This study analyzes the molecular mechanisms underlying reprogramming of gametophore leaf cells to stem cells in the moss Physcomitrella patens. Cyclin-dependent kinase A is found to link cell cycle reactivation with the generation of stem cell characters.
Abstract
During regeneration, differentiated plant cells can be reprogrammed to produce stem cells, a process that requires coordination of cell cycle reactivation with acquisition of other cellular characteristics. However, the factors that coordinate the two functions during reprogramming have not been determined. Here, we report a link between cell cycle reactivation and the acquisition of new cell-type characteristics through the activity of cyclin-dependent kinase A (CDKA) during reprogramming in the moss Physcomitrella patens. Excised gametophore leaf cells of P. patens are readily reprogrammed, initiate tip growth, and form chloronema apical cells with stem cell characteristics at their first cell division. We found that leaf cells facing the cut undergo CDK activation along with induction of a D-type cyclin, tip growth, and transcriptional activation of protonema-specific genes. A DNA synthesis inhibitor, aphidicolin, inhibited cell cycle progression but prevented neither tip growth nor protonemal gene expression, indicating that cell cycle progression is not required for acquisition of protonema cell-type characteristics. By contrast, treatment with a CDK inhibitor or induction of dominant-negative CDKA;1 protein inhibited not only cell cycle progression but also tip growth and protonemal gene expression. These findings indicate that cell cycle progression is coordinated with other cellular changes by the concomitant regulation through CDKA;1.
INTRODUCTION
A stem cell is defined as an undifferentiated cell that has the capacity for self-renewal and that can give rise to more specialized cells (Lajtha, 1979; Gilbert, 2006; Slack, 2008). Land plants have meristems localized at the tips of their bodies harboring stem cells with continuous cell division activity. Stem cells are initiated at an early stage of development and maintained during the growth period, providing cells that give rise to most parts of the plant body. In addition, stem cells are repeatedly formed from differentiated cells during development and growth, such as the formation of rhizoids (Sakakibara et al., 2003; Menand et al., 2007b) and side branches (Harrison et al., 2009) in bryophytes. Furthermore, under the appropriate inductive conditions, differentiated cells can be reprogrammed to form stem cells. In vascular plants, dissected or wounded tissues can proliferate when treated with exogenous phytohormones to form callus, which can be fated to form shoot or root meristematic tissue bearing stem cells (Skoog and Miller, 1957; Raghavan, 1989). In ferns (Raghavan, 1989) and bryophytes (Chopra and Kumra, 1988), a differentiated cell that faces wounded cells is reprogrammed to form a stem cell called an apical cell without forming callus. It is thought that the differentiated cells of land plants are more competent for reprogramming into stem cells than those of metazoan cells, although artificial expression of two transcription factors, Oct4 and Sox2, along with other factors made it possible to reprogram differentiated somatic cells into pluripotent stem cells in mice and humans (reviewed in Masip et al., 2010). However, the molecular mechanisms of reprogramming remain elusive (Vogel, 2005; Birnbaum and Sánchez Alvarado, 2008).
In angiosperms, reprogramming of differentiated cells into stem cells is accompanied by reentry into the cell cycle from a nonproliferative state to the G1 phase (den Boer and Murray, 2000). In response to chemicals, including Suc and phytohormones, D-type cyclin (CYCD) is induced and binds to the A-type cyclin-dependent kinase (CDKA) to form a CDKA/CYCD complex. An active form of this complex regulates three proteins, which are themselves in a complex, namely, E2 promoter binding factor (E2F), retinoblastoma-related (RBR), and dimerization partner (DP) cell cycle regulators, resulting in the transition from G1 to S phase, with activation of S-phase genes (Inzé and De Veylder, 2006). In addition to the reentry into the cell cycle, other cellular characteristics, including gene expression patterns and consequent cellular morphology and growth, are changed during reprogramming (Che et al., 2002, 2006a, 2006b; Sugimoto et al., 2010). The factors responsible for coordinating these processes during reprogramming are still unknown.
Cell division and other cellular characteristics are well coordinated not only in reprogramming but throughout development. This coordination appears to depend at least in part on certain cell cycle regulators themselves. For example, in Arabidopsis thaliana, reduction of RBR expression levels in the shoot meristem caused diminished expression of CLAVATA3 and WUSCHEL regulating stem cell identity (Borghi et al., 2010). Similarly, the induction of a kinase-negative form of CDKA in the shoot meristem caused a portion of the meristem cells to expand and exhibit endoreduplication and thus resemble differentiated cells (Gaamouche et al., 2010). These results could be taken to mean that RBR and CDKA regulate both cell division and other cellular characteristics, but on the other hand, cellular characteristics could have been affected as a consequence of the altered cell division, for example, because of abnormal intercellular communication. It is difficult to distinguish between these alternatives in a complex multicellular meristem.
To analyze the molecular mechanism of coordination between cell cycle regulation and other cellular state changes during reprogramming from differentiated cells to stem cells, we used the moss Physcomitrella patens. This moss forms a hypha-like body, called a protonema, and a shoot-like body, called a gametophore, in the gametophyte generation. Two types of protonemata, named chloronemata and caulonemata, are recognized according to their differences in cellular morphology and growth. A single stem cell is situated at the tip of each protonemal filament and at the apex of each gametophore, which are named a protonema apical cell and a gametophore apical cell, respectively (Cove et al., 2006; Rensing et al., 2008; Prigge and Bezanilla, 2010). When part of a gametophore leaf of the moss is excised and cultivated for a few days on culture medium without phytohormone supplementation, leaf cells facing the cut edge change into cells that are indistinguishable from the apical cells of chloronemata (Figures 1A and 1B; see Supplemental Movie 1 online; Chopra and Kumra, 1988; Prigge and Bezanilla, 2010). During this reprogramming, the leaf cells reenter the cell cycle and acquire protonema-specific cellular characteristics, including tip growth.
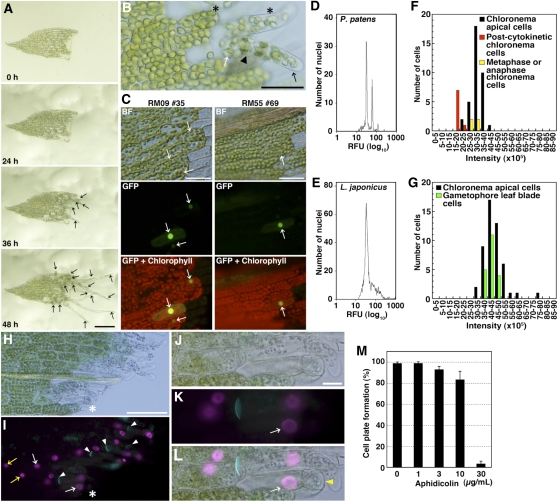
Figure 1.
Formation of Chloronema Apical Cells in Excised Leaves.
(A) Sequential bright-field micrographs of chloronemata apical cell formation in an excised leaf. Snapshots at 0, 24, 36, and 48 h after excision were taken from a time-lapse movie (see Supplemental Movie 1 online). Arrows denote chloronema apical cells. Bar = 100 μm.
(B) A magnified micrograph showing cells facing the cut at 36 h after excision. Asterisks indicate leaf cells showing tip growth before their cell division. An arrowhead indicates a septum between a distal chloronema apical cell (black arrow) and a proximal cell without mitotic activity (white arrow). Bar = 50 μm.
(C) Promoter activities of protonema-specific genes RM09 and RM55. Leaves excised from gametophores of protonema-specific marker lines (RM09 #35 and RM55 #69; see Supplemental Figures 2 and 3 online) were incubated on BCDAT medium. Bright-field images (BF), sGFP fluorescence (green), and autofluorescence of chlorophyll (red) images were recorded at 30 h after excision. Arrows indicate cells expressing sGFP before cell division. Bars = 50 μm.
(D) and (E) Nuclear DNA content of gametophore leaves of P. patens (D) and leaves of L. japonicus (Gifu strain) (E) quantified with flow cytometry. RFU, relative fluorescence units.
(F) and (G) Nuclear DNA contents quantified by microscopy images.
(F) Comparison of nuclear DNA contents of chloronema interphase apical cells (black; n = 36) with those of postcytokinetic chloronema cells as a standard for 1C nuclei (red: n = 8) and those of metaphase or anaphase chloronema cells as a standard for 2C nuclei (yellow; n = 4).
(G) Comparison of nuclear DNA contents of chloronema interphase apical cells (black; n = 50) with those of leaf blade cells (green; n = 20). Amounts of DNA are shown as intensity calculated from fluorescent images of DAPI-stained nuclei using ImageJ.
(H) to (L) A bright-field image ([H] and [J]) and a fluorescent image ([I] and [K]) of an excised leaf incubated with 10 μM EdU for 40 h after excision. The leaf was stained with aniline blue to detect callose (cyan), which is present in newly synthesized cell plates (white arrowheads in [I]). White arrows indicate EdU-labeled nuclei in leaf cells facing the cut and acquiring tip growth before its cell division. Yellow arrows indicate EdU-labeled nuclei in leaf cells that do not face the cut, which are likely endoreduplicated. Bars = 100 μm in (H) and 20 μm in (J).
(J) to (L) A magnified micrograph including a leaf cell that acquired tip growth before its cell division, indicated by asterisks in (H) and (I).
(L) A merged image of (J) and (K). The yellow arrowhead indicates a protrusion employing tip growth.
