that an organ-wide coordination of cell proliferation and postmitotic cell expansion underlies leaf organogenesis and regulates leaf size (Tsukaya, 1998, 2002, 2005, 2006, 2008; Horiguchi et al., 2006a). Such organ-wide coordination is suggested by an intriguing phenomenon that we called compensation (Tsukaya, 1998, 2002) where decreased cell number in leaf primordia triggers an unusual enhancement of postmitotic cell expansion that, in extreme cases, results in a greater than twofold increase in cell cross area (Mizukami and Fischer, 2000; De Veylder et al., 2001; Horiguchi et al., 2005; Ferjani et al., 2007; Fujikura et al., 2009).
Recently, we demonstrated that two qualitatively different modes, namely non-cell-autonomous and cell-autonomous modes, are involved in the coordination of cell proliferation and postmitotic cell expansion in developing leaves (Kawade et al., 2010). These findings should provide novel insight into the mechanism for organ size control in plants. To that end, compensation offers a model case to investigate links between cell proliferation and postmitotic cell expansion at the organ level, which may, in turn, help disentangle size regulatory mechanisms (Tsukaya, 1998, 2002, 2005, 2006). Of the several compensation-exhibiting mutants that we have isolated and characterized (Ferjani et al., 2007), here, we report our findings on the fugu5 mutants, which are defective in V-PPase function.
RESULTS
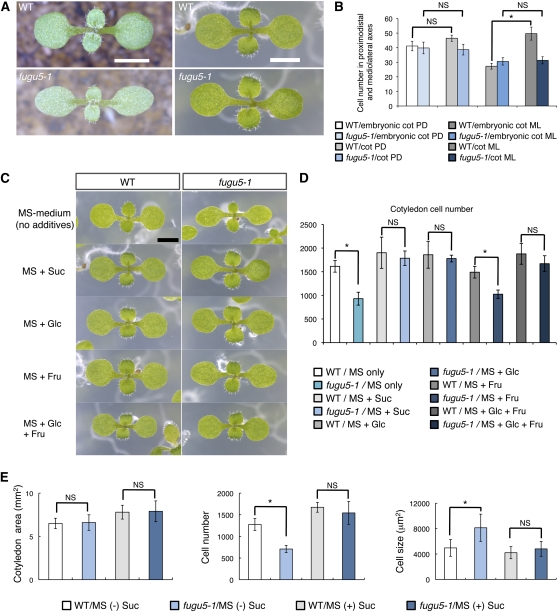
fugu5 Mutants Have Altered Cotyledons
In Arabidopsis, cotyledons are formed during embryogenesis, and embryo cell proliferation is reactivated immediately after germination (Tsukaya et al., 1994; Stoynova-Bakalova et al., 2004). The mobilization of storage compounds supports a short period of heterotrophic growth until the photosynthetic apparatus is established. Cotyledons of fugu5 mutants are rectangular and contain fewer (~60%) and larger cells (~175%) than the wild type when grown on inorganic media (rockwool; Figure 1A; see Supplemental Figure 1A online; Ferjani et al., 2007). Comparison of the cell numbers in the proximodistal and mediolateral axes of embryonic and mature cotyledons revealed that cell numbers in fugu5-1 remained constant in both directions (Figure 1B). However, cell numbers along medio-lateral axes in mature cotyledons almost doubled in the wild type (Figure 1B). These results suggest that in fugu5-1, cell division is almost totally inhibited in cotyledons postembryonically. Also, compensation occurs in the first pair of leaves in fugu5-1, but to a lesser extent than in cotyledons (see Supplemental Figures 1A and 1B online). Thus, we focus our analysis primarily on the cotyledon.
Figure 1.
Morphological and Cellular Phenotypes of the fugu5 Mutant.
(A) Effect of growth media composition on the gross phenotype of the fugu5 mutant. Wild-type (WT) and fugu5-1 mutant seedlings were grown for 8 DAS either on rockwool (left panels) or on standard MS medium (right panels). Bars = 2 mm.
(B) Cell proliferation is compromised in fugu5 cotyledons after germination. Number of palisade mesophyll tissue cells along proximodistal and mediolateral axes of either embryonic cotyledons (dissected from imbibed dry seeds) or mature cotyledons was determined after growth on rockwool for 25 DAS. Data are means and sd (n = 8). Cot, cotyledons; PD, proximodistal; ML, mediolateral.
(C) Effect of various carbohydrates on the gross phenotype of fugu5 cotyledons. The effect of exogenously supplied Suc, Glc, and Fru (58 mM each) on cotyledon phenotype is shown. The effect of simultaneous addition of Glc and Fru was also tested. Eight-day-old seedlings of the wild type (left panels) and fugu5-1 (right panels) are shown. Bar = 2 mm.
(D) Effect of different kinds of carbohydrates on fugu5-1 cotyledon cell number. Palisade mesophyll tissue cell numbers in seedlings of either the wild type or fugu5-1 mutants were determined 8 DAS. Data are means and sd (n = 8). NS, no significant difference between the two genotypes (the wild type and fugu5-1) under the indicated growth conditions.
(E) Effect of exogenously supplied Suc on compensated cell enlargement. Cotyledons were collected from the wild type and fugu5-1 mutant grown on MS media with or without 2% Suc for 21 DAS. Data are means and sd (n = 8). NS, no significant difference between the two genotypes (the wild type and fugu5-1). Asterisk indicates significant difference at P < 0.01.
Suc Is Sufficient to Rescue fugu5 Mutant Phenotypes
Importantly, we observed that fugu5-1 morphological phenotypes recovered on Murashige and Skoog (MS) medium (Figure 1A, right panels), indicating that fugu5-1 phenotypes are conditional and may be influenced by some component(s) of the growth medium. Which component(s) could this be? First, to identify the component(s) responsible for rescuing the fugu5-1 phenotype, we grew the plants on MS media with differing compositions of sugars and vitamins (see Supplemental Figure 2 online; Figures 1C and 1D) and then determined the leaf index and the cell number in cotyledons 8 d after sowing (DAS; see Supplemental Figure 2 online; Figures 1C and 1D). Interestingly, we found that Suc was sufficient to rescue the fugu5-1 gross phenotype (see Supplemental Figure 2 online; Figure 1C) and to reestablish cell number to normal level (Figure 1D). Next, we examined the growth of fugu5-1 on MS media containing similar concentrations of Glc, Fru, or both (Figures 1C and 1D). Glc mimicked the effects of Suc, but Fru did not (Figures 1C and 1D). Moreover, we found that sorbitol and 3-O-methylglucose (a nonmetabolizable analog of Glc) failed to rescue the fugu5-1 phenotype (see Supplemental Figure 3 online). On the other hand, by large-scale screening (Horiguchi et al., 2006b), we also identified two additional mutant alleles of FUGU5 gene, namely, fugu5-2 and fugu5-3 (see Supplemental Figure 4A online). These two alleles exhibited similar phenotypes to fugu5-1 that recovered when Suc was supplied in the growth media (see Supplemental Figures 4A to 4D online). Taken together, these results demonstrate that Suc is an essential metabolite for the proper resumption of postembryonic development in these fugu5 mutants.
Because decreased cell numbers trigger the unusual cell enlargement observed in compensation-exhibiting mutants (Ferjani et al., 2007, 2008; Tsukaya, 2008), we next assessed whether exogenously supplied Suc affects cell size in fully expanded cotyledons. Consistently, cell size was normal in fugu5-1 mutants grown in the presence of Suc (Figure 1E). Together, these findings demonstrate that Suc can reestablish cell number and, as a result, prevent excessive cell enlargement in the fugu5-1 mutant background.