(M) Percentage of leaves having cells with cell plate formation in excised leaves (n > 20) after a 72-h incubation with aphidicolin (0, 1, 3, 10, or 30 μg/mL). Error bars indicate sd from four biological replicates.
Here, we study the reprogramming of excised leaf cells. We find that intact leaf cells are arrested at the late S-phase, a stage that differs from the G1-phase arrest typical of angiosperms (den Boer and Murray, 2000). For reprogramming, we find that CDKA is necessary for cell cycle progression and that this kinase regulates cell cycle progression and acquisition of new cell characteristics in parallel. We thus present a factor that concomitantly regulates cell division and cellular change in reprogramming differentiated cells to become stem cells in plants.
RESULTS
Reprogramming Gametophore Cells to Become Chloronema Apical Cells
When the distal part was excised from a leaf and cultured, most of the cells facing the cut initiated tip growth at ~36 h after excision (Figure 1A; see Supplemental Movie 1 online). With continued tip growth, cells proceeded to M-phase and asymmetrically divided into an apical cell with mitotic activity and a basal, nondividing cell (Figure 1B). The apical cell appeared to be similar to a chloronema apical cell, in terms of growth rate, chloroplast morphology, and septum orientation.
To further examine whether leaf cells acquired chloronema apical cell characteristics, we generated protonema marker lines using genes encoding hypothetical proteins (RM09 [XP_001784484] and RM55 [XM_001784210]) that are expressed only in protonemata, including chloronemata, but not in gametophores, according to real-time quantitative RT-PCR (qRT-PCR; see Supplemental Figure 1 online). A 2-kb DNA fragment upstream of each coding region was fused to a DNA fragment of a synthetic nucleotide sequence for the SV40 nuclear localization signal (NLS; Kalderon et al., 1984), the sGFP gene encoding modified green fluorescent protein (GFP; Chiu et al., 1996), and the uidA gene encoding β-glucuronidase (GUS; Jefferson, 1987) and introduced into P. patens to form the RM09 and RM55 lines (see Supplemental Figure 2 online). In both lines, GFP was detected in all protonema cells but undetectable in gametophores, even young ones, suggesting that the promoter activities are protonema specific and independent of cell cycle activity (see Supplemental Figure 3 online). After leaf excision in both lines, GFP was detected in cells facing the cut, prior to the onset of tip growth (Figure 1C), indicating that the leaf cells are reprogrammed to acquire at least some protonemal cell characteristics before tip growth and also before mitosis.
Gametophore Leaf Cells Reenter the Cell Cycle in Late S-Phase
To identify the phase of the cell cycle for cells in intact leaves, we measured the DNA content of leaf cell nuclei. A previous report using flow cytometry showed that chloronema cells are arrested in G2 phase, which is different from cell cycle arrest in angiosperms, which typically takes place in G1 phase (den Boer and Murray, 2000; Schween et al., 2003). We measured DNA content with flow cytometry of propidium iodide–stained gametophore nuclei (Dolezel and Bartos, 2005), with nuclei from Lotus japonicus leaves serving as a standard (Figures 1D and 1E). As expected, nuclei from L. japonicus gave rise to a single peak, representing 2C DNA content. The nuclei from P. patens leaves had two peaks, with the predominant one at approximately the same relative fluorescence value as that of L. japonicus. Given that the P. patens genome size (490 Mb; Rensing et al., 2008) is about the same as that of L. japonicus (440 Mb; Kawasaki and Murakami, 2000) and that L. japonicus leaves are diploid whereas those of the moss are haploid, we infer that the nuclei from lotus are in G1 phase and those in the moss are in G2. Presumably, the second peak in the profile from P. patens reflects the presence of some 4C (i.e., endoreduplicated) nuclei.
Gametophore leaves contain several tissue types, including blade and vein tissues; therefore, to analyze the DNA content of blade cell nuclei specifically, we used microphotometry (Friedman, 1991). To define the 1C level, we used nuclei of chloronema apical cells just after cytokinesis (postcytokinetic cells); to define the 2C level, we used condensed chromosomes of chloronema cells at metaphase or anaphase. The DNA content of gametophore leaf blade cells overlapped with 2C chloronema cells (Figures 1F and 1G), supporting our inference that leaf cell nuclei are in G2. Also similar to the flow cytometry results, a few leaf nuclei had DNA contents higher than 2C, consistent with endoreduplication.
The results from flow cytometry and microphotometry consistently imply that gametophore blade cells are arrested in G2. However, unexpectedly, in cells induced to reprogram, but before cytokinesis, we found that 5-ethynyl-2′-deoxyuridine (EdU) was incorporated in leaf cell nuclei (Figures 1H to 1L). This compound is a terminal alkyne-containing analog of thymidine that is a marker for DNA synthesis (Salic and Mitchison, 2008). Its incorporation indicates that P. patens leaf cells progress through S-phase during reprogramming. EdU incorporation also occurred in cells that were distant from the cut (Figure 1I, yellow arrows) and that do not divide, suggesting that excision also induces endoreduplication. We also tested the necessity of DNA synthesis for cell cycle progression using aphidicolin, an inhibitor of DNA polymerase α and δ that prevents nuclear DNA replication but not endoreduplication (Planchais et al., 2000; Quelo et al., 2002). If leaf cells were arrested in G2-phase, they would be able to divide once after excision in the presence of aphidicolin. However, aphidicolin inhibited cell division of leaf cells after excision (Figure 1M). These results suggest that leaf cells reenter the cell cycle in late S-phase.
Expression of Cell Cycle Regulators during Reprogramming
Differentiated cells of angiosperms are usually arrested at G1-phase (De Veylder et al., 2007); thus, the arrest of P. patens gametophytes in late S-phase is unusual. Therefore, we analyzed transcript levels during reprogramming for putative orthologous genes to Arabidopsis cell cycle regulators, including CYCD (Pp CYCD;1 and Pp CYCD;2), CDKA (Pp CDKA;1 and Pp CDKA;2), E2F (Pp E2F;1, Pp E2F;2, Pp E2F;3, and Pp E2F;4), DP (Pp DP;1, Pp DP;2, and Pp DP;3), RBR (Pp RBR;1, Pp RBR;2, and Pp RBR;3), INHIBITOR/INTERACTOR OF CYCLIN-DEPENDENT KINASE (ICK)/KIP-RELATED PROTEIN (KRP) (Pp ICK/KRP), CYCLIN B (CYCB) (Pp CYCB;1 and Pp CYCB;2), and CDKB (Pp CDKB;1) (Banks et al., 2011; http://moss.nibb.ac.jp/treedb). To do so, gametophores were cut in a homogenizer to induce reprogramming synchronously in a sufficiently large sample for analysis by qRT-PCR (see Supplemental Figure 4 online). Here, the α-tubulin gene TUA1 was used as a reference, and it was shown to undergo little if any change in abundance during reprogramming (see Supplemental Figure 5 online).
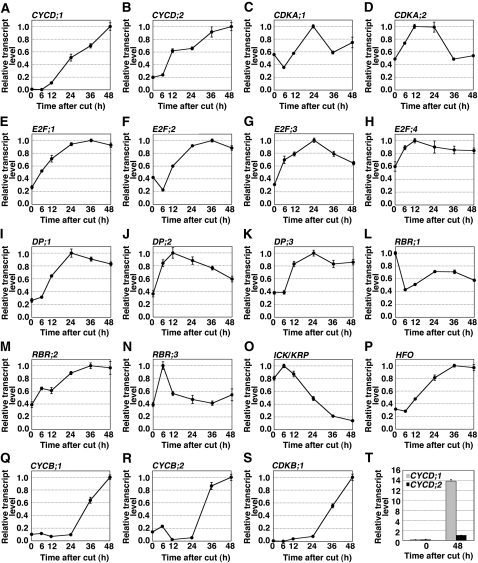
In a complex with CDKA, the D-type cyclin regulates the G1/S-phase transition (Oakenfull et al., 2002). Transcripts of Pp CYCD;1 and Pp CYCD;2 accumulated between 6 and 48 h after cutting (Figures 2A and 2B). However, the fold induction of Pp CYCD;1 was nearly 20 times more than that of Pp CYCD;2, and at 48 h, the absolute abundance of the Pp CYCD;1 transcript was greater than that of Pp CYCD;2 by more than 10 times (Figure 2T). Therefore, we analyzed only Pp CYCD;1 subsequently.
Figure 2.
Expression of Cell Cycle Regulators during Reprogramming.
(A) to (S) Accumulation patterns of transcripts encoding cell cycle regulators in gametophores cut in a homogenizer. CYCD;1 (A), CYCD;2 (B), CDKA;1 (C), CDKA;2 (D), E2F;1 (E), E2F;2 (F), E2F;3 (G), E2F;4 (H), DP;1 (I), DP;2 (J), DP;3 (K), RBR;1 (L), RBR;2 (M), RBR;3 (N), ICK/KRP (O), HFO (P), CYCB;1 (Q), CYCB;2 (R), and CDKB;1 (S). Vertical and horizontal axes of graphs indicate the relative transcript level and time after cut (h), respectively. Each transcript level determined by qRT-PCR analysis was normalized with that of TUA1 (see Supplemental Figure 5 online), and the highest value of each transcript was taken as 1.0. Error bars indicate se of the mean (n = 4).
(T) Absolute quantification of CYCD;1 and CYCD;2 transcripts after cut. The value of CYCD;2 transcripts at 48 h after cut was taken as 1.0. Error bars indicate se of the mean (n = 4).
In contrast with CYCD, the other member of the complex, CDKA was not strongly induced by cutting (Figures 2C and 2D). Both Pp CDKA;1 and Pp CDKA;2 transcripts were detected at time zero, and neither changed in abundance by more than twofold thereafter.
One of the targets of the CDKA/CYCD complex is the E2F/DP/RBR complex. A broadly similar pattern of induction was seen for the four E2F genes, three DP genes, and Pp RBR;2, which reached maximal levels between 12 and 36 h after cutting and then decreased by 48 h (Figures 2E to 2K and 2M). The patterns differed in the magnitudes of the increase and decrease. By contrast, Pp RBR;3 reached maximal abundance at 6 h after cutting and by 12 h had returned to essentially precutting levels (Figure 2N), and Pp RBR;1 levels decreased by 6 h and remained low thereafter (Figure 2L). The varied expression patterns of the E2Fs, DPs, and RBRs might reflect a functional divergence among E2F/DP/RBR complexes in the regulation of S-phase genes, a divergence that has been reported in Arabidopsis (Inzé and De Veylder, 2006).
The CDKA/CYCD complex is inhibited by ICK/KRP, and this inhibition must be relieved for the cell to enter S-phase (Inzé and De Veylder, 2006). The Pp ICK/KRP transcript, after a small and transient increase, was strongly repressed by cutting (Figure 2O), suggesting that Pp ICK/KRP plays a similar role to At ICK/KRP. Finally, during S-phase in flowering plants as well as in metazoans, expression of histone H4 is strongly upregulated (Osley, 1991; Sorrell et al., 2001; Menges et al., 2002). Therefore, as a marker for S-phase, we quantified levels of the P. patens histone H4 gene HFO (Figure 2P). The levels of this transcript peaked at 36 h after cutting, half a day later than the peak of the genes that regulate S-phase entry, a timing that is consistent with our functional annotation.
CYCB and CDKB function in the G2 to M phases in Arabidopsis (Inzé and De Veylder, 2006). The P. patens homologs of these genes were all induced by cutting; however, the induction began after 24 h and might not even have reached saturation by 48 h (Figures 2Q to 2S). This timing is consistent with the first mitotic figures being evident around 48 h after cutting (see Supplemental Figure 4D online) and follows the earlier induction of genes regulating S-phase entry. These results suggest that the function of CYCB and CDKB is conserved in P. patens. Taken together, the results in Figure 2 imply that there is strong functional conservation of these major cell cycle regulators between Arabidopsis and P. patens, although functional divergence among family members within each class probably has occurred as the two lineages evolved.
Activation of the CYCD;1 Promoter in Leaf Cells Facing a Cut
To collect enough tissue for qRT-PCR, we cut gametophores in a homogenizer; therefore, the samples contained a mixture of induced and noninduced cells (see Supplemental Figure 4 online). To determine whether the cell cycle regulators CYCD;1, CDKA;1, and CDKA;2, which were hypothesized above to drive reentry into S-phase, are expressed in the specific cells undergoing reprogramming, we generated transgenic reporter lines. These lines expressed a translational fusion in which the GUS gene (uidA) was inserted just before the stop codon of each gene (see Supplemental Figures 6 and 7 online).
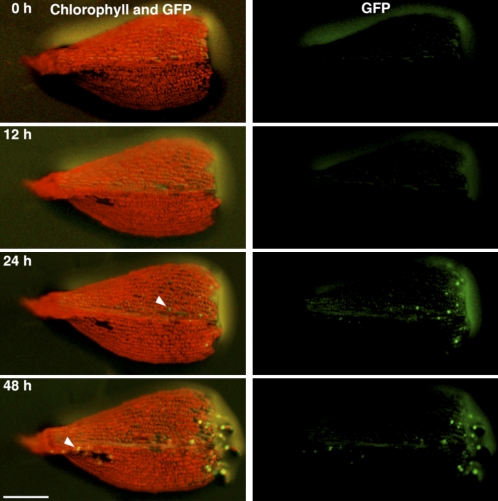
As described below, expression of CDKA;1-GUS and CDKA;2 GUS was readily observed; however, CYCD;1-GUS was not detectable in protonemata or gametophores (see Supplemental Figure 6C online). Therefore, we fused the NLS-GFP-GUS DNA fragment (as used in Figure 1C) to a 1-kb sequence upstream of the CYCD;1 start codon and created three transgenic lines expressing this reporter (ProCYCD;1:NLS-GFP-GUS; see Supplemental Figure 8 online). GUS was detected in all three lines but GFP only in two of them (#153 and #263), perhaps because these two lines harbored multiple sGFP sequences. The GUS-staining pattern in the three lines and the GFP expression pattern in the two lines were indistinguishable. When a distal part of a gametophore leaf was excised and incubated on agar medium, GFP became visible in cells facing a cut by 24 h (Figure 3). These kinetics are consistent with the qRT-PCR data (Figure 2A) and support our hypothesis that CYCD;1 helps drive the reactivation of the cell cycle.
Figure 3.
CYCD;1 Promoter Activity in Excised Leaves.
Fluorescent images of an excised leaf from the ProCYCD;1:NLS-GFP-GUS #263 line over time. Autofluorescence of chlorophyll (red) and sGFP fluorescence (green) were recorded at the indicated times (h) after excision. Arrowheads indicate the sGFP-expressing cells facing damaged cells in the excised leaf. Bar = 200 μm.
CDKA;1 and CDKA;2 Are Expressed before Cutting and Activated afterwards
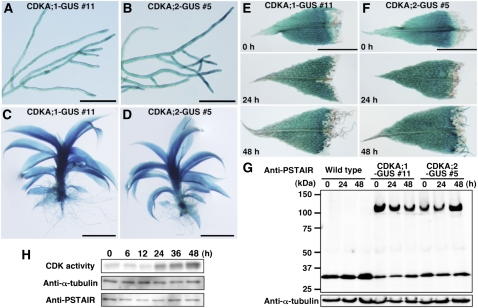
CYCD forms a complex with CDKA; thus, the presence of both is required for cell cycle progression. In Arabidopsis, the CDKA;1 gene is abundant in the shoot meristem but decreases in leaf cells (Hemerly et al., 1993). By contrast, we report here that CDKA;1-GUS and CDKA;2-GUS were detected throughout protonemata and gametophores, with no apparent maxima at or near stem cells (Figures 4A to 4D). Likewise, in excised leaves undergoing reprogramming at a cut, GUS activity was maintained in all gametophore leaf cells and was present throughout the induced chloronemata (Figures 4E and 4F). For both intact and excised samples, indistinguishable patterns were seen in the independent transgenic lines. We also investigated the levels of CDKA;1-GUS and CDKA;2-GUS fusion proteins in excised leaves using an antibody against the PSTAIR motif, which is conserved among CDKA homologs (Colasanti et al., 1991; Ferreira et al., 1991; Hirayama et al., 1991). The antibody detected a band running at the expected molecular mass (~115 kD), but the intensity of the band did not change appreciably following excision and culture (Figure 4G). Together, these results indicate that the amount of CDKA;1 and CDKA;2 protein remains mostly constant during reprogramming.
Figure 4.
Spatiotemporal Expression of CDKA;1-GUS and CDKA:2-GUS in Chloronemata, Gametophores, and Excised Leaves.
(A) to (D) Histochemical detection of GUS activity in chloronemata ([A] and [B]) and gametophores ([C] and [D]) of the CDKA;1-GUS #11 line ([A] and [C]) and the CDKA;2-GUS #5 line ([B] and [D]). Bars = 200 μm in (A) and (B) and 1 mm in (C) and (D).
(E) and (F) GUS activity in excised leaves of the CDKA;1-GUS #11 (E) and CDKA;2-GUS #5 (F) lines at the indicated time points after excision. Bars = 200 μm.
(G) Accumulation of CDKA;1-GUS and CDKA;2-GUS proteins in excised leaves. Total proteins from excised leaves of the wild type, CDKA;1-GUS #11 line, and CDKA;2-GUS #5 line at the indicated times were analyzed using the anti-PSTAIR antibody. The ~110- and 34-kD bands correspond to the CDKA;1-GUS fusion protein and CDKA;2 protein in the CDKA;1-GUS #11 line and to the CDKA;2-GUS fusion protein and CDKA;1 protein in the CDKA;2-GUS #5 line, respectively. Anti-α-tubulin antibody was used as a loading control.
(H) Histone H1 phosphorylation activity in cut leaves. CDK protein was isolated from gametophore leaves with p13suc1 beads at the indicated time points after cutting. CDK-bead complexes were examined for their ability to phosphorylate histone H1 (top) and for the total quantity of CDK protein (bottom). The starting protein extracts were probed with anti-α-tubulin (middle) as a loading control.
To examine whether CDKA is activated during reprogramming, we assayed kinase activity in a soluble extract based on phosphorylation of histone H1, a standard assay for CDK activity although the specificity is not absolute (Harashima et al., 2007). As for Figure 2, reprogramming was induced by cutting in a homogenizer. By 24 h after cutting in the middle of the reprogramming process, CDK activity was strongly increased and appeared to increase moderately thereafter (Figure 4H). Therefore, despite the constant protein levels, CDK appears to be activated by cutting, consistent with CDKA;1 and CDKA;2, in complex with CYCD;1, regulating cell cycle reentry and progression.