To ascertain how Suc acts to stimulate cell proliferation, we examined the cell cycling activity in the fugu5 CYCB1;1pro:GUS reporter line (GUS is β-glucuronidase). In wild-type cells grown on MS media without Suc, the GUS signals were detectable in cotyledons 48 h after the onset of imbibition; however, no signal was observed in fugu5-1 (cf. Supplemental Figures 5B and 5J online). By contrast, upon growth on MS medium containing 2% Suc, both the wild type and fugu5-1 showed the GUS signal in the cotyledons 72 h after imbibition (cf. Supplemental Figures 5G and 5O online). Surprisingly, RT-PCR analysis revealed that the mRNA levels of a cyclin gene CYCB1;1 and several other core cell cycle genes were comparable in the fugu5-1 mutant and wild type at 48 h after the onset of imbibition (see Supplemental Figure 5Q online), suggesting posttranscriptional regulation of the CYCB1;1 gene in the fugu5-1 background.
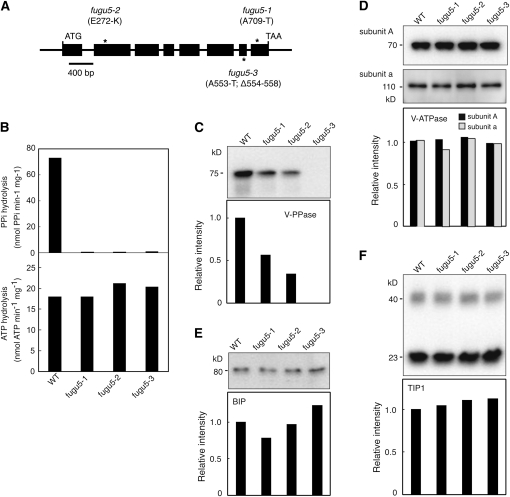
fugu5 Is Mutated in AVP1, Which Encodes a Key Enzyme in PPi Hydrolysis
To clone the gene mutated in fugu5, we used a map-based approach to identify a point mutation in At1g15690 that resulted in replacement of Ala-709 by Thr in the fugu5-1 mutant (Figure 2A; see Supplemental Figure 6 online). At1g15690 encodes the V-PPase, previously reported as AVP1 (Li et al., 2005). Subsequent sequencing of At1g15690 revealed that Glu-272 was replaced by Lys in the fugu5-2 mutant and that Ala-553 was replaced by Thr, plus residues from Leu-554 to Ala-558 were deleted at the transmembrane domain 12 in the fugu5-3 mutant (Figure 2A; see Supplemental Figure 6 online). The fugu5 mutation sites and the membrane topology of the V-PPase of Arabidopsis, as deduced from that of Streptomyces coelicolor (Mimura et al., 2004), are shown (see Supplemental Figure 6 online).
Figure 2.
V-PPase and V-ATPase Activities in fugu5 Mutant Alleles.
(A) Schematic representation of the AVP1/FUGU5 gene. Exons are shown as filled rectangles. The molecular lesion in each of the three loss-of-function fugu5 alleles is indicated by an asterisk. In fugu5-1, the Ala-709 residue is replaced by Thr. In fugu5-2, the Glu-272 residue is replaced by Lys. In fugu5-3, the Ala-553 residue is replaced by Thr, and the five residues from Leu-554 to Ala-558 are deleted.
(B) Substrate hydrolysis activity of V-PPase (top) and V-ATPase (bottom) in crude membranes prepared from wild-type (WT) and fugu5 mutants. Plants were grown in culture medium without Suc for 17 DAS. Crude membranes were prepared from shoots of more than 240 plants and used for enzyme assays as described in Methods.
(C) to (F) Protein levels of vacuolar membrane proton pumps, BIP, and aquaporin. Crude membrane aliquots (2 μg protein) of the membrane fractions were separated by SDS-PAGE and subsequently immunoblotted with anti-V-PPase (C), anti-subunit A and anti-subunit a of V-ATPase (D), anti-BIP (E), and anti-TIP1s (F). Apparent molecular masses of the immunostained bands are shown in each panel. The relative signal intensity of immunostained bands was quantitatively measured and calculated as the ratio to that of the wild type of each protein. V-PPase protein was not detected in the fugu5-3 mutant line.
Next, to evaluate the effect of the above mutations on the function of AVP1/FUGU5 protein, PPi hydrolysis activity was measured directly in crude membrane fractions. Importantly, no PPi hydrolysis activity was detected in any of the three fugu5 mutant alleles (Figure 2B, top). Thus, it appeared likely that these fugu5 mutants are null mutant alleles of V-PPase. Furthermore, when the substrate hydrolysis activity of V-ATPase was also measured, we observed no significant difference between the wild type and fugu5 mutants (Figure 2B, bottom). However, SDS-PAGE and subsequent immunoblotting with anti-V-PPase revealed significantly decreased protein levels in fugu5-1 and fugu5-2, while the V-PPase protein was not detected in the fugu5-3 mutant (Figure 2C). Additionally, protein levels of subunit A and V-ATPase, BIP, and aquaporin (TIP1) revealed no major difference in any fugu5 mutant allele compared with the wild type (Figures 2D to 2F). Thus, these results show that the phenotypes observed in fugu5 mutants are specifically caused by the total loss of V-PPase activity.
AVP1/FUGU5 Does Not Function as an Auxin-Related Regulator
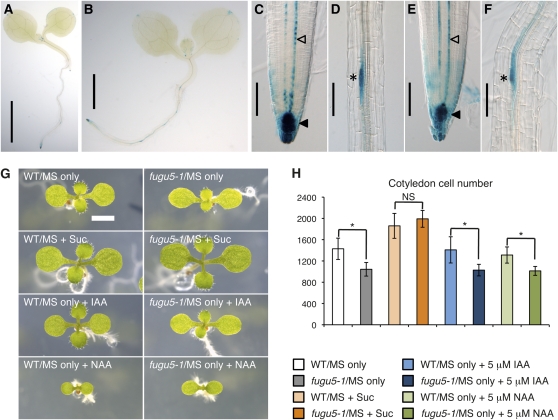
AVP1/FUGU5 has been reported to be essential for the regulation of auxin-mediated organ development, and the avp1-1 mutant allele was shown to exhibit reduced auxin transport (Li et al., 2005). To check such phenotypes, the expression pattern of DR5:GUS, a synthetic auxin response reporter gene (Ulmasov et al., 1997), was analyzed in both fugu5-1 and wild-type backgrounds (Figures 3A to 3F). Unexpectedly, our results revealed no difference in the distribution of DR5:GUS signals between fugu5-1 and the wild type (Figures 3A to 3F). Moreover, whereas avp1-1 grown with exogenously supplied auxin, such as indole-3-acetic acid and 1-naphthaleneacetic acid, were reported to produce a callus (Li et al., 2005), fugu5-1 growth under the same conditions was severely inhibited, to the same extent as the wild type, which is a typical response to high concentrations of auxins, and no callus formation occurred (Figures 3G and 3H). Actually, none of the reported avp1-1 mutant allele phenotypes, such as disrupted root and shoot development and infertility, were observed in any fugu5 mutant allele. In fact, while avp1-1 mutant plants were unable to set seeds due to severe growth and developmental defects (Li et al., 2005), all fugu5 mutant lines developed normal flowers and were as fertile as the wild type (see Supplemental Figure 7 online). Finally, we analyzed the null allele vhp1-1, which is a T-DNA insertion line of AVP1/FUGU5. In vhp1-1, the T-DNA is inserted in the sixth exon, and 27 bases are deleted at the insertion site. Consistently, we found that both the gross and cellular phenotypes of vhp1-1 were identical to those of the other fugu5 alleles (see Supplemental Figures 8A to 8D online). Together, these results strongly suggest that V-PPase is not involved in the regulation of auxin-mediated organ development and that the phenotypes reported in avp1-1 are likely to be allele specific.