CYCD;1 Binds to CDKA in Leaf Cells
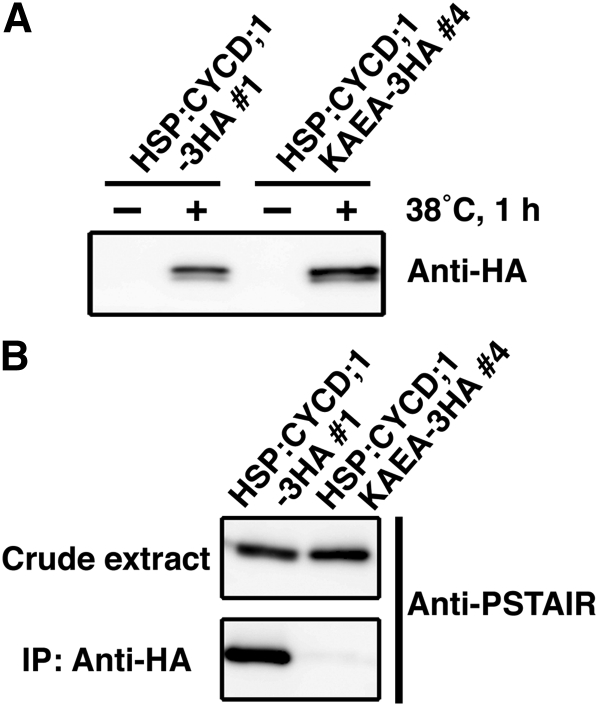
Our results show that the accumulation of CYCD;1 transcripts after cutting coincides with CDK activation (Figures 2A, 3, and 4H). In angiosperms, activation of CDKA requires binding to CYCD (Nakagami et al., 1999, 2002). Therefore, to determine whether a similar binding happens in P. patens, we examined the interaction between CYCD;1 and CDKA by coimmunoprecipitation using epitope-tagged CYCD;1 (Figure 5). As a negative control, we prepared a CYCD;1 mutant (CYCD;1KAEA) in which Ala replaced two amino acids (Lys-141 and Glu-171) previously identified in tobacco (Nicotiana tabacum) as being required for CYCD binding CDKA (Kawamura et al., 2006). Constructs containing the coding sequences driven by a heat shock–inducible promoter (HSP; Saidi et al., 2005) and followed by a triple hemagglutinin (HA)-tag were recombined into the PIG1 targeting region (Okano et al., 2009) to obtain the stable HSP:CYCD;1-3HA and HSP:CYCD;1KAEA-3HA transgenic lines (see Supplemental Figure 9 online). In the absence of heat shock, transgenic protein extracted from gametophores was undetectable with the anti-HA antibody (Figure 5A). After heat shock, total protein was immunoprecipitated with anti-HA and probed with the anti-PSTAIR antibody (Figure 5B). A band reacting with the anti-PSTAIR antibody was present in the proteins from the HSP:CYCD;1-3HA line but was absent in those from the HSP:CYCD;1KAEA-3HA line. These results suggest that during reprogramming of leaf cells facing a cut, CYCD;1 binds CDKA.
Figure 5.
Protein–Protein Interaction of CYCD;1 and CDKA.
(A) Protein expression of CYCD;1-3HA and CYCD;1KAEA-3HA upon induction with heat shock treatment. Anti-HA antibody was used to recognize the fusion proteins.
(B) Physical interaction of CYCD;1 and CDKA in gametophores. Total protein extracts from the HSP:CYCD;1-3HA #1 and HSP:CYCD;1KAEA-3HA #4 lines after heat shock treatment were prepared. After immunoprecipitation with the anti-HA antibody, crude extracts and immunoprecipitates were examined using the anti-PSTAIR antibody to recognize CDKA.
CDKA Activation Is Necessary for Cell Cycle Progression during Reprogramming
Next, we examined whether reentry into the cell cycle during reprogramming requires CDKA activation. We used a technique previously used in angiosperms, showing that expression of CDKA engineered to abolish kinase activity acts as a dominant-negative because it disrupts the productive association of the endogenous forms (CDKA;1 and CDKA;2) with CYCD;1 (Hemerly et al., 1995, 2000). To make a dominant-negative P. patens CDKA, we substituted Asn for Asp-147 in CDKA;1, a substitution that renders the kinase catalytically inactive, added a triple HA-tag just prior to the stop codon, and drove expression with the β-estradiol–inducible transcription system (Zuo et al., 2000). We generated transgenic lines expressing this construct (XVE:CDKA;1DN-3HA) and, as controls, lines with similarly inducible HA-tagged wild-type CDKA;1 (XVE:CDKA;1-3HA) and NLS-GFP-GUS (XVE:NLS-GFP-GUS) (see Supplemental Figures 10 and 11 online).
To examine the behavior of the inducible promoter, whole transgenic gametophores were incubated in liquid medium for 24 h. For both forms of the kinase, expression (as detected with the anti-HA antibody) was strictly dependent on the presence of β-estradiol (see Supplemental Figures 10C and 10D online). To assess the uniformity of induction, gametophore leaves were incubated as above, excised, and incubated in the same medium for a further 3 d. Expression of GFP (in XVE:NLS-GFP-GUS lines) appeared uniformly throughout the excised gametophore leaves treated with β-estradiol and was undetectable for those incubated without it (see Supplemental Figure 11C online). Taken together, these results indicate that the β-estradiol system regulates the expression of these genes faithfully and without prominent spatial or temporal heterogeneity.
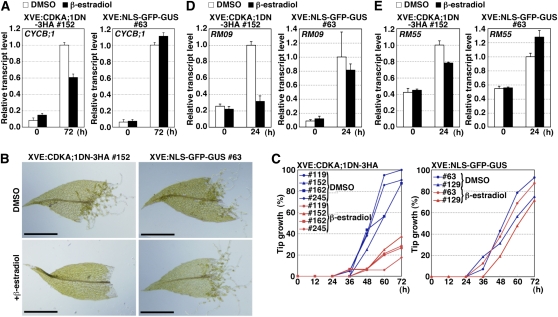
To examine the effect of the dominant-negative form of CDKA on cell cycle progression, we compared accumulation of the G2/M cyclin (CYCB;1) transcript between the transgenic lines. At 72 h after excision, induction of CDKA;1DN-3HA inhibited the accumulation of CYCB;1 transcripts, whereas that of NLS-GFP-GUS did not (Figure 6A), indicating that the dominant-negative CDKA;1 inhibited cell cycle progression.
Figure 6.
Inhibition of Cell Cycle Progression and Other Cellular Changes by Induction of a Kinase-Negative Form of CDKA.
(A), (D), and (E) Inhibition of accumulation of CYCB;1 (A), RM09 (D), and RM55 (E) transcripts by induction of the kinase-negative CDKA;1 form. Four-week-old gametophores of the XVE:CDKA;1DN-3HA #152 and XVE:NLS-GFP-GUS #63 lines were incubated in BCDAT liquid medium with or without 1 μM β-estradiol to induce transgene expression for 24 h. Thereafter, the distal half of leaves in the fifth to tenth positions were excised with a razor blade from the incubated gametophores and further incubated with or without 1 μM β-estradiol. The leaves were collected at 24 and 72 h for qRT-PCR analyses. Each transcript level was normalized with the TUA1 transcript, and the value of the transcript at 72 h (A) and 24 h ([D] and [E]) without β-estradiol (DMSO) was taken as 1.0. Error bars indicate se of the mean (n = 4).
(B) and (C) Effect of the induction of CDKA;1DN-3HA and NLS-GFP-GUS proteins on tip growth.
(B) Bright-field images of excised leaves of XVE:CDKA;1DN-3HA #152 and XVE: NLS-GFP-GUS #63 lines incubated for 72 h with or without 1 μM β-estradiol. Bars = 500 μm.
(C) Percentage of leaves with at least one cell acquiring tip growth in examined excised leaves (n > 15) with (red lines) or without (blue lines) 1 μM β-estradiol. Left: XVE:CDKA;1DN-3HA #119, #152, #162, and #245 lines. Right: XVE:NLS-GFP-GUS #63 and #129 lines.
CDKA Regulates Protonema-Specific Genes and Tip Growth
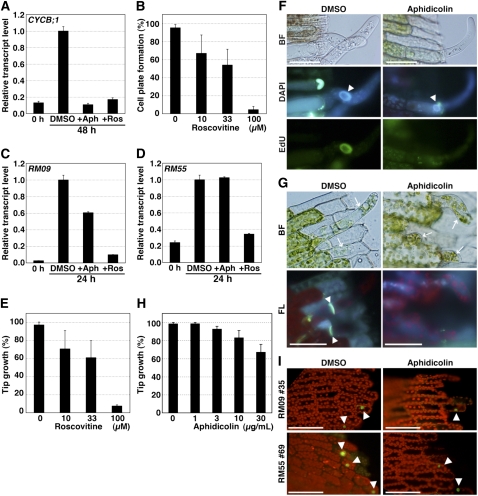
In addition to arresting the cell cycle, expression of the dominant-negative CDKA also slowed or prevented tip growth (Figures 6B and 6C). This suggests that CDKA regulates not only cell cycle progression but also other cellular changes. Consistently, the dominant-negative CDKA reduced the amount of RM09 and RM55 transcripts (Figures 6D and 6E). As an alternative to the dominant-negative CDKA, we used the chemical roscovitine, which inhibits CDKs (Planchais et al., 1997, 2000). Roscovitine inhibited the accumulation of CYCB;1 (Figure 7A), cytokinesis (Figure 7B), the accumulation of RM09 (Figure 7C) and RM55 (Figure 7D) transcripts, and tip growth (Figure 7E). In fact, roscovitine inhibited gene expression and tip growth more strongly than did expression of the dominant-negative CDKA. This is probably because the extent of the inhibition achieved by the dominant-negative strategy is dependent upon the balance between endogenous CDKA;1 and exogenous CDKA;1DN-3HA, whereas roscovitine inhibits endogenous CDKA activity generally. This result rules out the possibility that the engineered CDKA caused neomorphic effects and instead supports our interpretation that active CDKA regulates both cell cycle reentry and other cellular changes in the reprogramming of moss gametophore cells.