Figure 3.
Expression of DR5:GUS and Effects of Exogenous Auxins.
(A) to (F) Expression pattern of the DR5:GUS reporter gene in young seedlings. Expression of DR5:GUS in the seedlings of the wild type (A) and fugu5-1 mutant (B) at 6 DAS. Expression of DR5:GUS in wild-type root and lateral root primordium at 6 DAS, respectively ([C] and [D]). Expression of DR5:GUS in fugu5-1 mutant root and lateral root primordium at 6 DAS, respectively ([E] and [F]). DR5:GUS signals around root apical meristem (black arrowheads), in the vascular system (open arrowheads), and in the emerging lateral root primordia (asterisk). Bars = 2 mm in (A), 1 mm in (B), and 100 μm in (C) to (F).
(G) Effect of exogenous auxin on the growth of fugu5 mutant. Seedlings of the wild type (WT) and fugu5-1 grown on MS alone, MS + 2% Suc, MS + indole-3-acetic acid (5 μM), and MS + 1-naphthaleneacetic acid (5 μM) 8 at DAS. Bar = 2 mm.
(H) Effect of exogenous auxin on cell number in the cotyledons. Average cell numbers of samples described in (G) were determined. Data are means and sd (n = 8). NS, no significant difference between the two genotypes (the wild type and fugu5-1). Asterisk indicates significant difference at P < 0.01.
Mobilization of Storage Proteins Is Not Affected in fugu5
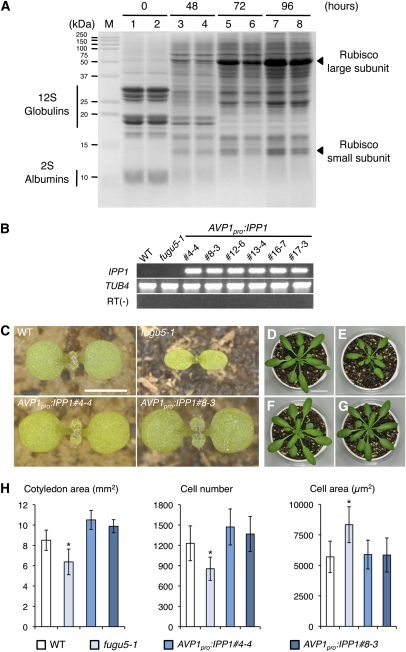
So, what is the major function of AVP1/FUGU5 in plant development? The V-PPase has two functions, namely, the hydrolysis of cytosolic PPi and the vacuolar acidification (Martinoia et al., 2007). In higher plants, seed storage proteins are deposited in protein storage vacuoles and represent a source of nitrogen for sustaining growth after seed germination (Müntz, 1998). Vacuoles are the major sites of cellular proteolysis, and this specific function predominates at the time of germination when amino acids have to be mobilized in the protein storage vacuoles, by vacuolar acidification, to nourish the embryo (Müntz, 2007). To examine the contribution of V-PPase as a proton pump in the mobilization of storage proteins, the amounts of storage proteins were evaluated in dry seeds and young seedlings as well. Our results showed that the initial amounts of 12S globulins and 2S albumins in dry seeds, the two major forms of storage proteins in Arabidopsis, were comparable in the wild type and fugu5-1 (Figure 4A, lanes 1 and 2). At 48 h, their amounts were significantly reduced (Figure 4A, lanes 3 and 4), then completely exhausted in both the wild type and fugu5-1 mutant at 72 h after the onset of imbibition (Figure 4A, lanes 5 and 6). These results clearly showed that the degradation of storage proteins is not affected by the loss of the proton-pumping activity of the V-PPase.
Figure 4.
Degradation of Protein Bodies in fugu5 and the Morphological and Cellular Phenotype of AVP1Pro:IPP1 Transgenic Lines.
(A) Effect of V-PPase dysfunction on mobilization of seed storage proteins. Protein samples were collected at 0, 48, 72, and 96 h (time after transfer to growth temperature) from either 30 seeds or seedlings of the wild type and fugu5-1 mutants. Proteins were separated using SDS-PAGE and subsequently stained with Coomassie blue. Lanes 1, 3, 5, and 7 are wild-type samples. Lanes 2, 4, 6, and 8 are the fugu5-1 mutant samples. Results were reproducible in three independent experiments.
(B) The heterologous expression of IPP1 in the fugu5-1 mutant background. RT-PCR analyses of IPP1 were performed in six independent AVP1Pro:IPP1 transgenic lines that we constructed. RT(−) = no reverse transcriptase. WT, wild type.
(C) The heterologous expression of IPP1 rescues fugu5 gross phenotypes. Gross morphology of seedlings of the wild type, fugu5-1, and two representative lines of AVP1Pro:IPP1 transgenic plants at 7 DAS. Bar = 2 mm.
(D) to (G) The heterologous expression of IPP1 rescues delayed growth of fugu5. Gross morphology of the wild type, fugu5-1, and AVP1Pro:IPP1#4-4 and AVP1Pro:IPP1#8-3 transgenic plants, respectively, at 28 DAS. Bar = 2 cm.
(H) The heterologous expression of IPP1 gene totally rescues fugu5 cellular phenotypes. Average area, cell number, and cell size of cotyledons of the wild type, fugu5-1, and two representative lines of AVP1Pro:IPP1 grown on rockwool for 25 DAS. Data are means and SD (n = 8). Asterisk indicates significant difference at P < 0.01 compared with the wild-type.
Specific Removal of PPi by IPP1 Rescued fugu5 Phenotypes
Next, we attempted to evaluate the contribution of PPi hydrolysis alone, the other function of V-PPase, and its relationship with the fugu5 phenotypes. To do this, we introduced the cytosolic IPP1 gene of S. cerevisiae (Perez-Castineira et al., 2002) under the control of the AVP1/FUGU5 promoter into the fugu5-1 mutant and wild-type background. The soluble inorganic pyrophosphatase IPP1 only hydrolyzes cytosolic PPi without interfering with vacuolar acidification, thus providing a tool of choice to separately investigate the two functions of the plant V-PPase. RT-PCR analysis of the IPP1 gene in the six independent AVP1Pro:IPP1 transgenic lines that we obtained confirmed that the IPP1 gene was properly expressed (Figure 4B). Among them, AVP1Pro:IPP1#4-4 and AVP1Pro:IPP1#8-3 were selected as representative lines for further analysis. Interestingly, our results showed that the fugu5-1 mutant gross phenotypes, such as cotyledon shape and size (Figure 4C), and the growth delay observed in fugu5-1 mutant plants recovered by the introduction of the AVP1Pro:IPP1 transgene (Figures 4D to 4G).
To further assess the effects of IPP1 expression on organ size, cell number, and cell size, these parameters were determined in fully expanded cotyledons (25 DAS). Consistently, we found that cotyledon area, cell number, and cell size in the AVP1Pro:IPP1 transgenic lines recovered to wild-type levels (Figure 4H). On the other hand, when IPP1 was introduced into the wild type, there was no effect on cell number or cell size in the cotyledons (see Supplemental Figure 9A online) and first leaves (see Supplemental Figure 9B online). Altogether, these results clearly indicate that the removal of PPi from the cytosol of fugu5 mutant by the action of IPP1 is necessary and sufficient to rescue their phenotypes by promoting cell proliferation, thus keeping a normal final cell and organ sizes.
Seed Lipid Reserves Are Properly Mobilized in fugu5
Why does fugu5 needs Suc for normal growth? During postgerminative growth of seedlings, fatty acids released from triacylglycerols (TAGs) stored in the lipid bodies of oilseeds are metabolized to produce Suc (Hayashi et al., 1998; Eastmond et al., 2000; Rylott et al., 2001; Penfield et al., 2005; Arai et al., 2008). The biochemical pathways required for carbon utilization from seed TAG stores are multistep processes that involve components of peroxisomal fatty acid β-oxidation, the glyoxylate cycle, and gluconeogenesis (Hayashi et al., 1998; Eastmond et al., 2000; Rylott et al., 2001; Penfield et al., 2005; Arai et al., 2008). Defects in β-oxidation appear to inhibit the conversion of seed lipid reserves into Suc, which is required as a carbon source for heterotrophic growth before photosynthesis begins. To determine whether lipid reserves in fugu5-1 were properly degraded, lipid contents in dry seeds as well as in young etiolated seedlings were quantified (Arai et al., 2008). We found that there was no significant difference in the degradation of lipid reserves between the wild type, fugu5-1, and AVP1Pro:IPP1#8-3 (see Supplemental Figure 10A online).
It has been reported that in the peroxisomes, β-oxidation transforms the nontoxic compound 2,4-dichlorophenoxybutyric acid into the toxic auxin analog 2,4-D, a feature that was exploited in a genetic screen to isolate mutants defective in β-oxidation (Hayashi et al., 1998). Then, to further examine the functionality of peroxisomes, growth in the presence of toxic levels of 2,4-dichlorophenoxybutyric acid was examined and revealed that fugu5-1 growth was inhibited to the same level as that of the wild type (see Supplemental Figure 10B online). Hence, these results consistently indicate that fatty acid β-oxidation is functional in fugu5-1.
V-PPase Dysfunction Partially Compromises Gluconeogenesis in fugu5 Mutants
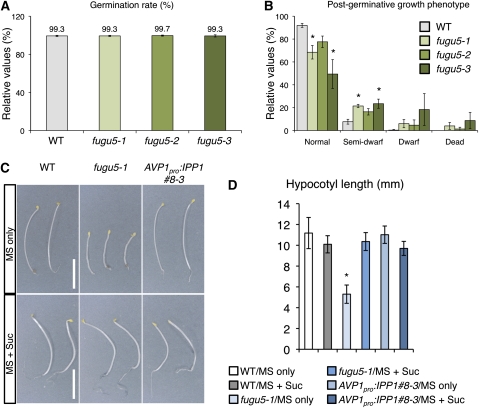
Mutants with defects in β-oxidation are either lethal or exhibit germination arrest after radicle emergence; they require Suc for growth in both the light and the dark (Penfield et al., 2005). The growth of other mutants, defective in the glyoxylate cycle or gluconeogenesis, is compromised in hypocotyl elongation only in the dark, unless supplied with an alternative carbon source (Penfield et al., 2005). The germination rate was not affected at all in fugu5 mutants (Figure 5A), consistent with functional β-oxidation. Furthermore, the majority of plants grew normally, while a certain level of growth inhibition was scored in fugu5 mutants (Figure 5B). Thus, under light conditions and even in the absence of Suc supply, fugu5 mutants can grow normally. Interestingly, we found that the hypocotyl elongation of the fugu5-1 mutant in the darkness was severely compromised in the absence of Suc (Figures 5C and 5D). Surprisingly, the growth of AVP1Pro:IPP1#8-3 was restored to wild-type levels in the dark even in the absence of Suc (Figures 5C and 5D), and fugu5-1 hypocotyl growth inhibition was restored when Suc was supplied in the dark (Figures 5C and 5D). Supplying Suc did not enhance AVP1Pro:IPP1#8-3 hypocotyl growth (Figure 5D), suggesting that Suc and IPP1 act in the same pathway. Thus, it appears that the removal of PPi by the action of V-PPase is required for proper glyoxylate cycle or gluconeogenesis that supports the reactivation of cell proliferation after germination.
Figure 5.
Germination Rate, Postgerminative Growth, and Seedling Growth Phenotype of fugu5 Mutant in the Dark.
(A) The germination rates are not affected in three fugu5 mutant alleles. One hundred seeds of each genotype (three sets) were sown on rockwool, and plants were grown in a 16/8-h light/dark cycle. The germination rates are relative mean values and sd from three independent experiments. WT, wild type.
(B) fugu5 mutants exhibit a slight postgerminative growth delay. The germination rate and postgerminative growth phenotype of plants were scored at 12 and 21 DAS, respectively. Data are relative mean values and sd from three independent experiments. Asterisk indicates significant difference at P < 0.05 compared with the wild type. Normal, plants without significant growth delay; semi-dwarf, plant size reduced by ~50% compared with normal plants; dwarf, plant size reduced by ~75% compared with normal plants.
(C) and (D) Hypocotyl elongation is severely inhibited in fugu5-1 in the dark in the absence of Suc.
(C) Photographs of hypocotyls of etiolated seedlings of the wild type, fugu5-1, and AVP1Pro:IPP1#8-3 grown for 4 DAS in the dark on either MS alone (top panel) or MS + Suc media (bottom panel). Bars = 5 mm.
(D) Hypocotyl length of etiolated seedlings was determined at 4 DAS. Data are means and sd (n = 20). Asterisk indicates significant difference at P < 0.001 compared with the wild type.
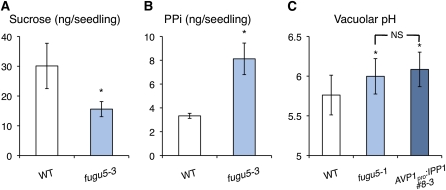
Next, to discriminate between the above two possibilities, the amounts of Suc and PPi were quantified in the wild type and fugu5 mutants. Etiolated seedlings grown on MS plates without Suc were used to evaluate only the amount of Suc produced from TAGs. Very importantly, in fugu5 mutants, the PPi contents were ~2.5-fold higher and Suc contents were only 50% that of the wild type (Figures 6A and 6B). These results unambiguously demonstrated that it is gluconeogenesis, rather than the glyoxylate cycle, that is inhibited due to the elevated levels of cytosolic PPi. In most cases reported, the observed PPi contents in various plant tissues ranged between 10 and 50 nmoles/g fresh weight (summarized and discussed in Heinonen, 2001). The PPi levels shown in Figure 6B are equivalent to 51.0 ± 3.4 nmoles/g fresh weight and 102 ± 8.8 nmoles/g fresh weight in the wild-type and fugu5-3 seedlings, respectively. Therefore, the PPi contents determined in this study agree well with those reported previously.
Figure 6.
Effects of the Loss of V-PPase Activity on Suc and PPi Contents and the Vacuolar pH.
(A) The amounts of Suc are significantly decreased in the fugu5-3 mutant. The amounts of Suc in the wild type (WT) and the fugu5-3 mutant were determined during postgerminative growth in 3-d-old etiolated seedlings grown on MS-only medium (four hundred etiolated seedlings per experiment) as described in Methods. Data are means and sd from three independent experiments. Asterisk indicates significant difference at P < 0.05 compared with the wild type.
(B) The amounts of PPi are significantly increased in the fugu5-3 mutant. Seedlings grown under the same conditions and collected at the same stage as described in (A) were used for the quantification of PPi. Data are means and sd from three independent experiments. Asterisk indicates significant difference at P < 0.05 compared with the wild type.
(C) Loss of the V-PPase activity causes a slight alkalinization of vacuolar pH in the fugu5-1 mutant. The vacuolar pH of 10-d-old seedlings grown on MS medium without Suc was determined using the fluorescent cell-permeant dye BCECF-AM as described (Krebs et al., 2010). The loss of V-PPase activity increases the vacuolar pH in root cells by 0.25 pH units. Data are means and sd of 23 measurements from 10 seedlings (the wild type), 25 measurements from nine seedlings (fugu5-1), and 26 measurements from 10 seedlings (AVP1pro:IPP1#8-3). Asterisk indicates significant difference at P < 0.005 compared with the wild type. NS, no significant difference between the two genotypes (fugu5-1 and AVP1pro:IPP1#8-3).