Figure 7.
Effects of Cell Cycle Blocking Reagents on Cell Cycle Progression and Other Cellular Changes.
(A) Effects of roscovitine and aphidicolin on the accumulation of CYCB;1 transcripts in excised leaves. Leaves excised from wild-type gametophores were incubated in BCDAT liquid medium with DMSO, 30 μg/mL aphidicolin (+Aph), or 100 μM roscovitine (+Ros) and collected at 0 and 48 h. The CYCB;1 transcript level was normalized with TUA1 transcript, and the highest value of the transcript was taken as 1.0. Error bars indicate se of the mean (n = 4).
(B) Percentage of leaves having at least one cell with cell plate formation in excised leaves (n > 20) after a 72-h incubation with roscovitine (0, 10, 33, or 100 μM). Error bars indicate sd from four biological replicates.
(C) and (D) Effect of roscovitine and aphidicolin on the accumulation of RM09 (C) and RM55 (D) transcripts in excised leaves. Leaves excised from wild-type gametophores were incubated in BCDAT liquid medium with DMSO, 30 μg/mL aphidicolin (+Aph), or 100 μM roscovitine (+Ros) and collected at 0 and 24 h. The RM09 and RM55 transcript levels were normalized with TUA1 transcript, and the highest value of the transcript was taken as 1.0. Error bars indicate se of the mean (n = 4).
(E) Percentage of leaves having at least one cell with tip growth in excised leaves (n > 20) after a 72-h incubation with roscovitine (0, 10, 33, or 100 μM). Error bars indicate sd from four biological replicates.
(F) Bright-field (BF) and fluorescent images of an excised leaf incubated with 10 μM EdU for 48 h after excision in the absence or the presence of 30 μg/mL aphidicolin. The leaves were stained with DAPI to detect nuclei (blue). White arrowheads indicate the DAPI-labeled nuclei in leaf cells facing the cut and acquiring tip growth. Bars = 50 μm.
(G) Bright-field (BF) and fluorescent (FL) images of excised leaves incubated with or without 30 μg/mL aphidicolin for 72 h and stained with aniline blue to detect newly synthesized cell plates. Arrows and arrowheads indicate cells with tip growth and newly synthesized cell plates, respectively.
(H) Percentage of leaves having at least one cell with tip growth in excised leaves (n > 20) after a 72-h incubation with aphidicolin (0, 1, 3, 10, or 30 μg/mL). Error bars indicate sd from four biological replicates.
(I) Effect of aphidicolin on promoter activities of protonemal cell-specific genes. Leaves excised from RM09 #35 and RM55 #69 gametophores were incubated in liquid BCDAT medium with or without 30 μg/mL aphidicolin. Fluorescent images of excised leaves of RM09 and RM55 were taken at 24 h after excision. Fluorescence images are overlays of chlorophyll autofluorescence (red) and GFP (green; arrowheads). Bars = 50 μm.
CDKA Regulates Cell Cycle Progression and Other Cellular Changes in Parallel
Thus far, we have shown that cell cycle progression, the induction of protonema-specific genes, and the acquisition of tip growth all require CDKA activation. However, it is not clear whether CDKA regulates all of these processes in parallel or in series. Specifically, it is conceivable that cell cycle progression is necessary for those other cellular changes to occur. Consistent with a parallel regulation, aphidicolin inhibited cell cycle progression and nuclear DNA synthesis (Figures 7A and 7F) but nevertheless cells facing the cut began tip growth without dividing (Figures 7G and 7H). Likewise, aphidicolin did not inhibit RM09 and RM55 transcription (Figures 7C, 7D, and 7I). These results show that the cellular changes do not require ongoing cell cycle progression and, together with the above results, imply that CDKA;1 is a reprogramming regulator, targeting both cell cycle status and cell fate.
DISCUSSION
CDKA;1 Links Cell Cycle Progression with Other Cellular Changes during Reprogramming
Growth and development of multicellular organisms depends on the strict regulation of the cell cycle. The central cell cycle regulator is CDKA, whose activity is precisely regulated both spatially and temporally (Gutierrez, 2005). Previously, not only has CDKA been shown to regulate cell cycle progression, but it has also been implicated in the specification of cell fate in Arabidopsis (Hemerly et al., 2000; Gaamouche et al., 2010). However, the phenotypes that have implicated CDKA in specifying cell fate include considerably aberrant division; therefore, it is not clear whether the observed changes in cell state are under direct control by CDKA or instead reflect secondary consequences contingent on the misregulated cell cycle.
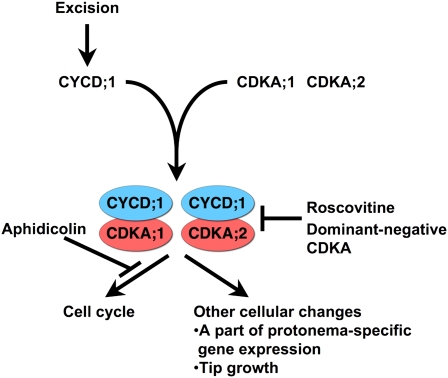
Here, for P. patens, we analyzed CDKA function during the reprogramming of leaf cells. Reprogramming was induced by cutting gametophores, and leaf cells facing the cut edge reentered the cell cycle and changed their cell fate to become chloronema apical cells. Induction of a dominant-negative form of CDKA;1 inhibited cell cycle progression and also inhibited tip growth and the expression of two protonema-specific genes, RM09 and RM55. By contrast, cell cycle arrest with aphidicolin did not inhibit these cellular changes, indicating that CDKA;1 regulates cellular traits independently of cell cycle progression (Figure 8). Evidently, CDKA;1 itself coordinates both cell cycle progression and other cellular changes during reprogramming.
Figure 8.
A Model Showing the Dual Roles of CDKA during Reprogramming.
Leaf excision induces CYCD;1 expression in the leaf cells facing the cut edge, which in turn activates CDKA kinase activity through interaction of CYCD;1 with CDKA;1 and CDKA;2. The activated CDKA;1 and CDKA;2 not only regulate the cell cycle but also affect other cellular changes reflected in part by protonema-specific gene expression and tip growth.
[See online article for color version of this figure.]
In budding yeast, a CDKA ortholog, Cdk1, encoded by CDC28, functions to connect cell cycle progression and cellular growth through activating a GTPase to polarize the actin cytoskeleton and initiate bud emergence (McCusker et al., 2007). Insofar as both actin and microtubules are involved in P. patens tip growth (Doonan et al., 1988; Finka et al., 2007; Perroud and Quatrano, 2008; Spinner et al., 2010), we might find that similar molecular mechanisms are shared by the two distantly related taxa. Cell cycle regulators also function pleiotropically in asymmetric cell divisions of Drosophila melanogaster and Caenorhabditis elegans (Berger et al., 2005; Tilmann and Kimble, 2005; Knoblich, 2008; Budirahardja and Gonczy, 2009). The D. melanogaster neuroblast is a unipotent stem cell that forms a neuroblast cell and a ganglion mother cell, which differentiates into a neuron. The asymmetric division of the neuroblast is conditioned by two cell cycle regulators, Aurora-A and Polo kinase, that phosphorylate a set of polarity-determining proteins and thereby dictate their localization (Wang et al., 2007; Wirtz-Peitz et al.., 2008). Similarly, chloronema apical cells in P. patens are polarized, with tip growth at one end, as well as polarized localization of chloroplasts and vacuoles (Menand et al., 2007a; Perroud and Quatrano, 2008). Therefore, we can speculate that principal targets of CDKA;1, in addition to cell division machinery, are involved in the establishment of polarity.
Acquisition of Chloronema-Specific Characteristics at S-phase
In Arabidopsis, molecular mechanisms of reprogramming have been investigated for the regeneration of shoot and root meristems from callus (Gordon et al., 2007; Sena et al., 2009; Sugimoto et al., 2010). However, in these experiments, it has been unclear at which cell cycle stage cells acquire a new fate. Here, we show that gametophore cells have an ~2C DNA content, which because the organism is haploid indicates that S-phase is essentially complete. However, we show that cells undergoing reprogramming incorporate EdU before cytokinesis, which implies that those cells reactivate the cell cycle in late S-phase, when the reprogramming process begins. In addition to reentering the cell cycle, we suspect that the acquisition of chloronema-specific traits (tip growth, specific gene expression, etc.,) likewise begins at late S-phase.