[See online article for color version of this figure.]
Although seedling growth, before the start of photosynthesis, relies on Suc produced by gluconeogenesis from seed lipid reserves, Suc is presumably available in photosynthetically active tissues, such as leaves produced at higher nodes. Interestingly, we found that in contrast with cotyledons and first leaves, where compensation was induced in fugu5-1, cell numbers and sizes in the third and fifth leaves were normal, compared with the wild type (see Supplemental Figure 1C online). Together, these results indicate that the loss of V-PPase activity is critical for cellular functions in organs formed at the earliest stages of postgerminative development but not in those photosynthetically active organs formed at the later stages.
It is well established that the tonoplast is energized by two distinct proton pumps, the V-ATPase and the V-PPase; however, both proton pumps seem to play differential role during plant development (Krebs et al., 2010). Although the effect of loss of function of some V-ATPase subunits on vacuolar pH has been clearly demonstrated (Krebs et al., 2010), the contribution of V-PPase alone to the vacuolar acidification has remained elusive. We thus investigated the vacuolar pH in the wild type, fugu5-1, and AVP1Pro:IPP1#8-3. Whereas vacuoles in wild-type root cells had a pH of 5.76, fugu5-1 vacuoles were found shifted to pH 5.99 (Figure 6C). On the other hand, the vacuoles in the AVP1Pro:IPP1#8-3 had a pH of 6.08, confirming that the heterologous expression of the cytosolic IPP1 does not interfere with vacuolar pH. Together, these findings clearly showed, on one hand, that the loss of the V-PPase activity caused a slight alkalinization of vacuolar pH, pointing to the contribution of the V-PPase to vacuolar acidification. However, on the other hand, when we compare the vacuolar pH in fugu5 and AVP1Pro:IPP1#8-3, it is evident that acidification of the vacuole is indeed not the major reason for the fugu5 phenotypes.
DISCUSSION
In the last four decades, a large body of data has been published showing that PPi can affect many biochemical as well as physiological reactions; however, a direct demonstration of the biological effects of PPi in vivo remained elusive (summarized and discussed in Heinonen, 2001).
Arabidopsis has three genes for H+-PPases: a single gene for the type I enzyme, At VHP1;1 (also called AVP1); and two genes for the type II enzyme, At VHP2;1 and At VHP2;2 (Drozdowicz and Rea, 2001; Segami et al., 2010). A comparative biochemical characterization between AVP1/FUGU5 and AVP2 (VHP2;1; a type II H+-PPase), after heterologous expression in yeast, revealed that AVP2 is also competent in both PPi hydrolysis and H+-translocation (Drozdowicz et al., 2000), therefore suggesting a role of both H+-PPase types in PPi homeostasis, yet this has to be demonstrated directly in plants (Rea and Sanders, 1987). In our recent study, the total protein amount of the type II enzymes was quantified to be <2.0 ng/mg of protein in the microsomal fraction (Segami et al., 2010). This amount is <0.2% of that of the type I enzyme. Also, the type II enzymes are localized not in the vacuolar membrane, but in the Golgi apparatus. Therefore, the physiological contribution of the type II H+-PPases in the vacuolar function and the PPi state in the cytoplasm may be negligible due to its extremely low expression level.
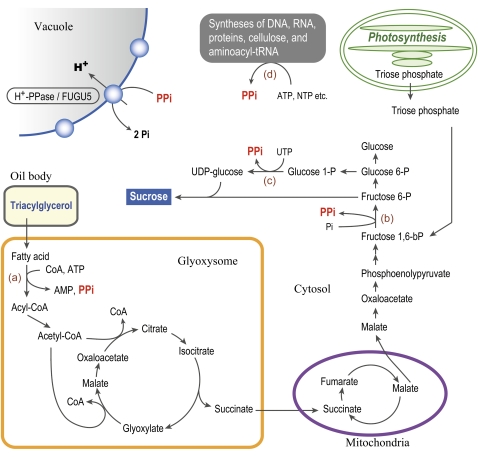
Here, our identification and analysis of the Arabidopsis avp1/fugu5 loss-of-function mutants provides a substantial advancement to our knowledge about the role of the V-PPase in vivo in plants. This study has also clarified that dysfunction of PPi removal in fugu5 mutant causes repression of cell proliferation and triggers compensation. Based on our findings, we consider the following model for the events that occur in the fugu5 mutant background and propose the implications of V-PPase early in plant development. Suc synthesis de novo was partially inhibited in fugu5 mutants, although they were able to mobilize TAGs up to a certain step downstream of β-oxidation (Figure 7). This could be explained by an inhibition of biochemical reactions that follow the β-oxidation, such as the gluconeogenesis, which seems likely to be a major target of PPi inhibition. This is very reasonable because PPi accumulates in the cytosol where all gluconeogenesis reactions take place. Alternatively, at this stage, the possibility that the amounts of other key metabolites and enzyme activities immediately upstream of gluconeogenesis might be affected in fugu5 cannot be ruled out.
Figure 7.
Suc Synthesis and PPi Generation Processes during Germination in Oilseeds.
The metabolic pathways shown here were deduced from Taiz and Zeiger (2010). Fatty acids from TAG are converted to acetyl-CoA by β-oxidation and then to succinate by the glyoxylate cycle operating in glyoxysomes. PPi is generated by the reaction of fatty acyl-CoA synthase (a). Succinate is transferred into mitochondria and converted to malate by the citric acid cycle reactions. Malate exported into the cytosol is oxidized to oxaloacetate and converted to phosphoenolpyruvate. During gluconeogenesis from phosphoenolpyruvate, PPi is generated by the reaction of PPi-dependent phosphofructokinase (b). PPi is also produced by the reaction of UDP-Glc pyrophosphorylase (c), which provides UDP-Glc. Syntheses of macromolecules, such as cellulose, also generate PPi as a by-product (d). PPi in the cytosol is consumed by V-PPase/AVP1/FUGU5.
In active gluconeogenesis in germinating oilseeds, PPi-dependent phosphofructokinase catalyzes the conversion of Fru-1,6-bisphosphate to Fru-6-phosphate (Fru-6-P) and produces PPi (Lim et al., 2009). Synthesis of Suc consumes Fru-6-P and UDP-Glc. UDP-Glc is converted from Glc-1-phosphate by UDP-Glc pyrophosphorylase, and PPi is generated concurrently (Figure 7). Thus, accumulation of PPi at excess levels in the cytosol of the fugu5 mutant might suppress these two reactions and, as a result, stop Suc synthesis in cotyledons. Interestingly, Stitt (1989) has demonstrated that in vitro Pi inhibits the reaction in the direction of Fru-6-P phosphorylation (glycolysis) and PPi is inhibitory to the opposite reaction (gluconeogenesis) using PPi:Fru-6-P phosphotransferase purified from potato (Solanum tuberosum) tuber. The concentrations of the reactants are close to equilibrium in plant cytoplasm (Kubota and Ashihara, 1990); therefore, the reaction is readily reversible if the concentrations change.
Several other supporting evidence for the importance of PPases in Suc metabolism have been reported in the literature, but their connecting relationship remained elusive. For example, transgenic tobacco (Nicotiana tabacum) plants overexpressing the E. coli ppa gene showed a dramatic change in photoassimilate partitioning, marked by high accumulation of Suc in source leaves (Sonnewald, 1992). Again, this indicates a fundamental relationship between PPase activity and Suc metabolism. Conversely, Suc supplied in the growth medium for fugu5 mutant might be used as a main carbon source for normal germination and development. Also, consumption of exogenous Suc may contribute to decrease the PPi level in growing seedlings. If any, these plausible roles of V-PPase totally differ from the actual understandings, such as auxin transport regulation. Henceforth, this report urges us to reconsider our knowledge of the role of V-PPase in vivo.