During the regeneration of Drosophila imaginal discs, cells in which leg identity was previously determined to switch to having wing identity, and this transdetermination requires extra time in S-phase to reset the epigenetic marks of the previous cellular memory (Sustar and Schubiger, 2005). As chromatin in S-phase is in an open conformation, structural proteins and epigenetic marks can be readily reorganized, making S-phase a suitable time for resetting cellular memory. Certainly, marking chromatin epigenetically is known to be accomplished by targets of CDK regulation. For example, mammalian CDK1 and CDK2 phosphorylate a Polycomb group protein, enhancer of zeste homolog 2, which has an essential role in promoting histone H3 Lys-27 trimethylation and thereby in the silencing of developmental regulators (Chen et al., 2010). In bryophytes, reprogramming of wounded gametophore cells is physiologically relevant. We hypothesize that arrest in late S-phase, rather than in G2-phase, is adaptive because it facilitates the remodeling of chromatin needed for cell fate change, thereby allowing chloronemata to emerge rapidly and synchronously. This hypothesis can be tested by identifying the direct phosphorylation targets of CDKA as well as by characterizing the epigenetic state of chromatin as cells are induced to reprogram.
Cell Cycle Reactivation of Differentiated Cells in Land Plants
During development, some mature cells remain competent for division but only divide when encountering a correct signal. For example, Arabidopsis pericycle cells are activated by auxin to form a lateral root meristem (De Smet et al., 2006; Dubrovsky et al., 2008). In this system, competency for cell cycle reactivation is related to the expression of cell cycle regulators, specifically At CDKA;1, which is expressed in the pericycle, whereas At CDKA;1 remains unexpressed in neighboring cells (Hemerly et al., 1993; Himanen et al., 2002). At CDKA;1 is regulated by At KRP2, which binds the kinase and inhibits its activity (Wang et al., 1998; De Veylder et al., 2001). This is seen, for example, by auxin treatment reducing the level of At KRP2 transcript in the pericycle and by overexpression of At KRP2 reducing lateral root formation (Himanen et al., 2002).
The temporal expression patterns of cell cycle regulators in auxin-treated Arabidopsis roots are similar to those we report here for P. patens cut gametophores. Pp CDKA;1 and Pp CDKA;2 are expressed in differentiated gametophores (Figure 4), which might mean that reliable reactivation of the cell cycle in gametophore cells is related to this constitutive expression. After cutting, transcripts of Pp KRP, apparently the sole KRP family member in the P. patens genome, conspicuously decrease, while transcripts of Pp CYCD;1 and Pp CYCD;2 increase (Figure 2). These results suggest that both Arabidopsis pericycle cells and P. patens leaf cells use similar molecular mechanisms for cell cycle reactivation. The distinct evolution of such common mechanisms in each linage may contribute to a distinct mode of postembryonic development and regeneration, which require the fine-tuning of cell proliferation and differentiation.
METHODS
Plant Materials and Growth Conditions
The Gransden-2004 strain of Physcomitrella patens (Rensing et al., 2008) was used as the wild-type strain and cultured on BCDAT medium under continuous white light at 25°C (Nishiyama et al., 2000). Polyethylene glycol–mediated transformation was performed as described previously (Nishiyama et al., 2000).
Preparation of Excised Leaves and Cut Gametophores
To observe gametophore leaf cell reprogramming to chloronema apical cells, a mat of chloronemata and caulonemata ~1 mm in diameter was inoculated on BCDAT medium and cultivated for 4 weeks to produce gametophores. The distal halves of the fifth to tenth youngest gametophore leaves were excised with a razor blade and cultivated.
For qRT-PCR and immunoblot analyses, a mixture of chloronemata and caulonemata was cut using a Polytron PT2100 homogenizer with a DA2120/2 generator shaft (Kinematica) for 10 s at middle speed and vegetatively propagated on BCDAT medium to produce gametophores. Four-week-old gametophores were collected with tweezers, and the attached rhizoids and protonemata were removed in water within 1 h of collection. Isolated gametophores were chopped with the Polytron homogenizer at maximum speed for 10 s a few times. Cut gametophores were washed with liquid BCDAT medium twice. Cut leaves were isolated from stems and further cultivated.
Plasmid Construction of pPIG1bNGGII
Primers used for plasmid construction are shown in Supplemental Table 3 online. For promoter reporter analysis in P. patens, the pPIG1b-NGGII (accession number AB537478) plasmid was constructed (see Supplemental Figures 2 and 8 online). The plasmid contained the following complex insert: multiple cloning sites (XbaI, BamHI, and SmaI), a reporter gene (NLS-GFP-GUS) composed of the synthetic nucleotide sequence encoding SV40 NLS (Kalderon et al., 1984), sGFP (Chiu et al., 1996), and uidA (GUS; Jefferson, 1987), and the blasticidin S deaminase (Tamura et al., 1995) expression cassette (BSD) derived from p35S-loxP-BSD (accession number AB537973) to give blasticidin S resistance. The above construct was inserted between PIG1bR and PIG1bL in the pPIG1b vector for gene targeting by homologous recombination (Okano et al., 2009). The BSD contains the cauliflower mosaic virus 35S promoter (Odell et al., 1985), the bsd gene (Tamura et al., 1995), and a fragment containing the cauliflower mosaic virus polyadenylation signal (Guerineau et al., 1990).
Plasmid Construction of the XVE-Inducible Vector pPGX6
To establish an estrogen-inducible system for P. patens, pPGX6 (accession number AB537481) was constructed (see Supplemental Figures 10 and 11 online). The LexA operator, the minimal 35S promoter, the terminator of pea (Pisum sativum) rbcS3A, and a DNA fragment encoding the XVE fusion protein were derived from pER8 (Zuo et al., 2000). The GX6 promoter for constitutive expression of XVE in P. patens was amplified with P005135f1 5′-TCATTGTTCTTCATTGTTTTCTATCA-3′ and P005135r1 5′-TTCGCCTCCACTCGAAACTCCA-3′ primers using P. patens genomic DNA as template. These fragments, the gateway rfcA cassette (Invitrogen), and the modified aminoglycoside phosphotransferase IV cassette (Hiwatashi et al., 2008) were inserted between PIG1bR and PIG1bL DNA fragments in the pPIG1b vector for gene targeting to a PIG1 locus of P. patens (Okano et al., 2009).
Plasmid Construction for Spatial Expression Analysis
Primers used for plasmid construction are given in Supplemental Table 3 online. To insert the uidA gene in frame with CYCD;1-, CDKA;1-, and CDKA;2-coding sequences, a genomic DNA fragment of each gene extending from the middle to the last codon was PCR amplified from wild-type genomic DNA. The amplified fragment was inserted into the 5′-end of the coding region of the uidA gene in the pTN83 (accession number AB538275) plasmid in frame. Genomic fragments containing the 3′-flanking region of each gene were inserted into the 3′-region of the nptII expression cassette of the plasmids. The generated constructs were digested by suitable restriction enzymes for gene targeting.
For the ProCYCD;1:NLS-GFP-GUS, RM09, and RM55 constructs, DNA fragments including partial sequences of each promoter were amplified with primers shown in Supplemental Table 3 online and inserted into the SmaI site of pPIG1bNGGII. The generated construct was digested with the restriction enzyme PmeI for gene targeting and introduced into wild-type P. patens.
Plasmid Construction for HSP:CYCD;1-3HA and HSP:CYCD;1KAEA-3HA
The open reading frame of CYCD;1 lacking the stop codon was cloned into the pENTR/D-TOPO vector (Invitrogen) to generate the plasmid pENTR CYCD;1-T. Mutation of CYCD;1 at putative sites that interact with CDK (CYCD;1KAEA) was performed with primers 5′-GCATATCTCTCGCTGCGGCAATGGAGGAATCCGACG-3′ and 5′-ACACTATTCAGAGGATGGCACTCTTAGTCTTGTCCAC-3′ to generate plasmid pENTR CYCD;1KAEA-T, using the QuikChange Multi site-directed mutagenesis kit (Stratagene). The resultant plasmids pENTR CYCD;1-T and pENTR CYCD;1KAEA-T were subjected to the LR reaction using the destination vectors pPHG-HA3 (accession number AB538231) to express the C-terminal three-repeated HA-tagged CYCD;1 and CYCD;1KAEA under control of the heat shock promoter, respectively (Okano et al., 2009). The generated constructs were digested with the restriction enzyme PmeI for gene targeting and introduced into wild-type P. patens.
Plasmid Construction for the XVE-Inducible NLS-GFP-GUS, CDKA;1-3HA, and CDKA;1DN-3HA
For the XVE:NLS-GFP-GUS construct, a fragment corresponding to NLS-GFP-GUS was cloned into pENTR/D-TOPO and subjected to the LR reaction using the destination vector pPGX6 to express the protein under the control of the XVE system (Zuo et al., 2000). For the XVE:CDKA;1-3HA and XVE:CDKA;1DN-3HA constructs, the open reading frame of CDKA;1 lacking the stop codon was fused to a fragment corresponding to a triple HA-tag and cloned into the pENTR/D-TOPO vector (Invitrogen) to generate the vector pENTR CDKA;1-3HA. To mutate CDKA;1 at the amino acid residue critical for kinase activity, PCR was performed with pENTR CDKA;1-3HA as template and the primer pair 5′-GCATATCTCTCGCTGCGGCAATGGAGGAATCCGACG-3′ and 5′-ACACTATTCAGAGGATGGCACTCTTAGTCTTGTCCAC-3′ to generate the plasmid pENTR CDKA;1DN-3HA. The resultant plasmids, pENTR CDKA;1-3HA and pENTR CDKA;1DN-3HA, were subjected to the LR reaction using the destination vector pPGX6. The generated constructs were digested with PmeI for gene targeting and introduced into wild-type P. patens.
Microscopy Analysis
Excised leaves were embedded on a glass-base Petri dish (Iwaki 3911-035; Asahi Techno Glass) in BCD medium, which is BCDAT medium lacking ammonium tartrate, with 1% agar (Nacalai Tesque). Images of leaves reprogramming were recorded at 2-min intervals with a commercial digital camera (Nikon D1) using an inverted microscope (Nikon Diaphot). The images were reconstructed to create a movie with ImageJ (http://rsb.info.nih.gov/ij/).
The amount of nuclear DNA in chloronema apical cells and gametophore leaf cells was calculated from microscopic images of fluorescent dye–stained nuclei. Protonemata and gametophore leaves were fixed with a mixture of ethanol and acetic acid (1:1) and stained with 0.1 μg/mL 4',6-diamidino-2-phenylindole (DAPI). DAPI-stained nuclei were excited with the Olympus filter set WU. An additional barrier filter (FF02-447/60-25; Semrock) was used to eliminate nonspecific fluorescence. Fluorescent images of nuclei were captured using a fluorescence microscope (BX60; Olympus) equipped with a CCD camera (DFC 350 FX; Leica). The acquired 16-bit grayscale images were analyzed using ImageJ. The intensity of an individual nucleus was calculated as the difference between the sum of the pixel values of the nuclear area and that of the neighboring area because background and/or cytoplasmic fluorescence varied among cells. For analyses of chloronemata, interphase apical cells were chosen randomly from the cell population. Metaphase or anaphase cells and postcytokinetic cells of chloronemata in the same preparation were used as controls representing 2C and 1C DNA content, respectively. Postcytokinetic cells were easily distinguished because the two daughter nuclei were close together.
For detection of DNA synthesis, the excised leaves were incubated in liquid BCDAT containing 10 μM EdU (Invitrogen) for 40 h, fixed with 3.7% formaldehyde, and treated with 1% Triton X-100. EdU-labeled nuclei were detected with Click-iT reaction cocktail (Invitrogen). To visualize newly synthesized cell plates in reprogrammed cells, the excised leaves were soaked in a solution containing 0.1% aniline blue and 0.1% K3PO4, pH 12.5. Fluorescent images from the staining or from sGFP of wild-type and transgenic plants were observed under fluorescence microscopes (BX51 and SZX16; Olympus).
The histochemical detection of GUS activity followed a previous report (Nishiyama et al., 2000).
RNA Preparation and qRT-PCR Analysis
Total RNA was purified from leaves with the RNeasy micro kit (Qiagen). First-strand cDNA was synthesized using Ready-To-Go You Prime first-strand beads (GE Healthcare) with oligo-d(T)12-18 primers (Invitrogen). qRT-PCR was performed using an ABI PRISM 7500 (Applied Biosystems) with the QuantiTect SYBR Green PCR kit (Qiagen). The sequences of primers for qRT-PCR are described in Supplemental Table 1 online. Results were analyzed using the comparative critical threshold method (Livak and Schmittgen, 2001). The quantification of each sample was performed in quadruplicate. Two biological replicates were analyzed for transcript accumulation.
DNA Gel Blot Analysis
Approximately 3 μg of genomic DNA was digested with restriction enzymes, run on 0.7% (w/v) SeaKemGTG agarose (BME), and transferred to a Hybond N+ nylon membrane (Amersham-Pharmacia Biotech). Probe labeling, hybridization, and detection were performed using the AlkPhos direct labeling and detection system with CDP-Star (GE Healthcare) according to the supplier’s instructions.
Flow Cytometry to Measure DNA Content
Flow cytometric analysis was performed as described previously (Okano et al., 2009). In short, adult leaves of Lotus japonicus (Gifu) and gametophore leaves of P. patens were chopped with a razor blade in extraction buffer (10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.1% Triton X-100, and 100 mg/mL RNase A) and stained with 1 mg/mL propidium iodide.
Protein Expression Using the Chemical-Inducible System in P. patens
For protein expression via the XVE-inducible system (Zuo et al., 2000), a mixture of chloronemata and caulonemata from transgenic lines was inoculated on BCDAT medium and cultivated for 4 weeks. Gametophores with rhizoids were picked from the 4-week-old colonies with tweezers, soaked in 1 mL BCDAT liquid medium with or without 1 μM β-estradiol (Wako) in a 24-well plate (Asahi Techno Glass), and cultivated for 24 h under continuous white light at 25°C. The distal half of the fifth to tenth youngest gametophore leaves was excised with a razor blade, soaked in fresh BCDAT liquid medium with or without 1 μM β-estradiol, and cultivated under the same conditions.
Protein Experiments
Protein extracts prepared from cut leaves were analyzed with SDS-PAGE. Proteins in a gel were transferred to an Immobilon-P membrane (Millipore) and detected with monoclonal anti-PSTAIR (P7962; Sigma-Aldrich), monoclonal anti-α-tubulin (DM1A; Sigma-Aldrich), or polyclonal anti-HA antibodies (sc-805; Santa Cruz Biotechnology). Purification of a CDK fraction from cut leaves and in vitro kinase reactions were performed as described previously (Araki et al., 2004).
For protein–protein interactions in vivo, 4-week-old gametophores of the HSP:CYCD;1-3HA and HSP:CYCD;1KAEA-3HA lines were incubated at 38°C for 1 h and subsequently kept at 25°C for 2 h. Total protein extracts from the lines were prepared with immunoprecipitation buffer (25 mM Tris-HCl, pH 7.5, 75 mM NaCl, 15 mM MgCl2, 15 mM EGTA, 0.1 mM Nonidet P-40, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM sodium ο-vanadate, and 1× complete protease inhibitor cocktail [Roche]) and were immunoprecipitated with anti-HA antibody. The immunoprecipitates were examined using the anti-PSTAIR antibody.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL/DDBJ data libraries under the following accession numbers: P. patens cell cycle–related genes, HFO, and TUA1 (see Supplemental Table 2 online); vectors for moss transformation, pTN83 (AB538275), pPIG1NGGII (AB537478), pPHG-HA3 (AB538231), and pPGX6 (AB537481).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of Protonema-Specific Genes.
Supplemental Figure 2. Construction of RM09 and RM55 Lines.
Supplemental Figure 3. Promoter Activities of the RM09 and RM55 Constructs.
Supplemental Figure 4. Formation of Chloronema Apical Cells in a Gametophore Leaf Cut with a Homogenizer.
Supplemental Figure 5. Constitutive Expression of α-Tubulin Gene TUA1 during the Reprogramming Process.
Supplemental Figure 6. Construction of the CYCD;1-GUS Lines and Expression of the CYCD;1-GUS Protein.
Supplemental Figure 7. Construction of the CDKA;1-GUS and CDKA;2-GUS Lines.
Supplemental Figure 8. Construction of the ProCYCD;1:NLS-GFP-GUS Lines.
Supplemental Figure 9. Construction of the HSP:CYCD;1-3HA and HSP:CYCD;1KAEA-3HA Lines.
Supplemental Figure 10. Construction of the XVE:CDKA;1-3HA and XVE:CDKA;1DN-3HA Lines.
Supplemental Figure 11. Construction of the XVE:NLS-GFP-GUS Line.
Supplemental Table 1. Primer Sequences for qRT-PCR.
Supplemental Table 2. List of the Physcomitrella patens Cell Cycle Regulators and Other Genes Used in This Study.
Supplemental Table 3. Primers for Plasmid Construction.
Supplemental Movie 1. Formation of Chloronemal Apical Cells in an Excised leaf.
Supplementary Material
Acknowledgments
We thank T. Kurata and K. Miyawaki for technical advice about RM09 and RM55 marker lines; N.-H. Chua for providing the pER8 vector; M. Kawaguchi for L. japonicus leaves; and M. Obara, T. Nishi, C. Honda, M. Mawatari, and other members of the Exploratory Research for Advanced Technology Reprogramming Evolution Project for technical assistance. We also thank the Functional Genomics Facility, Model Plant Research Facility, and Data Integration and Analysis Facility of National Institute for Basic Biology and T. Baskin for English editing.
AUTHOR CONTRIBUTIONS
M.I., M.K., and M.H. designed this work and mainly wrote the manuscript. M.I. performed most of the experiments, and A.A. and Y.O. helped his experiments. T.M. and W.E.F. performed the microscopy analysis to quantify nuclear DNA amounts. Y.S., M.K., and N.S. performed the flow cytometric analysis and improved the protocol for detection of DNA synthesis in the moss. T.N. contributed to genome analyses. Y.H. performed transformation. A.I. generated protonema marker lines.