METHODS
Plant Materials and Growth Conditions
The wild type used in this study was Columbia-0 (Col-0), and all mutants and transgenic plants were in the Col-0 background. Isolation of fugu5 mutants was reported previously (Horiguchi et al., 2006b; Ferjani et al., 2007). The vhp1-1 mutant line was selected from a large T-DNA insertion library of Arabidopsis thaliana, which was prepared by the Kazusa DNA Research Institute. Prior to analyses, all of the mutants were backcrossed to Col-0 at least three times. Seeds were sown on rockwool (Nitto Boseki), watered daily with 0.5 g L−1 Hyponex solution (Hyponex Japan), and grown in a growth room with a 16/8-h light/dark cycle with white light fluorescent lamps at ~50 μmol m−2 s−1 at 22°C. To determine the effect of growth medium composition, sterilized seeds were sown on MS medium (Wako) or MS medium with 2% (w/v) Suc, Gamborg's B5 vitamins, or other compounds where indicated and solidified using 0.5% (w/v) gellan gum (Murashige and Skoog, 1962; Gamborg et al., 1968). The seeds were then incubated at 4°C in darkness for 3 d. After cold treatment, the seedlings were grown for the indicated times.
Microscopy Observations and Phenotypic Analysis
To measure leaf areas and cell numbers, leaves were fixed with formalin/acetic acid/alcohol and cleared with chloral solution (200 g chloral hydrate, 20 g glycerol, and 50 mL deionized water) as described previously (Tsuge et al., 1996). Whole leaves and leaf cells were observed using a stereoscopic microscope (MZ16a; Leica Microsystems) and a microscope equipped with Nomarski differential interference contrast (DMRX E; Leica Microsystems), respectively. Cell numbers along the transverse axis were counted at the widest point of the cotyledon, and those along the longitudinal cotyledon axis were counted at a short distance from the midvein. The cell size was determined as the average cell area of 20 palisade cells per leaf (n = 8), observed from the paradermal view. Leaf index values were calculated as the ratio of cotyledon length to cotyledon width. Statistical analysis was conducted using two-tailed Student’s t test. Wild-type and mutant plants carrying a CYCB1;1pro:GUS gene or DR5:GUS gene were subjected to GUS staining as described (Donnelly et al., 1999). The GUS-stained plants were cleared in the chloral solution for observation.
Genetic Mapping of AVP1/FUGU5
A mapping population was generated by crossing the fugu5-1 mutant with Landsberg erecta. Genomic DNA from F2 plants that showed the fugu5-1 mutant phenotype were selected and subjected to map-based cloning. The AVP1/FUGU5 locus was genetically mapped to the upstream region of chromosome 1 using various genetic markers (simple sequence length polymorphism, cleaved amplified polymorphisms, and small insertion/deletions) according to the sequence information available at The Arabidopsis Information Resource database (http://www.Arabidopsis.org/index.jsp).
Generation of Transgenic Plants
The 4.4-kb full-length promoter region of AVP1/FUGU5 was amplified by PCR using the B4F-pAVP1-FW/B1R-pAVP1-RV primer set (see Supplemental Table 1 online). The PCR-amplified fragments were cloned into the pDONRP4-P1R vector by performing BP recombination (Invitrogen). The coding region of IPP1 from Saccharomyces cerevisiae was amplified by PCR using the IPP1-FW/IPP1-RV primer set (see Supplemental Table 1 online). The products of the first PCR reaction were subjected to a second round of PCR with the B1/B2 primer set (see Supplemental Table 1). The products of the second reaction were fused into pDONR201 by performing BP recombination (Invitrogen). The resulting vectors were subjected to the LR reaction with the R4 gateway binary vector R4pGWB501 (Nakagawa et al., 2008). The resultant final construct was used to transform wild-type and fugu5-1 mutant plants by the floral dip method (Clough and Bent, 1998). Several independent T3 homozygous lines expressing the IPP1 gene under the AVP1/FUGU5 promoter from a single T-DNA insertion locus were identified in both wild-type and fugu5-1 backgrounds. Line numbers AVP1Pro:IPP1#4-4 and #8-3 (in fugu5 mutant background) and line numbers AVP1Pro:IPP1#2-15 and #6-4 (in wild-type background) were chosen for further study.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted using the RNeasy plant mini kit (Qiagen). RNA samples were then treated with DNase I (Life Technologies), followed by reverse transcription using a transcription first-strand cDNA synthesis kit (Invitrogen). RT-PCR was performed with an ABI 9700 (Life Technologies). Most sequences for RT-PCR primers for core cell cycle genes were found at http://atrtprimer.kaist.ac.kr/ (Han and Kim, 2006). The primer sequences used are listed in Supplemental Table 1 online.
Crude Membrane Preparation
Shoots from wild-type and fugu5 mutant plants (240 to 270 plants) were homogenized in 25 mL of 50 mM Tris-acetate, pH 7.5, 0.25 M sorbitol, 1% polyvinylpyrrolidone, 1 mM EGTA, 2 mM DTT, and 20 μM p-(amidinophenyl) methanesulphonyl fluoride hydrochloride with a mortar and pestle. The homogenate was filtrated through three layers of cheesecloth and centrifuged (10,000g, 10 min). After centrifugation of the supernatant again (100,000g, 40 min), the pellet obtained was suspended in 20 mM Tris-acetate, pH 7.5, 0.25 M sorbitol, 1 mM EGTA, 1mM MgCl2, and 2 mM DTT and used as crude membrane. Protein content was determined using a protein assay kit (Bio-Rad).
SDS-PAGE and Immunoblotting Analysis
Protein samples were analyzed using SDS-PAGE and immunoblotting. After electrophoresis, the proteins were transferred to an Immobilon-P membrane (Millipore). The primary antibodies used were prepared previously: antibodies to V-PPase (antigen peptide sequence, DLVGKIERNIPEDDRN; Kobae et al., 2006), V-ATPase subunit A (TKAREVLQREDDLNEI; Kobae et al., 2006), V-ATPase subunit a (ELVEINANNDKLQRSYNEL; Kobae et al., 2004), BIP (SKDNKALGKLRREC; Kobae et al., 2004), and TIP1s (antibody to the purified protein; Higuchi et al., 1998). Antigen on the membrane was visualized with horseradish peroxidase–coupled protein A and chemiluminescent reagents ECL (GE Healthcare). Images and the intensity of immunostained bands were quantitatively measured by densitometry with a cooled CCD camera system (Light-Capture II; Atto Co.).
Enzyme Assays
PPi hydrolysis by V-PPase was measured at 30°C as described previously (Maeshima and Yoshida, 1989). The reaction medium contained 1 mM Na4PPi, 50 mM KCl, 1 mM Na-molybdate, 0.2% Triton X-100, 1 mM MgSO4, 0.5 mM KF, and 30 mM Tris-Mes, pH 7.2. ATP hydrolysis activity, by V-ATPase, was measured at 30°C as described previously (Schumacher et al., 1999) in a reaction medium containing 1 mM NaN3, 0.1 mM sodium molybdate, 0.5 mM sodium vanadate, 0.03% Triton X-100, 3 mM ATP, 3 mM MgSO4, and 30 mM Tris-Mes, pH 7.2. The activity was determined in the presence or absence of KCl or NaNO3. The nitrate-sensitive and chloride-stimulated activity was defined as the V-ATPase activity.