References
- Araki S., Ito M., Soyano T., Nishihama R., Machida Y. (2004). Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J. Biol. Chem. 279: 32979–32988 [DOI] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Pallavi S.K., Prasad M., Shashidhara L.S., Technau G.M. (2005). A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat. Cell Biol. 7: 56–62 [DOI] [PubMed] [Google Scholar]
- Birnbaum K.D., Sánchez Alvarado A. (2008). Slicing across kingdoms: Regeneration in plants and animals. Cell 132: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L., Gutzat R., Futterer J., Laizet Y., Hennig L., Gruissem W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budirahardja Y., Gonczy P. (2009). Coupling the cell cycle to development. Development 136: 2861–2872 [DOI] [PubMed] [Google Scholar]
- Che P., Gingerich D.J., Lall S., Howell S.H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Lall S., Nettleton D., Howell S.H. (2006a). Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Love T.M., Frame B.R., Wang K., Carriquiry A.L., Howell S.H. (2006b). Gene expression patterns during somatic embryo development and germination in maize Hi II callus cultures. Plant Mol. Biol. 62: 1–14 [DOI] [PubMed] [Google Scholar]
- Chen S., Bohrer L.R., Rai A.N., Pan Y., Gan L., Zhou X., Bagchi A., Simon J.A., Huang H. (2010). Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat. Cell Biol. 12: 1108–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W.-l., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Chopra R.N., Kumra P.K. (1988). Biology of Bryophytes. (New Delhi, India: Wiley Eastern; ). [Google Scholar]
- Colasanti J., Tyers M., Sundaresan V. (1991). Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc. Natl. Acad. Sci. USA 88: 3377–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D., Bezanilla M., Harries P., Quatrano R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57: 497–520 [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60: 871–887 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., Naudts M., Inzé D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inze D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665 [DOI] [PubMed] [Google Scholar]
- den Boer B.G., Murray J.A. (2000). Triggering the cell cycle in plants. Trends Cell Biol. 10: 245–250 [DOI] [PubMed] [Google Scholar]
- Dolezel J., Bartos J. (2005). Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. (Lond.) 95: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J.H., Cove D.J., Lloyd C.W. (1988). Microtubules and microfilaments in tip growth — Evidence that microtubules impose polarity on protonemal growth in Physcomitrella patens. J. Cell Sci. 89: 533–540 [Google Scholar]
- Dubrovsky J.G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M.G., Friml J., Shishkova S., Celenza J., Benkova E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.C., Hemerly A.S., Villarroel R., Van Montagu M., Inzé D. (1991). The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A., Schaefer D.G., Saidi Y., Goloubinoff P., Zryd J.P. (2007). In vivo visualization of F-actin structures during the development of the moss Physcomitrella patens. New Phytol. 174: 63–76 [DOI] [PubMed] [Google Scholar]
- Friedman W.E. (1991). Double fertilization in Ephedra trifurca, a nonflowering seed plant: the relationship between fertilization events and the cell cycle. Protoplasma 165: 106–120 [Google Scholar]
- Gaamouche T., Manes C.L., Kwiatkowska D., Berckmans B., Koumproglou R., Maes S., Beeckman T., Vernoux T., Doonan J.H., Traas J., Inzé D., De Veylder L. (2010). Cyclin-dependent kinase activity maintains the shoot apical meristem cells in an undifferentiated state. Plant J. 64: 26–37 [DOI] [PubMed] [Google Scholar]
- Gilbert S.F. (2006). Developmental Biology. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Gordon S.P., Heisler M.G., Reddy G.V., Ohno C., Das P., Meyerowitz E.M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Guerineau F., Brooks L., Meadows J., Lucy A., Robinson C., Mullineaux P. (1990). Sulfonamide resistance gene for plant transformation. Plant Mol. Biol. 15: 127–136 [DOI] [PubMed] [Google Scholar]
- Gutierrez C. (2005). Coupling cell proliferation and development in plants. Nat. Cell Biol. 7: 535–541 [DOI] [PubMed] [Google Scholar]
- Harashima H., Shinmyo A., Sekine M. (2007). Phosphorylation of threonine 161 in plant cyclin-dependent kinase A is required for cell division by activation of its associated kinase. Plant J. 52: 435–448 [DOI] [PubMed] [Google Scholar]
- Harrison C.J., Roeder A.H., Meyerowitz E.M., Langdale J.A. (2009). Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. Curr. Biol. 19: 461–471 [DOI] [PubMed] [Google Scholar]
- Hemerly A., Engler Jde A., Bergounioux C., Van Montagu M., Engler G., Inzé D., Ferreira P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A.S., Ferreira P., de Almeida Engler J., Van Montagu M., Engler G., Inzé D. (1993). cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5: 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A.S., Ferreira P.C., Van Montagu M., Engler G., Inzé D. (2000). Cell division events are essential for embryo patterning and morphogenesis: studies on dominant-negative cdc2aAt mutants of Arabidopsis. Plant J. 23: 123–130 [DOI] [PubMed] [Google Scholar]
- Himanen K., Boucheron E., Vanneste S., de Almeida Engler J., Inzé D., Beeckman T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Imajuku Y., Anai T., Matsui M., Oka A. (1991). Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 105: 159–165 [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y., Obara M., Sato Y., Fujita T., Murata T., Hasebe M. (2008). Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 20: 3094–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Kalderon D., Richardson W.D., Markham A.F., Smith A.E. (1984). Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311: 33–38 [DOI] [PubMed] [Google Scholar]
- Kawamura K., Murray J.A., Shinmyo A., Sekine M. (2006). Cell cycle regulated D3-type cyclins form active complexes with plant-specific B-type cyclin-dependent kinase in vitro. Plant Mol. Biol. 61: 311–327 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Murakami Y. (2000). Genome analysis of Lotus japonicus. J. Plant Res. 113: 497–506 [Google Scholar]
- Knoblich J.A. (2008). Mechanisms of asymmetric stem cell division. Cell 132: 583–597 [DOI] [PubMed] [Google Scholar]
- Lajtha L.G. (1979). Stem cell concepts. Differentiation 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Masip M., Veiga A., Izpisua J.C., Simon C. (2010). Reprogramming with defined factors: From induced pluripotency to induced transdifferentiation. Mol. Hum. Reprod. 16: 856–868 [DOI] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T.A., Yates J.R., III, Gygi S.P., Kellogg D.R. (2007). Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 9: 506–515 [DOI] [PubMed] [Google Scholar]
- Menand B., Calder G., Dolan L. (2007a). Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J. Exp. Bot. 58: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. (2007b). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A. (2002). Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Kawamura K., Sugisaka K., Sekine M., Shinmyo A. (2002). Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sekine M., Murakami H., Shinmyo A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Hiwatashi Y., Sakakibara I., Kato M., Hasebe M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Oakenfull E.A., Riou-Khamlichi C., Murray J.A. (2002). Plant D-type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell J.T., Nagy F., Chua N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313: 810–812 [DOI] [PubMed] [Google Scholar]
- Okano Y., Aono N., Hiwatashi Y., Murata T., Nishiyama T., Ishikawa T., Kubo M., Hasebe M. (2009). A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 106: 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M.A. (1991). The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60: 827–861 [DOI] [PubMed] [Google Scholar]
- Perroud P.F., Quatrano R.S. (2008). BRICK1 is required for apical cell growth in filaments of the moss Physcomitrella patens but not for gametophore morphology. Plant Cell 20: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S., Glab N., Inzé D., Bergounioux C. (2000). Chemical inhibitors: A tool for plant cell cycle studies. FEBS Lett. 476: 78–83 [DOI] [PubMed] [Google Scholar]
- Planchais S., Glab N., Trehin C., Perennes C., Bureau J.M., Meijer L., Bergounioux C. (1997). Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 12: 191–202 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Bezanilla M. (2010). Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137: 3535–3543 [DOI] [PubMed] [Google Scholar]
- Quelo A.H., Bryant J.A., Verbelen J.P. (2002). Endoreduplication is not inhibited but induced by aphidicolin in cultured cells of tobacco. J. Exp. Bot. 53: 669–675 [DOI] [PubMed] [Google Scholar]
- Raghavan V. (1989). Developmental Biology of Fern Gametophytes. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Saidi Y., Finka A., Chakhporanian M., Zrÿd J.-P., Schaefer D.G., Goloubinoff P. (2005). Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat-shock promoter: A tool for plant research and biotechnology. Plant Mol. Biol. 59: 697–711 [DOI] [PubMed] [Google Scholar]
- Sakakibara K., Nishiyama T., Sumikawa N., Kofuji R., Murata T., Hasebe M. (2003). Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130: 4835–4846 [DOI] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schween G., Gorr G., Hohe A., Reski R. (2003). Unique tissue-specific cell cycle in Physcomitrella. Plant Biol. 5: 50–58 [Google Scholar]
- Sena G., Wang X., Liu H.Y., Hofhuis H., Birnbaum K.D. (2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 54: 118–130 [PubMed] [Google Scholar]
- Slack J.M. (2008). Origin of stem cells in organogenesis. Science 322: 1498–1501 [DOI] [PubMed] [Google Scholar]
- Sorrell D.A., Menges M., Healy J.M., Deveaux Y., Amano C., Su Y., Nakagami H., Shinmyo A., Doonan J.H., Sekine M., Murray J.A. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol. 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner L., Pastuglia M., Belcram K., Pegoraro M., Goussot M., Bouchez D., Schaefer D.G. (2010). The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Jiao Y., Meyerowitz E.M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sustar A., Schubiger G. (2005). A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell 120: 383–393 [DOI] [PubMed] [Google Scholar]
- Tamura K., Kimura M., Yamaguchi I. (1995). Blasticidin S deaminase gene (BSD): A new selection marker gene for transformation of Arabidopsis thaliana and Nicotiana tabacum. Biosci. Biotechnol. Biochem. 59: 2336–2338 [DOI] [PubMed] [Google Scholar]
- Tilmann C., Kimble J. (2005). Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev. Cell 9: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. (2005). How does a single somatic cell become a whole plant? Science 309: 86. [DOI] [PubMed] [Google Scholar]
- Wang H., Ouyang Y., Somers W.G., Chia W., Lu B. (2007). Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature 449: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qi Q., Schorr P., Cutler A.J., Crosby W.L., Fowke L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J.A. (2008). Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.