Quantitative Analyses of Total TAGs
The amounts of seed lipid reserves contained in dry seeds and in 1-, 2-, 3-, and 4-d-old etiolated seedlings were measured by determining the amount of total TAG using the assay kit Triglyceride E-Test (Wako). Either 20 dry seeds or 20 seedlings were homogenized in a mortar in 100 μL of sterile distilled water. Homogenates were mixed with 0.75 mL of reaction buffer provided in the kit as described previously (Arai et al., 2008). The concentration of TAG in the sample was determined according to the manufacturer’s protocol.
Quantitative Analyses of Suc and PPi
The amounts of Suc and PPi in 3-d-old etiolated seedlings were measured by determining the amount of total Suc and PPi with an ion chromatography system (DX-500; Dionex Co.) using a CarboPAC PA1 column for Suc and an IonPac AS11 for PPi, respectively. For the quantification of PPi, we carefully extracted organic and inorganic ions from seedlings. Each etiolated seedling was collected from the plates and immediately put into liquid nitrogen to avoid hydrolysis of PPi in the tissues. Four hundred frozen seedlings were homogenized in chilled 80% ethanol, vortexed for 10 min, treated at 80°C for 30 min, and then centrifuged at 20,000g for 5 min. The precipitate was resuspended in chilled 80% ethanol and centrifuged. The first and second supernatants were combined and dried with a centrifugal evaporator. An aliquot of water was added into the tube, and water-soluble compounds were extracted. The extract was filtered with a Sep-Pak Light C18 filter (Waters) and then with a Dismic-13cp (Advantec). During the process, phosphatases have no opportunity to hydrolyze PPi in the tissues and extracts. Typically, the fate of internal standards, such as sugar and inorganic ion, was followed and checked to verify that recovery was more than 85%. To determine the PPi content, the sample was pretreated using an ASRS-300 anion self-regenerating suppressor column (Dionex) before the IonPac AS11 anion-exchange column to reduce the background noise. In this experiment, we could clearly distinguish PPi from Pi, polyphosphates, inorganic acid, or organic acid.
Measurement of Vacuolar pH
The vacuolar pH of 10-d-old seedlings grown on MS plates without Suc was determined using the fluorescent cell-permeant dye BCECF-AM (Invitrogen) as described (Krebs et al., 2010). Loading of the dye was performed in 10 μM BCECF-AM and 0.02% Pluronic F-127 (Invitrogen). After 1 h of staining in darkness with gentle agitation, the seedlings were washed once in distilled water. BCECF fluorescence was detected using an Olympus FV1000-D confocal laser scanning microscope. The images were obtained using the Fluoview Confocal software and a UPLSAPO ×60 water immersion objective. The fluorophore was excited at 488 and 440 nm, respectively, and the emission was detected between 530 and 550 nm. All the images were recorded in the fully elongated region of root cells without root hairs. Ratio images were generated using the ratio/concentration tool of the Olympus Fluoview software, and the images were processed using Adobe Photoshop software. Then the ratio values were obtained using the Olympus Fluoview software. The integrated pixel density was measured and the values of the 488-nm excited images were divided by the values of the 440-nm excited images. The chromatic aberration of the objective was carefully checked and considered. The ratio was then used to calculate the pH values on the basis of a calibration curve. In situ calibration of BCECF was performed with 10-d-old Arabidopsis seedlings loaded with the dye. The seedlings were incubated in pH equilibration buffers (De Angeli et al., 2006) 15 min prior to measurement. The buffers contained 50 mM MES-BTP, pH 5.38 to 6.4, or 50 mM HEPES-BTP, pH 6.8, and 50 mM ammonium acetate. The calibration curve was obtained by plotting the average ratio values of eight seedlings against the pH.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: Arabidopsis AVP1/FUGU5 (also called VHP1;1) , At1g15690; VHP2;1, At1g78920; VHP2;2, At1g16780; CYCB1;1, At4g37490; CYCB1;2, At5g06150; CDKA;1, At3g48750; CDKB1;1, At3g54180; CDKB1;2, At2g38620; and TUBULINβ 4, At5g44340; S. cerevisiae IPP1, YBR011C; vhp1-1 T-DNA insertion mutant, Kazusa reference number KG8420.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Compensation in fugu5 Is Restricted to Cotyledons and First Leaves.
Supplemental Figure 2. Phenotype of Wild-Type and fugu5 Mutant Young Seedlings Grown on MS Media with Different Supplements.
Supplemental Figure 3. Effect of a Glc Analog and Osmotic Stress on the fugu5 Phenotype.
Supplemental Figure 4. Effect of Suc on the Cotyledon Phenotype of Different Alleles of the fugu5 Mutant.
Supplemental Figure 5. Effect of Suc on the Expression of the CYCB1;1pro:GUS Reporter and Several Core Cell Cycle Genes.
Supplemental Figure 6. Mutation Sites and Membrane Topology of V-PPase.
Supplemental Figure 7. Gross Morphology of Wild-Type and fugu5 Mutant Allele Plants.
Supplemental Figure 8. The vhp1-1 Mutant, a T-DNA Insertion Line Allele of fugu5, Exhibits Compensation.
Supplemental Figure 9. Phenotype of AVP1pro:IPP1 (Wild-Type) Transgenic Plants.
Supplemental Figure 10. Mobilization of Seed Lipid Reserves and Effect of 2,4-DB on fugu5 Mutant.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank J. Celenza (Boston University) for donating CYCB1;1pro:GUS line seeds, T.J. Guilfoyle (University of Missouri) for providing DR5:GUS line seeds, T. Nakagawa (Shimane University) for the R4pGWB501 binary vector, and Y. Kamada (National Institute for Basic Biology) for providing S. cerevisiae genomic DNA. We also thank M. Asaoka and C. Sato (Nagoya University) for their expert help with the PPi and Suc quantification, A. Miyaka (Nagoya University) for measurement of vacuolar pH, and Y. Nakanishi and M. Inagaki (Nagoya University) for isolation of vhp1-1 mutant line. We thank H. Iida and Y. Nakayama (Tokyo Gakugei University) for providing laboratory facilities and technical support for SDS-PAGE analyses and E. (Takabe) Itoh (University of Tokyo) for technical support. This work was supported by Grants-in-Aid from the Japan Society for the Promotion of Science (Grant 16-04179 to A.F.), Grant-in-Aid for Young Scientists (B) (to A.F.), Creative Scientific Research (to H.T.), Scientific Research (A, to H.T. and G.H.; B, to M.M.), Scientific Research on Priority Areas (to H.T.), Young Scientists (B) and Exploratory Research (to G.H.), and the Ministry of Education, Culture, Sports, Science, and Technology of Japan, as well as grants from the Bio-Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (to H.T.) and the Toray Science Foundation (to H.T.).
AUTHOR CONTRIBUTIONS
A.F. conceived the project, designed the study, performed the experiments, and wrote the article. S.S. performed experiments and analyzed the data. G.H. performed experiments directed the study and wrote the article. Y.M. performed experiments and analyzed the data. M.M. designed and directed the study and wrote the article. H.T. designed and directed the study and wrote the article.
References
- Anastasiou E., Lenhard M. (2007). Growing up to one’s standard. Curr. Opin. Plant Biol. 10: 63–69 [DOI] [PubMed] [Google Scholar]
- Arai Y., Hayashi M., Nishimura M. (2008). Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis. Plant Cell 20: 3227–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Brevet A., Fromant M., Lévêque F., Schmitter J.M., Blanquet S., Plateau P. (1990). Pyrophosphatase is essential for growth of Escherichia coli. J. Bacteriol. 172: 5686–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Angeli A., Monachello D., Ephritikhine G., Frachisse J.M., Thomine S., Gambale F., Barbier-Brygoo H. (2006). The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., Naudts M., Inzé D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Drozdowicz Y.M., Kissinger J.C., Rea P.A. (2000). AVP2, a sequence-divergent, K(+)-insensitive H(+)-translocating inorganic pyrophosphatase from Arabidopsis. Plant Physiol. 123: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdowicz Y.M., Rea P.A. (2001). Vacuolar H(+) pyrophosphatases: From the evolutionary backwaters into the mainstream. Trends Plant Sci. 6: 206–211 [DOI] [PubMed] [Google Scholar]
- Eastmond P.J., Germain V., Lange P.R., Bryce J.H., Smith S.M., Graham I.A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Horiguchi G., Yano S., Tsukaya H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Yano S., Horiguchi G., Tsukaya H. (2008). Control of leaf morphogenesis by long- and short-distance signaling: Differentiation of leaves into sun or shade types and compensated cell enlargement. In Plant Cell Monographs: Plant Growth Signaling, Bögre L., Beemster G.T.S., (Berlin, Heidelberg, Germany: Springer Berlin Heidelberg; ), pp. 47–62 [Google Scholar]
- Fujikura U., Horiguchi G., Ponce M.R., Micol J.L., Tsukaya H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59: 499–508 [DOI] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Han S., Kim D. (2006). AtRTPrimer: Database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T., Suga S., Tsuchiya T., Hisada H., Morishima S., Okada Y., Maeshima M. (1998). Molecular cloning, water channel activity and tissue specific expression of two isoforms of radish vacuolar aquaporin. Plant Cell Physiol. 39: 905–913 [DOI] [PubMed] [Google Scholar]
- Heinonen J.K. (2001). Biological Role of Inorganic Pyrophosphate. (Boston/Dordrecht/London: Kluwer Academic Publishers; ). [Google Scholar]
- Horiguchi G., Ferjani A., Fujikura U., Tsukaya H. (2006a). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Fujikura U., Ferjani A., Ishikawa N., Tsukaya H. (2006b). Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J. 48: 638–644 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Ingram G.C., Waites R. (2006). Keeping it together: Co-ordinating plant growth. Curr. Opin. Plant Biol. 9: 12–20 [DOI] [PubMed] [Google Scholar]
- Kawade K., Horiguchi G., Tsukaya H. (2010). Non-cell-autonomously coordinated organ size regulation in leaf development. Development 137: 4221–4227 [DOI] [PubMed] [Google Scholar]
- Ko K.M., Lee W., Yu J.R., Ahnn J. (2007). PYP-1, inorganic pyrophosphatase, is required for larval development and intestinal function in C. elegans. FEBS Lett. 581: 5445–5453 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Mizutani M., Segami S., Maeshima M. (2006). Immunochemical analysis of aquaporin isoforms in Arabidopsis suspension-cultured cells. Biosci. Biotechnol. Biochem. 70: 980–987 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Uemura T., Sato M.H., Ohnishi M., Mimura T., Nakagawa T., Maeshima M. (2004). Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 45: 1749–1758 [DOI] [PubMed] [Google Scholar]
- Krebs M., Beyhl D., Görlich E., Al-Rasheid K.A., Marten I., Stierhof Y.D., Hedrich R., Schumacher K. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A. (2009). Making bigger plants: Key regulators of final organ size. Curr. Opin. Plant Biol. 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Kubota K., Ashihara H. (1990). Identification of non-equilibrium glycolytic reactions in suspension-cultured plant cells. Biochim. Biophys. Acta 1036: 138–142 [DOI] [PubMed] [Google Scholar]
- Li J., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Lim H., Cho M.H., Jeon J.S., Bhoo S.H., Kwon Y.K., Hahn T.R. (2009). Altered expression of pyrophosphate: Fructose-6-phosphate 1-phosphotransferase affects the growth of transgenic Arabidopsis plants. Mol. Cells 27: 641–649 [DOI] [PubMed] [Google Scholar]
- Lundin M., Baltscheffsky H., Ronne H. (1991). Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J. Biol. Chem. 266: 12168–12172 [PubMed] [Google Scholar]
- Maeshima M. (2000). Vacuolar H(+)-pyrophosphatase. Biochim. Biophys. Acta 1465: 37–51 [DOI] [PubMed] [Google Scholar]
- Maeshima M., Yoshida S. (1989). Purification and properties of vacuolar membrane proton-translocating inorganic pyrophosphatase from mung bean. J. Biol. Chem. 264: 20068–20073 [PubMed] [Google Scholar]
- Martinoia E., Maeshima M., Neuhaus H.E. (2007). Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Micol J.L. (2009). Leaf development: Time to turn over a new leaf? Curr. Opin. Plant Biol. 12: 9–16 [DOI] [PubMed] [Google Scholar]
- Mimura H., Nakanishi Y., Hirono M., Maeshima M. (2004). Membrane topology of the H+-pyrophosphatase of Streptomyces coelicolor determined by cysteine-scanning mutagenesis. J. Biol. Chem. 279: 35106–35112 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntz K. (1998). Deposition of storage proteins. Plant Mol. Biol. 38: 77–99 [PubMed] [Google Scholar]
- Müntz K. (2007). Protein dynamics and proteolysis in plant vacuoles. J. Exp. Bot. 58: 2391–2407 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. (2008). Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci. Biotechnol. Biochem. 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Penfield S., Graham S., Graham I.A. (2005). Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Perez-Castineira J.R., Lopez-Marques R.L., Villalba J.M., Losada M., Serrano A. (2002). Functional complementation of yeast cytosolic pyrophosphatase by bacterial and plant H+-translocating pyrophosphatases. Proc. Natl. Acad. Sci. USA 99: 15914–15919 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rea P.A., Sanders D. (1987). Tonoplast energization: Two H+ pumps, one membrane. Physiol. Plant. 71: 131–141 [Google Scholar]
- Rylott E.L., Hooks M.A., Graham I.A. (2001). Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem. Soc. Trans. 29: 283–287 [DOI] [PubMed] [Google Scholar]
- Schumacher K., Vafeados D., McCarthy M., Sze H., Wilkins T., Chory J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S., Nakanishi Y., Sato M.H., Maeshima M. (2010). Quantification, organ-specific accumulation and intracellular localization of type II H(+)-pyrophosphatase in Arabidopsis thaliana. Plant Cell Physiol. 51: 1350–1360 [DOI] [PubMed] [Google Scholar]
- Sonnewald U. (1992). Expression of E. coli inorganic pyrophosphatase in transgenic plants alters photoassimilate partitioning. Plant J. 2: 571–581 [PubMed] [Google Scholar]
- Stitt M. (1989). Product inhibition of potato tuber pyrophosphate:fructose-6-phosphate phosphotransferase by phosphate and pyrophosphate. Plant Physiol. 89: 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoynova-Bakalova E., Karanov E., Petrov P., Hall M.A. (2004). Cell division and cell expansion in cotyledons of Arabidopsis seedlings. New Phytol. 162: 471–479 [Google Scholar]
- Taiz L., Zeiger E. (2010). Plant Physiology, 5th ed (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Tsuge T., Tsukaya H., Uchimiya H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (1998). Relationship between shape of cells and shape of the organ. In Phytohormones and Cell Shape Imazeki H., Shibaoka H., (Tokyo: Gakkai-shuppann Center; ), pp. 177–184 [In Japanese] [Google Scholar]
- Tsukaya H. (2002). Interpretation of mutants in leaf morphology: Genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int. Rev. Cytol. 217: 1–39 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2005). Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 49: 547–555 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2008). Controlling size in multicellular organs: Focus on the leaf. PLoS Biol. 6: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H., Tsuge T., Uchimiya H. (1994). The cotyledon: A superior system for studies of leaf development. Planta 195: 309–312 [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T., Horiguchi G., Yano S., Tsukaya H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- White D.W.R. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.